Abstract
Frameshift mutations can be suppressed by a variety of differently acting external suppressors. The +1 frameshift mutation hisC3072, which has an extra G in a run of Gs, is corrected by the external suppressor mutation sufF44. We have shown that sufF44 and five additional allelic suppressor mutations are located in the gene argU coding for the minor tRNAArgmnm5UCU and alter the secondary and/or tertiary structure of this tRNA. The C61U, G53A, and C32U mutations influence the stability, whereas the C56U, C61U, G53A, and G39A mutations decrease the arginylation of tRNAArgmnm5UCU. The T-10C mutant has a base substitution in the -10 consensus sequence of the argU promoter that reduces threefold the synthesis of tRNAArgmnm5UCU . The lower amount of tRNAArgmnm5UCU or impaired arginylation, either independently or in conjunction, results in inefficient reading of the cognate AGA codon that, in turn, induces frameshifts. According to the sequence of the peptide produced from the suppressed -GGG-GAA-AGA- frameshift site, the frameshifting tRNA in the argU mutants is tRNAmnm5s2UUCGlu, which decodes the GAA codon located upstream of the AGA arginine codon, and not the mutated tRNAArgmnm5UCU. We propose that an inefficient decoding of the AGA codon by a defective tRNAArgmnm5UCU stalls the ribosome at the A-site codon allowing the wild-type form of peptidyl- tRNAmnm5s2UUCGlu to slip forward 1 nucleotide and thereby re-establish the ribosome in the 0-frame. Similar frame-shifting events could be the main cause of various phenotypes associated with environmental or genetically induced changes in the levels of aminoacylated tRNA.
Keywords: frameshifting, arginine tRNA, suppression
INTRODUCTION
The genetic code contains 61 sense codons for only 20 amino acids. Therefore, one amino acid can be coded by several synonymous codons. However, codon usage in genes is not random (Nakamura et al. 2000) and correlates with the relative quantities of individual transfer RNAs. In genes encoding abundant proteins, codons corresponding to the major tRNAs are used (Ikemura 1981b). This arrangement of biased codon usage was suggested to maximize the speed and fidelity of translation. The presence of rare codons throughout the messenger RNA lowers its translation rate to some extent, but does not influence the expression level of the protein (Kurland 1991). Instead, protein synthesis is determined by the rate of translation initiation (Kurland 1991). In cases in which rare codons are close to the translation initiation site, they modulate protein synthesis by stalling the ribosome and preventing entry of incoming ribosomes (Chen and Inouye 1990; Rosenberg et al. 1993). This mechanism has been suggested to regulate the expression of a group of essential genes for various cellular functions (Chen and Inouye 1994). Alternatively, if the level of a minor aminoacyl-tRNA is limiting, the cognate rare codon could lower the synthesis of the mature protein by triggering frameshift errors. Indeed, occurrence of two rare codons in tandem induces ribosomes to shift frequently (Spanjaard and van Duin 1988). This paper addresses how the changed concentration of a minor tRNA may induce frameshifts even at a single rare codon.
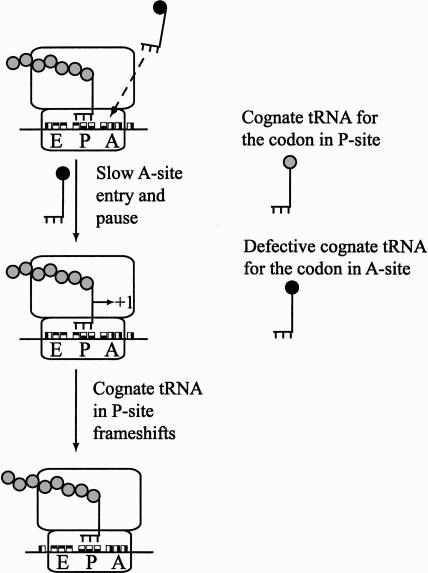
The genetic analysis of ribosomal reading frame maintenance is founded on the analysis of suppression of frame-shift mutations. Frameshift mutations are created by either addition or deletion of nucleotides in the gene sequence, resulting in an out-of-frame translation of the mRNA and production of an aberrant protein. Correction of the phenotype is possible if the ribosome shifts the reading frame back to the original, as shown in Figure 1 ▶. Slow entry of a cognate tRNA to the A-site may induce a ribosomal pause, allowing the peptidyl-tRNA in the P-site to shift 1 nt either to the left (−1 frameshifting) or to the right (+1 frameshifting) (Farabaugh 1996; Atkins et al. 2000; Gallant et al. 2000). The direction is determined by the ability to form at least two Watson-Crick base pairs with the codon in the new frame (Farabaugh and Björk 1999), although landing sites with less or no base-pairing can also be used (Herr et al. 2004). Peptidyl-tRNA can also change the reading frame because of the slow action of a release factor at a stop codon in the A-site (Weiss et al. 1990). Spontaneous frameshifting events are very infrequent, probably <10−5 per codon (Kurland et al. 1996). However, the frequency can be affected by certain sequence contexts and/or by certain physiological conditions (Fu and Parker 1994; Barak et al. 1996; Schwartz and Curran 1997; Stahl et al. 2004); for example, upon entrance into stationary phase, frameshifting increases threefold to sixfold in Escherichia coli (Wenthzel et al. 1998). It has also been observed that both the deprivation of a particular aa-tRNA species and accumulation of abnormally high concentrations of other tRNA species increases frame-shifting levels (Farabaugh 1996; Kurland et al. 1996; Atkins et al. 2000; Gallant et al. 2000).
FIGURE 1.
Frameshifting model. A defective cognate tRNA enters the A-site slowly, thereby inducing a stalled ribosome that allows the wild-type cognate peptidyl-tRNA to slip into the +1 frame. The pattern of the nucleotides in the E-, P-, and A-sites corresponds to the triplets found in the wild-type allele of the frameshift mutant.
Alteration in either tRNA structure or extent of modification can also cause translating ribosomes to frameshift (Björk et al. 1989; Farabaugh and Björk 1999; Urbonavicius et al. 2001) and thereby suppress frameshift mutations. In 1970 Riddle and Roth presented a collection of Salmonella enterica Serovar Typhimurium frameshift suppressors (Riddle and Roth 1970). The suppressors were classified by the frameshift mutations they suppressed, by their location on the chromosome, and by their dominance or recessiveness to the wild-type alleles (Riddle and Roth 1972a). Suppressor mutations in the sufA and sufB genes were dominant and affected the structure of two proline tRNAs later identified as tRNACGGPro and tRNAGGGPro, respectively (Riddle and Roth 1972b; Sroga et al. 1992). Mutations in sufD were also dominant and affected tRNACCCGly (Riddle and Roth 1972b; Riddle and Carbon 1973). Since the recessive suppressor sufF corrected some of the same frameshift mutations as the dominant suppressor sufD, sufF was also inferred to elicit a change in a glycine tRNA. Consistent with this suggestion was the fact that glycine tRNA from the sufF44 mutant had altered chromatographic properties (Riddle and Roth 1972b). Together with the recessiveness of sufF44 to the wild-type allele, this evidence suggested that sufF might encode a tRNA-modifying enzyme acting on a glycine tRNA. It is now known that tRNAmnm5UCCGly in E. coli has the modified uridine 5-methylaminomethyluridine (mnm5U) as the wobble nucleoside (T. Suzuki, pers. comm.) and is similar to the homologous glycine tRNA in Bacillus subtilis (Sprinzl et al. 1998). Since deficiency in the related nucleoside 5-methylaminomethyl-2-thiouridine (mnm5s2U) induces frameshifts (Urbonavicius et al. 2001), we hypothesized that sufF may encode an enzyme involved in the synthesis of mnm5U. However, in contrast to our presumptions, we present evidence that the product of sufF is not a protein but a minor arginine tRNA. A mechanism by which this mutated tRNA suppresses a frameshift mutation located upstream of the codon AGA is proposed.
RESULTS
The frameshift mutation hisC3072 is a G insertion in a run of Gs
All the frameshift suppressors described earlier (Riddle and Roth 1970) were grouped according to their suppressing specificity of different frameshift mutations. The sufF class only suppressed mutations hisC3072 and hisC3736. The hisC gene from the hisC3072 and hisC3736 mutants was sequenced (see Table 1 ▶ for strains used), and it was found that they both had a G insertion (underlined) in a run of Gs: GGG GAA AGA (N)87 UGA causing a shift in the reading frame (presented is the reading frame formed following the insertion). Thus, as anticipated, the mutation created a sequence consistent with a possible slip forward by a defective tRNAGly. In the mutants, the first 102 amino acids of the HisC protein were coded correctly (the full protein is 359 amino acids long), but due to the frameshift mutation, translation proceeded in the wrong frame until a stop codon was encountered. Cells carrying such a truncated HisC had a His− phenotype.
TABLE 1.
Strains
| Strain | Genotype | Source |
| LT2 | Wild type | Riddle and Roth 1970 |
| GT885 | hisO1242 hisC3072 | Riddle and Roth 1970 |
| GT889 | hisO1242 hisC3736 | Riddle and Roth 1970 |
| GT400 | hisO1242 hisC3072 sufF44 (C61U) trpA8 | Riddle and Roth 1970 |
| GT5148 | hisO1242 hisC3072 argU2343 (G53A) | This work |
| GT5130 | hisO1242 hisC3072 purE::Tn10dTc | This work |
| GT5164 | hisO1242 hisC3072 argU2343 fimC2529::Tn10dTc | This work |
| GT5162 | hisO1242 hisC3072 purE8 argU2343 fimC2529::Tn10dTc | This work |
| GT5144 | hisO1242 hisC3072 argU2341 (C32U) | This work |
| GT5150 | hisO1242 hisC3072 argU2344 (C61U) | This work |
| GT5136 | hisO1242 hisC3072 argU2342 (T-10C) | This work |
| GT5140 | hisO1242 hisC3072 argU2346 (C56U) | This work |
| GT5141 | hisO1242 hisC3072 argU2345 (G39A) | This work |
| GT6863 | hisO1242 hisC3072 ttcA5<>kan | This work |
The sufF44 mutation is in the argU gene
The sufF44 mutation is 27% linked to mutation in the purE gene that induces purine auxotrophy (Riddle and Roth 1972a). A transposon Tn10dTc insertion was isolated (strain GT5164) and was shown to be linked to both the purE and sufF44 genes (data not shown). Direct sequencing of chromosomal DNA isolated from GT5164 using Tn10 specific primers revealed that the Tn10dTc element was inserted into the gene fimC, encoding a periplasmic chaperone involved in the formation of type 1 fimbriae.
Since the sufF44 mutation was recessive to the wild-type allele, it seemed possible to isolate a plasmid harboring the sufF+ allele. Such a plasmid should change the phenotype of the sufF44 mutant from His+ to His−. Thus, the S. enterica gene bank was introduced into His+ strain GT5164 (hisO1242 hisC3072 argU2343 fimC2529::Tn10dTc), and His− transformants were screened. Plasmid DNA was purified from several His− transformants, and the chromosomal inserts harbored by the plasmids were partially sequenced. The sequence obtained from clone GT5164/ pTH67 showed that the 16.7-kb-long insert contained DNA from a region close to the fimC gene and contained a gene coding for tRNAmnm5UCUArg. To further investigate the role of tRNAArgmnm5UCU, plasmid pMW210 containing the tRNAArgmnm5UCU gene from phage T4 cloned into the vector pACYC184 was used (Wikström et al. 1992). After introducing this plasmid into the His+ strain GT5148 (hisO1242 hisC3072 argU2343), all cells became His−, supporting the argument that the sufF44 mutation was in the argU (sometimes called fimU) tRNAArgmnm5UCU gene.
New mutants defective in tRNAArgmnm5UCU were isolated
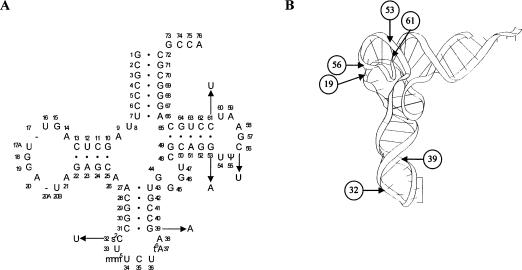
By using mutagens hydroxylamine or nitrosoguanidine, 10 new purE-linked frameshifting mutants were isolated that could suppress the hisC3072 mutation. The sequence of the argU gene from all mutants revealed that five different tRNAArgmnm5UCU mutations had been created (Fig. 2 ▶) and one mutation was located in the upstream region of the argU gene (position -10 according to the transcriptional start) (Saxena and Walker 1992).
FIGURE 2.
(A) Cloverleaf structure of tRNAArgmnm5UCU with the mutations discussed in this paper indicated. The C61U mutation was isolated in five independent cases, and one of them is the original mutant sufF44; all the other mutations were obtained once. (B) Tertiary structure of tRNAPhe from yeast (see Kim et al. 1974). The arrows are pointing to the positions of mutated nucleotides.
The mutations C61U and G53A in the TψC stem and G39A in the anticodon stem disrupted base-pair interactions in the tRNAArgmnm5UCU cloverleaf structure. Mutation C56U disrupted the G19–C56 pair, which locks the D and TψC loops together. Mutation C32U changed a residue in the anticodon loop, which also eliminated the possibility to form the 2-thiocytidine (s2C) modification at position 32 of tRNA. Each of those changes can cause defects in the tRNA structure and influence the function of tRNA. Mutation T-10C was a change in the argU promoter -10 consensus sequence altering the conserved sequence TATAAT to TACAAT, which could influence expression levels of tRNAArgmnm5UCU (Youderian et al. 1982; Hawley and McClure 1983).
The thio-group of s2C32 does not influence frameshifting in the hisC3072 mutant
Mutation C32U prevented s2C modification formation at tRNA position 32. Lack of s2C32 decreases the rate of A-site selection at the AGG codon, but not at the AGA codon when the frameshifting site is CCC-AGA/G-UAA (Jäger et al. 2004). Since the frameshifting site in the hisC3072 mutant was different (-GGG-GAA-AGA-N87-UGA), we tested whether tRNAArgmnm5UCU lacking the s2-group of s2C32 would be able to suppress the hisC3072 mutation. Therefore, a deletion of the gene ttcA, which codes for one of the steps in the formation of s2C (Jäger et al. 2004), was introduced into strain GT885 (hisO1242 hisC3072), creating strain GT6863 (hisO1242 hisC3072 ttcA5<>kan). However, this double mutant was still His−, indicating that lack of the sulfur modification s2C32 in tRNAArgmnm5UCU was not a cause of frameshifting (data not shown), a result consistent with earlier results (Jäger et al. 2004).
Several of the argU mutants have reduced levels of tRNAArgmnm5UCU
As the argU2342 (T-10C) mutation was located in the promoter region of the argU gene, it might alter the level of tRNAmnm5UCU Arg. Therefore, we compared the ratios between the yields of tRNAArgmnm5UCU and tRNAmnm5s2UUGGlu or tRNAcmnm5UmAALeu in total RNA isolated from the argU2342 (T-10C) mutant as well as from the wild type. Northern blot analysis of tRNAs fractionated on a polyacrylamide gel was performed by probing with different oligonucleotides homologous to tRNAArgmnm5UCU, tRNAmnm5s2UUGGlu or to tRNAcmnm5UmAALeu (Fig. 3 ▶). Bands corresponding to tRNA from different hybridization experiments were quantified, and their relative ratios were calculated. The results revealed that the argU2342 (T-10C) mutant had only 33% of the amount of tRNAArgmnm5UCU compared to wild-type cells, indicating reduced expression of the argU gene as a result of the mutation in the −10 sequence of its promoter.
FIGURE 3.
Relative levels of tRNAArgmnm5UCU in wild type and the promoter mutant argU2342 (T-10C). aThe bands specific for different deacylated tRNAs were quantified and the r atio between them calculated. The result is the percentage of the level observed in wild type ± standard error. The difference observed between the levels in mutant and in wild type is significant based on a t-test (p = 0.003).
Since mutations influencing the secondary structure of tRNAArgmnm5UCU might destabilize the tRNA, similar experiments were also performed with other mutants. The radioactive signal obtained from the probe binding to tRNAArgmnm5UCU was normalized to the signals obtained from tRNAmnm5s2UUG−Gln or tRNAcmnm5UmAALeu-binding probes in each strain. The comparison of tRNAArgmnm5UCU levels in the mutants versus the level in the wild type demonstrated that the argU2344 (C61U), argU2343 (G53A), and argU2341 (C32U) mutations reduced tRNAArgmnm5UCU concentrations more than twofold (Table 2 ▶). However, argU2345 (G39A) and argU2346 (C56U) had similar levels of tRNAArgmnm5UCU compared to the wild type.
TABLE 2.
Levels of tRNAArgmnm5UCU
| tRNAArgmnm5UCU | wt | C32U | G53A | C61U | C56U | G39A |
| Relative level of tRNAmnm5UCUArg a | 100 | 48 ± 10b | 32 ± 3b | 39 ± 6b | 126 ± 11 | 141 ± 31 |
aResults are the percentage of deacylated tRNAArgmnm5UCU levels compared to the wild-type level ± standard error.
bThe difference observed between the levels in the mutant and in wild type is significant based on a t-test (p < 0.01).
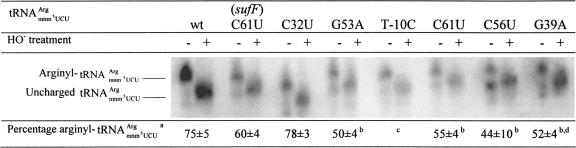
Except the C32U mutant, all mutants had a reduced arginylation of tRNAArgmnm5UCU
Changes in the tRNA structure could influence the efficiency of arginylation of tRNAArgmnm5UCU. We therefore compared the levels of charged tRNAArgmnm5UCU versus uncharged tRNAArgmnm5UCU in all of the mutants. To prevent deacylation of bound amino acids, tRNAs were isolated under acidic conditions. An aliquot of tRNAs from each preparation was deacylated by alkaline hydrolysis. Both deacylated and untreated aliquots were run on an acidic polyacrylamide gel, on which charged and uncharged tRNAs were separated. This gel was later used in Northern blot analysis and hybridized to a radioactively labeled probe specific for tRNAArgmnm5UCU (Fig. 4 ▶). Both charged and uncharged tRNAArgmnm5UCU forms isolated from argU2341 (C32U) migrated faster on the gel, which might be caused by lack of the s2C32 modification. In the wild type, most of the tRNAArgmnm5UCU was in the charged form, comprising 75% of total tRNAArgmnm5UCU. Previous works describe similar levels of aminoacylation of different tRNA species in wild-type cells under steady-state growth (Varshney et al. 1991; Kruger and Sorensen 1998; McClain et al. 1999). Visual analysis of the bands in argU2346 (C56U), argU2344 (C61U), argU2343 (G53A), and argU2345 (G39A) mutants clearly indicated that in those samples the amount of uncharged tRNAArgmnm5UCU form increased. To facilitate the comparison, we quantified the intensity of different bands. Due to blurriness, it was hard to determine the precise boundaries of each band; therefore, the whole experiment was repeated three times, and the results are presented in Figure 4 ▶. The analysis showed that the aminoacylation level in the argU2346 (C56U) decreased the most (44% of the tRNAArgmnm5UCU was charged). In argU2344 (C61U), argU2343 (G53A), and argU2345 (G39A), 50%–55% of the tRNAArgmnm5UCU was aminoacylated. Mutation C32U did not change the charging level of tRNAArgmnm5UCU . The quantify cation of charging in the argU2342 (T-10C) was uncertain because of the diminished amounts of tRNAArgmnm5UCU, although visual estimation indicated unaffected arginylation levels of tRNAArgmnm5UCU in this mutant. Such a result was the most expected, since the T-10C mutation did not change the structure of tRNAArgmnm5UCU
FIGURE 4.
Charging levels of different tRNAArgmnm5UCU mutants. Bands corresponding to arginyl- tRNAArgmnm5UCU and uncharged tRNAArgmnm5UCU are indicated. aThe percentage and standard error of arginyl- tRNAArgmnm5UCU [100× arginyl-tRNAArgmnm5UCU/(arginyl- tRNAArgmnm5UCU + uncharged tRNAArgmnm5UCU)] are the average of three independent experiments. bThe difference in charging observed between the mutant and the wild type is significant based on a t-test (p ≤ 0.03). cThe quantification was unreliable because of too high background and diminished amounts of tRNAArgmnm5UCU in the T-10C mutant (Fig. 3 ▶). dThe double bands of uncharged tRNA in G39A lanes are due to artifacts in this particular gel. The bands migrated as uniform bands in the other two experiments.
Wild-type tRNAmnm5s2UUCGlu and not the altered tRNAArgmnm5UCU is the frameshifting tRNA
One mutant, argU2343 (G53A), was chosen for analysis to clarify the nature of the slippage event at the frameshift sequence of hisC3072. For that purpose, plasmid pUST274 was constructed (Fig. 5A ▶). The frameshift sequence was inserted between the genes encoding the glutathione-S-transferase (GST) and the maltose-binding protein (MBP) with six histidine residues (6×His) at the carboxy terminus in the gst-malE fusion gene. malE is in the +1 frame relative to gst, explaining why the complete GST-MBP-6×His fusion protein is only synthesized when +1 frameshifting occurs. If +1 frameshifting does not occur, translation terminates at the UAA stop codon found downstream of the gst gene, and only GST is produced (Figs. 5A ▶, 6 ▶). The complete fusion protein was purified from strain GT5164 [argU2343 (G53A)/pUST274] using the GST and 6×His affinity tags. To liberate the slippage junction fused to the MBP-6×His, the frameshift product was treated with PreScission Protease. This protease cuts the protein at the specific protease site between the GST moiety and the rest of the peptide. After digestion the sample was analyzed by mass spectrometry, and the observed values were compared to the predicted masses of peptides having different possible amino acid composition (Fig. 6 ▶). The major peak observed in a mass spectrum in the 41500–46000 Da range was at 43374.50 Da (data not shown). This correlated best with the expected 43374.15-Da mass of a peptide putatively produced when the frameshifting tRNA was glutamate tRNA. To verify this hypothesis further, the N terminus of the slippage junction fused to MBP-6×His was sequenced. The first 12 amino acids of the peptide were determined as GPLGIPGEDAIL, where E (glutamate) was the last amino acid inserted in 0 frame, suggesting that the frameshifting tRNA at the frameshift site GGG-GAA-AGA in hisC3072 was wild-type tRNAmnm5s2UUCGlu and not the mutated tRNAArgmnm5UCU
FIGURE 5.
Plasmid pUST274 construct used for measuring +1 frameshifting. (A) The malE gene is in the +1 frame relative to the gst gene. In the 3′-terminus of gst, the sequence coding for a PreScission protease recognition site is indicated as a black box; the first stop codon encountered in the 0 frame in malE is marked with an *; 6×His, the six-histidine tag is indicated as a white box. (B) Frameshifting (FS) and termination (Term) products from the wild type and argU2343 (G53C) mutant separated on an SDS gel and were detected by anti-GST antibodies in Western blot analysis.
FIGURE 6.
Various alternatives of fusion proteins produced by slippage by different tRNAs. After PreScission Protease digestion the GST part is cleaved away and the molecular mass of the resulting peptides is analyzed. (I) The peptide part liberated after the cleavage if no frame-shifting occurred; the coding sequence is also presented. The critical AGA codon for R (arginine) is indicated. (II) Fragments of different peptides liberated after the cleavage if different tRNAs were slipping. The amino acid inserted by the frameshifting tRNA and the sequence where this tRNA frameshifts are in bold. The sequence translated and amino acids inserted in the +1 frame are in italic. On the right side are the expected masses of those hypothetical peptides.
Efficiency of frameshifting
Our data suggested a correlation between the frameshifting, which induces His+ phenotype, and the decrease in arginyl- tRNAArgmnm5UCU levels. To receive a quantitative estimate for the frequency of frameshifting in tRNAArgmnm5UCU mutants, we used the above-mentioned plasmid pUST274 containing the GGG-GAA-AGA sequence. This plasmid was introduced into the wild type and argU2343 (G53A) mutant, which had the lowest level of arginyl- tRNAArgmnm5UCU. Total protein extracts from GT5164/pUST274 and LT2/pUST274 strains were fractionated by gel electrophoreses. The termination product and complete fusion proteins were detected by Western blot analysis using anti-GST antibody (Fig. 5B ▶). The extract prepared from the mutant strain contained two clear bands (26 kDa and 70 kDa), while the wild type had only the termination product. Comparison of the frame-shifting and termination peptide abundances indicated that for tRNAArgmnm5UCU with alteration in position 53, the frequency of frameshifting at the GGG-GAA-AGA sequence was ~5%. We expect a lower frequency of frameshifting induced by the other mutants, since they reduce the level of charged tRNAArgmnm5UCU less (Table 2 ▶; Fig. 4 ▶).
DISCUSSION
tRNAArgmnm5UCU is a tRNA, found in low quantities in E. coli (Ikemura 1981a; Dong et al. 1996). We describe five different alterations in this tRNA or in the promoter for its structural gene (argU) that induce frameshifts at the sequence GGG-GAA-AGA-(NNN)29-UAA in the hisC3072 mutated gene. We suggest that the mutations decreased the concentration of charged tRNAArgmnm5UCU and thereby were allowing tRNAmnm5s2UUCGlu, which decodes GAA upstream of the arginine codon AGA, to frameshift. This suggestion is consistent with the frameshifting model presented in Figure 1 ▶.
Alterations influencing the structure of a tRNA can destabilize this tRNA and increase its degradation—why it could become less abundant. In addition, changed tRNA structure can disturb its aminoacylation by cognate amino-acyl-tRNA synthetase. Indeed, the argU2343 (G53A) and argU2344 (C61U) mutants exhibited decreased amounts and reduced charging of tRNAArgmnm5UCU (Table 2 ▶; Fig. 4 ▶). The concentration of arginyl- tRNAArgmnm5UCU in those mutants was only 21% and 28% of the wild-type level, respectively, considering both the level of arginylation and the total level of tRNAArgmnm5UCU. The G53A and C61U mutations altered bases in the TψC stem and disrupted the conserved C61–G53 base pair, which presence is critical for the characteristic TψC loop structure. Replacement of C61 by U61 induces destabilization of this structure, as revealed by changed reactivity of the phosphate 60, which makes a hydrogen bond with the amino group of C61 (Dirheimer et al. 1995). Interestingly, a single alteration of C to U at position 61 in another tRNA, namely, Salmonella tRNAGly (sufS617), induces suppression of a -1 frameshift mutation (O’Mahony et al. 1989).
The argU2341 (C32U) mutation lowered the abundance of tRNAArgmnm5UCU twofold (Table 2 ▶). The residue at position 32 forms a noncanonical bifurcated hydrogen bond with residue 38 at the stem/loop junction and shapes the anti-codon loop (Auffinger and Westhof 1999). Phylogenetic data revealed that the presence of pyrimidines in position 32 was preferred in the majority of tRNAs; 56% of the cases containing C32 and 39% U32. The distribution of the 32–38 pair is biased toward C-A, since it is found in 42%, s2C-A in 1%, and U-A in 5% of all sequenced tRNAs. Although both C32–A38 and U32–A38 pairs adopt the same configuration in crystal structures, substitution of a C-A pair by U-A affects the discriminating ability of tRNA transcripts and their frameshifting efficiency (Lustig et al. 1993; O’Connor 1998; Auffinger and Westhof 1999). Our results also demonstrate a lowered tRNAArgmnm5UCU stability by an s2C32-to-U32 change. We also show that this effect could not be attributed to the lack of the s2-group, but rather to the change of the base in position 32. The argU2341 (C32U) mutation did not affect the aminoacylation of tRNAArgmnm5UCU , indicating a normal interaction with the arginyl-tRNA synthetase (ArgRS). Similar unchanged arginylation in vitro was reported for tRNAICGArg lacking s2C32 (Kruse et al. 1978). We conclude that the base and not the s2-modification stabilizes tRNA structure, whereas neither of them interferes with tRNAArgmnm5UCU and ArgRS interaction.
The C56U and G39A alterations did not affect the stability of tRNAArgmnm5UCU (Table 2 ▶), but decreased its aminoacylation (Fig. 4 ▶). The argU2346 (C56U) mutant had the most pronounced decrease (twofold) when compared to the wild type. Base C56 is a highly conserved nucleotide in tRNAs and is involved in forming the G19–C56 pair, which locks the D and TψC loops together (Dirheimer et al. 1995). Changing the G19–C56 pair into a weaker G19–U56 pair most likely destabilizes the D–TψC interaction, though probably not strongly enough to influence the stability of tRNAArgmnm5UCU (Table 2 ▶). In contrast, the structural rigidity of the tRNAmnm5UCCGly was drastically reduced by the C56A mutation that prevented base-pairing to G19 (Herr et al. 1999). Changed bonding between bases 19 and 56 in our C56U mutant could influence the interference with the adjacent nucleotide A20, which together with C35, U/G36, and A/G73 are the major positive recognition elements in tRNAArg:s for ArgRS (McClain and Foss 1988; McClain et al. 1990; Giegé et al. 1998). The A at position 20 is conserved in tRNAArg:s from all organisms except yeast. Alteration of this A to U affects both the suppression efficiency and the acceptor identity of suppressor tRNAArg (McClain and Foss 1988). Therefore, our observation that the C56U alteration induced less arginylation is consistent with other observations (McClain and Foss 1988; McClain et al. 1990; Delagoutte et al. 2000) and implicates this region of the tRNA being, indeed, important for an optimal tRNA–ArgRS interaction.
The T-10C base substitution in the promoter region of the argU2342 mutant greatly reduced the abundance of tRNAArgmnm5UCU (Fig. 3 ▶). The promoter of argU encoding tRNAArgmnm5UCU has similar features to strong rRNA promoters such as an upstream activator sequence, a -35 sequence very similar to the δ70 consensus sequence, and a -10 TATAAT consensus sequence (Lisser and Margalit 1993). However, argU expression is severely inhibited by - sequences downstream of the transcription start point, rendering tRNAArgmnm5UCU a minor tRNA (Saxena and Walker 1992). The conserved promoter elements are devoted to stabilizing the RNA polymerase at the promoter, directing the holoenzyme-dependent melting of the DNA, and allowing a productive initiation of transcription (Helmann and deHaseth 1999). The general rule concerning changes in the conserved −10 promoter region suggests that decreasing the homology to the consensus sequence will reduce gene expression (Hawley and McClure 1983). A single base substitution at the −10 position from T to C (TATAAT to TACAAT) as in the argU2342 (T-10C) mutant has been previously isolated in the bacteriophage P22 ant gene promoter. That mutation caused a mild reduction of ant promoter activity (Youderian et al. 1982). A similar single substitution in the rrnB promoter P1 induced an ~10-fold decrease in its activity (Gaal et al. 1989). In the argU2342 case, the T-10C mutation reduced threefold the synthesis of tRNAArgmnm5UCU.
In summary, the isolated mutants had either decreased amounts of tRNAArgmnm5UCU and/or reduced charging of this tRNA due to compromised secondary or tertiary structure, and in one case due to impaired transcription of the argU gene. The A61U and G53A mutations in the T-stem alter the face of tRNA (consisting of T-stem and acceptor-stem helices), to which the elongation factor Tu (EF-Tu)·GTP binds (Nissen et al. 1995, 1999). This could affect the formation of the ternary complex (EF-Tu·GTP·aa-tRNA) required for tRNA selection to the A-site codon. The G1-to-A1 change in the acceptor-stem of the same tRNAArgmnm5UCU was demonstrated to impair the EF-Tu · GTP binding (Sakamoto et al. 2004).
The earlier mentioned G1A mutant in tRNAArgmnm5UCU [argU10(Ts)] and some other tRNA mutants, such as the divE having an A10G change in the major tRNAcmo5UGASer and feeB having a C77A change in the tRNAU*AGLeu, have one common phenotype—they all induce a cell division defect (Tamura et al. 1984; Garcia et al. 1986; Chen et al. 1991). When the argU10(Ts) mutant is shifted to nonpermissive temperature, it ceases to grow in ~1 h, although the rate of total protein synthesis, measured as TCA-precipitable material, is reduced only to two-thirds of the wild-type rate and continues for at least 2 h. In contrast, the synthesis of the λ cI repressor containing five AGA codons read by tRNAArgmnm5UCU is severely reduced. This observation suggests that it is not the general protein synthesis but rather the synthesis of some specific protein(s) pivotal in controlling cell division that is affected (Chen et al. 1990). Note, however, that monitoring TCA-precipitable material does not reveal changes in the frequency of frameshifting. The argU10(Ts) mutant has a severely decreased amount of tRNAArgmnm5UCU , a reduced level of arginylation, and impaired binding of this tRNA to EF-Tu·GTP (Sakamoto et al. 2004). Therefore, it was inferred that decoding of the cognate AGA codon in this mutant is inefficient and inhibits the synthesis of specific protein(s) containing AGA codons in their mRNA. However, less efficient reading of a codon does not affect the overall expression of a protein (Kurland 1991).
Our data suggest an alternative mechanism eliciting the defects in cell division. Amino acid sequence analysis of the frameshift product in the argU2343 (G53C) mutant revealed that while the mutated tRNA is tRNAArgmnm5UCU, the tRNA that frameshifts is tRNAmnm5s2UUCGlu . When the ribosome arrives at the -GGG-GAA-AGA- site and the AGA codon is in the ribosomal A-site, the tRNAmnm5s2UUCGlu reading the GAA codon resides in the P-site (see Fig. 1 ▶). Due to decreased levels of charged tRNAArgmnm5UCU , the level of the ternary complex loaded with the arginyl- tRNAArgmnm5UCU is reduced, which slows down the AGA reading and stalls the ribosome. Besides, ribosome stalling could be prolonged by inefficient A-site entry by the ternary complex containing altered tRNAArgmnm5UCU . Such ribosome pausing destabilizes cognate peptidyl- tRNAmnm5s2UUCGlu in the P-site and facilitates its shift forward by 1 nt (Fig. 1 ▶; Qian et al. 1998; Farabaugh and Björk 1999; Gallant et al. 2000; Hansen et al. 2003). We also cannot exclude the alternative that ribosome pausing stimulates out-of-frame binding of an incoming aspartyl-tRNA cognate for the codon in the +1 frame (Stahl et al. 2002). The wild-type levels of tRNAArgmnm5UCU are among the lowest in the cell (Dong et al. 1996). Observed frameshifting occurred when the arginyl- tRNAArgmnm5UCU levels were decreased only by twofold. We assume that reduction down to “trace” levels of arginylated tRNAArgmnm5UCU , as in the argU10(Ts) mutant (Sakamoto et al. 2004), will elicit extensive frameshifting, at least at -GAA-AGA- sites. The peptidyl- tRNAmnm5s2UUCGlu , which frameshifts at the -GAA-AGA- site, “takes off” from the GAA codon and lands at the AA-A codon. For frequent frameshifting, weak base-pairing at the “take-off” site is important (Gallant et al. 2004), whereas the ability to form Watson-Crick base pairs at the landing site is less critical, even though strong base-pairing is preferred (Herr et al. 2004). As a take-off site present on the 5′-side of the hungry codon, the GAA codon is moderately proficient, while some other codons, like UUU, are up to sevenfold more proficient (Gallant et al. 2004). Therefore, the frameshifting frequency would be probably higher at the UUU-AGA site then at the GAA-AGA site.
Based on such considerations and the results presented here, we suggest that cell division defects in various tRNA mutants are caused by frameshifting errors rather than inefficient translation in the zero frame of some specific mRNAs. A survey of the entire E. coli genome determined the frequency of different codon pairs (Boycheva et al. 2003). The arginine codon pairs AGA-AGA, AGG-AGG, AGA-AGG, and AGG-AGA were found in 45, 4, 17, and 14 locations, respectively. The shifty UUU-AGA and UUU-AGG codon pairs were found in 30 and nine locations, respectively. Even though the frequency of occurrence of those codon pairs is rather low, still it is quite possible that some of them are residing in the mRNAs coding for the proteins required for cell division. Frameshifting reduces the total number of ribosomes that traverse the whole mRNA and thereby decreases the synthesis of a mature protein(s). Such frameshifting events may be the main cause of many phenotypes associated with environmental and genetically induced changes in the level of aminoacylated tRNA. The argU mutants described in this paper did not induce any cell division defects at high temperature (data not shown). The argU10(Ts) mutant having only 5% of the wild-type amount of the arginyl-tRNAArgmnm5UCU at permissive temperature has no growth defects then, but stops dividing at high temperature, when the amount of tRNAArgmnm5UCU is reduced to trace level (Sakamoto et al. 2004). Among the mutants described in this paper, the most affected mutant [argU2343 (G53A)] has 21% of its arginyl-tRNAArgmnm5UCU left. Apparently, this amount is sufficient for normal cell division but low enough to induce frameshifting in hisC3072 mRNA.
The supply of arginyl-tRNA also becomes exhausted under the induced translation of tandem AGG or AGA arginine codons (Spanjaard and van Duin 1988; Chen and Inouye 1990; Rosenberg et al. 1993). At AGG-AGG or AGA-AGA sites, up to 50% of the ribosomes frameshift into the +1 frame, and this effect can be suppressed by introducing extra copies of the gene coding for tRNAArgmnm5UCU (Spanjaard and van Duin 1988; Spanjaard et al. 1990). Therefore, translation of genes having tandem AGG-AGG or AGA-AGA codons should be prone to trigger frameshifting. Several such genes exist in S. enterica, and one of them is the trmD gene, which codes for the tRNA (m1G37) methyl-transferase (TrmD) that modifies G37 to m1G37 (Björk et al. 1989; Li and Björk 1999). Although mutants G53C and C61U of tRNAArgmnm5UCU neither influenced the levels of TrmD peptide, nor the presence of m1G in the tRNAs (data not shown), this does not rule out that the expression of other genes having such tandem codons may be affected by mutations in the argU gene.
In contrast to Riddle and Roth (1972b) and our presumption that sufF is involved in modification of tRNAmnm5UCCGly, we demonstrate that this gene encodes tRNAArgmnm5UCU. We also show that it is not the tRNAmnm5UCCGly that frameshifts at the GGG-GAA-AGA site. Even so, Riddle and Roth’s suggestion that sufF influences tRNAmnm5UCCGly modification may still be correct as the effect could be indirect; e.g., the particular modification enzyme at sites prone to frameshift could contain the AGA codons and their translation by the mutated tRNAArgmnm5UCU could become impaired. To test this possibility, we purified tRNAmnm5UCCGly from the wild type and the sufF44 mutant, but no differences in the modification patterns were observed (data not shown). Yet an influence on tRNA modification by sufF was not ruled out since we do not know the migration properties of mnm5U in our HPLC system.
MATERIALS AND METHODS
Bacteria and growth conditions
The bacterial strains used were derivatives of S. enterica (Table 1 ▶). As rich media we used either Luria-Bertani (LB) (Bertani 1951) or the complex medium NAA (Difco nutrient broth [0.8%]; Difco Laboratories) supplemented with aromatic amino acids, aromatic vitamins, and adenine (Davis et al. 1980). The minimal medium was made from the basal medium E (Vogel and Bonner 1956) with 15 g of agar per liter and supplemented with 0.2% glucose and required amino acids or vitamins (Davis et al. 1980). TYS-agar (10 g of Trypticase Peptone, 5 g of yeast extract, 5 g of NaCl, and 15 g of agar per liter) was used as solid rich medium.
Genetic procedures
Transduction with phage P22 HT105/1 (int-201) (Schmieger 1972) was performed as previously described (Davis et al. 1980).
Plasmid pUST274 was constructed by cloning the DNA fragment containing the frameshift sequence into the BamHI and EcoRI sites of vector pGHM57 (Herr et al. 2001). The DNA fragment was made from the complementary oligonucleotides (5′-TTTGGATCCCGGGGGAAAGACGCCATTCTCTACTGTCCGAAT TCTTTT-3′ and 5′-AAAAGAATTCGGACAGTAGAGAATGGCGT CTTTCCCCCGGGATCCAAA-3′), and the ends were trimmed with BamHI and EcoRI endonucleases. Ligated plasmids were transformed into strain DH5α, analyzed by sequencing the insert, and retransformed by electroporation into different S. enterica strains. DNA sequencing was performed on chromosomal DNA, plasmids, or PCR products following the manual of the Applied Biosystems ABI PRISM Cycle Sequencing Ready Reaction Kit Big Dye.
Isolation of frameshift suppressors linked to purE
Phage P22 was grown on a culture of strain GT885 (hisO1242 hisC3072), and the phage stock was treated by hydroxylamine (Hong and Ames 1971). Alternatively, strain GT885 was treated with nitrosoguanidine, and phage P22 was grown on the mutagenized cells. Such phage stocks were used to infect strain GT5130 (hisO1242 hisC3072 purE::Tn10), and His+ and Pur+ transductants were selected on minimal medium. The obtained clone were tested for the linkage between His+ and Pur+ phenotypes by transductional analysis.
Cloning of the wild-type allele of the argU2343 frameshift suppressor
The S. enterica gene bank used in this study was kindly provided by Tord Hagervall (Umeå, Sweden). It contains 20,000 clones harboring chromosomal DNA fragments produced by partial Sau3A digestion and inserted into plasmid pLG399 (Stoker et al. 1982). This gene bank was introduced into the histidine prototrophic (His+) strain GT5164 (hisO1242 hisC3072 argU2343 fimC2529::Tn10dTc), and kanamycin-resistant (KmR) clones were screened for lost ability to grow without added histidine (His− phenotype).
Analysis of aminoacylation levels and cellular levels of tRNAArgmnm5UCU
Cells were grown at 37°C to a density of 4–6 × 108 cells/mL (100–150 Klett units) in LB medium. After harvesting by centrifugation, total RNA was prepared under acidic conditions as described by Varshney et al. (1991), and ~20 μg of the purified tRNA was used in further analysis. Half of it was deacylated in 0.5 M Tris-HCl (pH 9.0) at 37°C for 20 min, and both untreated and deacylated tRNA samples were separated on a 6.5% polyacrylamide gel containing 8 M urea. The tRNA was transferred to a Zeta-Probe GT membrane (BioRad) (Hagervall et al. 1998). Northern hybridization with tRNA-specific oligonucleotides was used for detection of different tRNA species. To avoid binding discrepancies induced by changed homologies due to mutations, the probe for tRNAArgmnm5UCU was complementary to the sequence, which was identical in all argU mutants and wild-type tRNA. The tRNA-specific oligonucleotides were complementary to nucleotides 14–31 of tRNAArgmnm5UCU (5′-CACGGCTTAGAAGGCCGTTG-3′), nucleotides 24–50 of tRNAmnm5s2UUGGln (5′-GGGAATGCCGGTATCAAAAACCGGTG-3′), and nucleotides 41–48 and binding to the Variable loop of tRNAcmnm5UmAALeu (5′-ACAGCGCGAACGCCGAGG-3′). The oligonucleotides were 5′-labeled using [γ-32P]-ATP (5000 Ci/mmol; Amersham Biosciences) and T4 polynucleotide kinase (USB). The membrane was prehybridized for 30 min and hybridied with the labeled oligonucleotide for another 30 min in hybridization buffer (1% crystalline BSA [fraction V], 1 mM EDTA, 7% SDS, 0.5 M sodium phosphate buffer at pH 7.2) at 70°C. After washing the radioactive bands were visualized and the radioactivity was quantified using a PhosphorImager and the ImageQuant analysis program.
Protein analysis
A previously described system was used to monitor frameshifting efficiency and to purify the frameshift product (Herr et al. 2001; Hansen et al. 2003). It employs a fusion protein consisting of maltose-binding protein (MBP) fused to glutathione-S-transferase (GST) at its N terminus and having six histidine residues (6×His) at the carboxy terminus (GST-MBP-6×His). A frameshift mutation in plasmid pUST274 has been inserted at the fusion point between the gst and malE genes (encoding the GST and MBP proteins, respectively). The resulting proteins produced from the fusion gene either following termination after the gst gene, or following a +1 frameshift and translation of the malE gene were purified by passing the protein extract over glutathionine-Sepharose (Amersham Biosciences). The presence of the two forms of protein was confirmed by Western blot analysis using anti-GST-HRP conjugate and ECL Plus Detection reagents (Amersham Biosciences). Bands specific for termination (26 kDa) and frameshift product (~70 kDa) were quantified and their relative ratio calculated. The full-length GST-MBP-6×His fusion protein was further purified by passing over Ni-NTA-agarose (QIAGEN). To strip the GST part from the frameshift product, the purified GST-MBP-6×His was digested by PreScission Protease (Amersham Pharmacia Biotech). Final cleanups and mass measurements of digestion products were performed as previously described (Hansen et al. 2003). In addition, digestion products were fractionated by gel electrophoresis and the 43-kDa peptide, corresponding to the slippage junction fused to MBP-6×His, was subjected to N-terminal sequencing by Edman degradation.
Acknowledgments
This work was supported by grants from the Swedish Cancer Foundation (Project 680) and Swedish Science Research council (Project BU-2930). We thank Kristina Nilsson and Kerstin Jacobsson for technical assistance, Per-Ingvar Ohlsson at the Department of Plant Physiology, Umeå University for protein sequencing and peptide mass determination, and Tord Hagervall and Mark Dopson for critical reading of the manuscript. We are grateful for the generous gift of the strain containing the original sufF44 mutation from John Roth, University of California at Davis, California, USA. Norma M. Wills and John F. Atkins are acknowledged for providing us with the vector pGHM57 and with the information on the assay system.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7256705.
REFERENCES
- Atkins, J.F., Herr, A.J., Massire, C., O’Connor, M., Ivanov, I., and Gesteland, R.F. 2000. Poking a hole in the sanctity of the triplet code: Inferences for framing. In The ribosome: Structure, function, and cellular interaction (eds. R.A. Garrett et al.), pp. 369–383. American Society for Microbiology, Washington, DC.
- Auffinger, P. and Westhof, E. 1999. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 292: 467–483. [DOI] [PubMed] [Google Scholar]
- Barak, Z., Gallant, J., Lindsley, D., Kwieciszewki, B., and Heidel, D. 1996. Enhanced ribosome frameshifting in stationary phase cells. J. Mol. Biol. 263: 140–148. [DOI] [PubMed] [Google Scholar]
- Bertani, G. 1951. Studies on lysogenesis. J. Bacteriol. 62: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk, G.R., Wikström, P.M., and Byström, A.S. 1989. Prevention of translational frameshifting by the modified nucleoside 1-methyl-guanosine. Science 244: 986–989. [DOI] [PubMed] [Google Scholar]
- Boycheva, S., Chkodrov, G., and Ivanov, I. 2003. Codon pairs in the genome of Escherichia coli. Bioinformatics 19: 987–998. [DOI] [PubMed] [Google Scholar]
- Chen, G.F. and Inouye, M. 1990. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 18: 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1994. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes & Dev. 8: 2641–2652. [DOI] [PubMed] [Google Scholar]
- Chen, K.S., Peters, T.C., and Walker, J.R. 1990. A minor arginine tRNA mutant limits translation preferentially of a protein dependent on the cognate codon. J. Bacteriol. 172: 2504–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.X., Bouquin, N., Norris, V., Casaregola, S., Seror, S.J. and Holland, I.B. 1991. A single base change in the acceptor stem of tRNA3Leu confers resistance upon Escherichia coli to the calmodulin inhibitor, 48/80. EMBO J. 10: 3113–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, W., Botstein, D., and Roth, J.R. 1980. A manual for genetic engineering. Advanced Bacterial Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Delagoutte, B., Moras, D., and Cavarelli, J. 2000. tRNA aminoacylation by arginyl-tRNA synthetase: Induced conformations during substrates binding. EMBO J. 19: 5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirheimer, G., Keith, G., Dumas, P., and Westhof, E. 1995. Primary, secondary, and tertiary structures of tRNAs. In tRNA: Structure, biosynthesis, and function (eds. D. Söll et al.), pp. 93–126. ASM, Washington, DC.
- Dong, H.J., Nilsson, L., and Kurland, C.G. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260: 649–663. [DOI] [PubMed] [Google Scholar]
- Farabaugh, P.J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60: 103–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh, P.J. and Björk, G.R. 1999. How translational accuracy influences reading frame maintenance. EMBO J. 18: 1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, C. and Parker, J. 1994. A ribosomal frameshifting error during translation of the argI mRNA of Escherichia coli. Mol. Gen. Genet. 243: 434–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal, T., Barkei, J., Dickson, R.R., deBoer, H.A., deHaseth, P.L., Alavi, H., and Gourse, R.L. 1989. Saturation mutagenesis of an Escherichia coli rRNA promoter and initial characterization of promoter variants. J. Bacteriol. 171: 4852–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant, J., Lindsley, D., and Masucci, J. 2000. The unbearable lightness of peptidyl-tRNA. In The ribosome: Structure, function, and cellular interaction (eds. R.A. Garrett et al.), pp. 385–396. American Society for Microbiology, Washington, DC.
- Gallant, J., Bonthuis, P., Lindsley, D., Cabellon, J., Gill, G., Heaton, K., Kelley-Clarke, B., MacDonald, L., Mercer, S., Vu, H., et al. 2004. On the role of the starved codon and the takeoff site in ribosome bypassing in Escherichia coli. J. Mol. Biol. 342: 713–724. [DOI] [PubMed] [Google Scholar]
- Garcia, G.M., Mar, P.K., Mullin, D.A., Walker, J.R., and Prather, N.E. 1986. The E. coli dnaY gene encodes an arginine transfer RNA. Cell 45: 453–459. [DOI] [PubMed] [Google Scholar]
- Giegé, R., Sissler, M., and Florentz, C. 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26: 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagervall, T.G., Pomerantz, S.C., and McCloskey, J.A. 1998. Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol. 284: 33–42. [DOI] [PubMed] [Google Scholar]
- Hansen, T.M., Baranov, P.V., Ivanov, I.P., Gesteland, R.F., and Atkins, J.F. 2003. Maintenance of the correct open reading frame by the ribosome. EMBO Rep. 4: 499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, D.K. and McClure, W.R. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11: 2237–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann, J.D. and deHaseth, P.L. 1999. Protein–nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry 38: 5959–5967. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Atkins, J.F., and Gesteland, R.F. 1999. Mutations which alter the elbow region of tRNA2Gly reduce T4 gene 60 translational bypassing efficiency. EMBO J. 18: 2886–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr, A.J., Nelson, C.C., Wills, N.M., Gesteland, R.F., and Atkins, J.F. 2001. Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J. Mol. Biol. 309: 1029–1048. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Wills, N.M., Nelson, C.C., Gesteland, R.F., and Atkins, J.F. 2004. Factors that influence selection of coding resumption sites in translational bypassing: Minimal conventional peptidyl-tRNA: mRNA pairing can suffice. J. Biol. Chem. 279: 11081–11087. [DOI] [PubMed] [Google Scholar]
- Hong, J.S. and Ames, B.N. 1971. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc. Natl. Acad. Sci. 68: 3158–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura, T. 1981a. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 146: 1–21. [DOI] [PubMed] [Google Scholar]
- ———. 1981b. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151: 389–409. [DOI] [PubMed] [Google Scholar]
- Jäger, G., Leipuviene, R., Pollard, M.G., Qian, Q., and Björk, G.R. 2004. The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica serovar Typhimurium. J. Bacteriol. 186: 750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., Suddath, F.L., Quigley, G.J., McPherson, A., Sussman, J.L., Wang, A.H., Seeman, N.C., and Rich, A. 1974. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185: 435–440. [DOI] [PubMed] [Google Scholar]
- Kruger, M.K. and Sorensen, M.A. 1998. Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol. 284: 609–620. [DOI] [PubMed] [Google Scholar]
- Kruse, T.A., Clark, B.F., and Sprinzl, M. 1978. The effect of specific structural modification on the biological activity of E. coli arginine tRNA. Nucleic Acids Res. 5: 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland, C.G. 1991. Codon bias and gene expression. FEBS Lett. 285: 165–169. [DOI] [PubMed] [Google Scholar]
- Kurland, C.G., Hughes, D., and Ehrenberg, M. 1996. Limitations of translation accuracy. In Escherichia coli and Salmonella. Cellular and molecular biology (eds. F.C. Neidhardt et al.), pp. 979–1004. American Society for Microbiology, Washington, DC.
- Li, J.N. and Björk, G.R. 1999. Structural alterations of the tRNA(m1G37)methyltransferase from Salmonella typhimurium affect tRNA substrate specificity. RNA 5: 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisser, S. and Margalit, H. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21: 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig, F., Borén, T., Claesson, C., Simonsson, C., Barciszewska, M., and Lagerkvist, U. 1993. The nucleotide in position 32 of the tRNA anticodon loop determines ability of anticodon UCC to discriminate among glycine codons. Proc. Natl. Acad. Sci. 90: 3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain, W.H. and Foss, K. 1988. Changing the acceptor identity of a transfer RNA by altering nucleotides in a “variable pocket.” Science 241: 1804–1807. [DOI] [PubMed] [Google Scholar]
- McClain, W.H., Foss, K., Jenkins, R.A., and Schneider, J. 1990. Nucleotides that determine Escherichia coli tRNAArg and tRNALys acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc. Natl. Acad. Sci. 87: 9260–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain, W.H., Jou, Y.Y., Bhattacharya, S., Gabriel, K., and Schneider, J. 1999. The reliability of in vivo structure-function analysis of tRNA aminoacylation. J. Mol. Biol. 290: 391–409. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Gojobori, T., and Ikemura, T. 2000. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 28: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B.F., and Nyborg, J. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270: 1464–1472. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Thirup, S., Kjeldgaard, M., and Nyborg, J. 1999. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Struct. Fold. Des. 7: 143–156. [DOI] [PubMed] [Google Scholar]
- O’Connor, M. 1998. tRNA imbalance promotes -1 frameshifting via near-cognate decoding. J. Mol. Biol. 279: 727–736. [DOI] [PubMed] [Google Scholar]
- O’Mahony, D.J., Mims, B.H., Thompson, S., Murgola, E.J., and Atkins, J.F. 1989. Glycine tRNA mutants with normal anticodon loop size cause -1 frameshifting. Proc. Natl. Acad. Sci. 86: 7979–7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, Q., Li, J.N., Zhao, H., Hagervall, T.G., Farabaugh, P.J., and Björk, G.R. 1998. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell 1: 471–482. [DOI] [PubMed] [Google Scholar]
- Riddle, D.L. and Carbon, J. 1973. Frameshift suppression: A nucleotide addition in the anticodon of a glycine transfer RNA. Nat. New Biol. 242: 230–234. [DOI] [PubMed] [Google Scholar]
- Riddle, D.L. and Roth, J.R. 1970. Suppressors of frameshift mutations in Salmonella typhimurium. J. Mol. Biol. 54: 131–144. [DOI] [PubMed] [Google Scholar]
- ———. 1972a. Frameshift suppressors. II. Genetic mapping and dominance studies. J. Mol. Biol. 66: 483–493. [DOI] [PubMed] [Google Scholar]
- ———. 1972b. Frameshift suppressors. III. Effects of suppressor mutations on transfer RNA. J. Mol. Biol. 66: 495–506. [DOI] [PubMed] [Google Scholar]
- Rosenberg, A.H., Goldman, E., Dunn, J.J., Studier, F.W., and Zubay, G. 1993. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J. Bacteriol. 175: 716–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K., Ishimaru, S., Kobayashi, T., Walker, J.R., and Yokoyama, S. 2004. The Escherichia coli argU10(Ts) phenotype is caused by a reduction in the cellular level of the argU tRNA for the rare codons AGA and AGG. J. Bacteriol. 186: 5899–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, P. and Walker, J.R. 1992. Expression of argU, the Escherichia coli gene coding for a rare arginine tRNA. J. Bacteriol. 174: 1956–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119: 75–88. [DOI] [PubMed] [Google Scholar]
- Schwartz, R. and Curran, J.F. 1997. Analyses of frameshifting at UUU-pyrimidine sites. Nucleic Acids Res. 25: 2005–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanjaard, R.A. and van Duin, J. 1988. Translation of the sequence AGG-AGG yields 50% ribosomal frameshift. Proc. Natl. Acad. Sci. 85: 7967–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanjaard, R.A., Chen, K., Walker, J.R., and van Duin, J. 1990. Frame-shift suppression at tandem AGA and AGG codons by cloned tRNA genes: Assigning a codon to argU tRNA and T4 tRNAArg. Nucleic Acids Res. 18: 5031–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sroga, G.E., Nemoto, F., Kuchino, Y., and Björk, G.R. 1992. Insertion (sufB) in the anticodon loop or base substitution (sufC) in the anticodon stem of tRNAProI2 from Salmonella typhimurium induces suppression of frameshift mutations. Nucleic Acids Res. 20: 3463–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, G., McCarty, G.P., and Farabaugh, P.J. 2002. Ribosome structure: Revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem. Sci. 27: 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, G., Salem, S.N., Chen, L., Zhao, B., and Farabaugh, P.J. 2004. Translational accuracy during exponential, postdiauxic, and stationary growth phases in Saccharomyces cerevisiae. Eukaryot. Cell 3: 331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker, N.G., Fairweather, N.F., and Spratt, B.G. 1982. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene 18: 335–341. [DOI] [PubMed] [Google Scholar]
- Tamura, F., Nishimura, S., and Ohki, M. 1984. The E. coli divE mutation, which differentially inhibits synthesis of certain proteins, is in tRNASer1. EMBO J. 3: 1103–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, U., Lee, C.P., and RajBhandary, U.L. 1991. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 266: 24712–24718. [PubMed] [Google Scholar]
- Vogel, H.J. and Bonner, D.M. 1956. Acetylornithinase of Escherichia coli: Partial purification and some properties. J. Biol. Chem. 218: 97–106. [PubMed] [Google Scholar]
- Weiss, R.B., Dunn, D.M., Atkins, J.F., and Gesteland, R.F. 1990. Ribosomal frameshifting from −2 to +50 nucleotides. Prog. Nucleic Acid Res. Mol. Biol. 39: 159–183. [DOI] [PubMed] [Google Scholar]
- Wenthzel, A.M., Stancek, M., and Isaksson, L.A. 1998. Growth phase dependent stop codon readthrough and shift of translation reading frame in Escherichia coli. FEBS Lett. 421: 237–242. [DOI] [PubMed] [Google Scholar]
- Wikström, P.M., Lind, L.K., Berg, D.E., and Björk, G.R. 1992. Importance of mRNA folding and start codon accessibility in the expression of genes in a ribosomal protein operon of Escherichia coli. J. Mol. Biol. 224: 949–966. [DOI] [PubMed] [Google Scholar]
- Youderian, P., Bouvier, S., and Susskind, M.M. 1982. Sequence determinants of promoter activity. Cell 30: 843–853. [DOI] [PubMed] [Google Scholar]