Abstract
RNA interference in mammalian cells is actively used to conduct genetic screens, to identify and to validate targets, and to elucidate regulators and modifiers of cellular pathways. To ensure the specificity and efficacy of the active 21mer siRNA molecules, it is pertinent to develop a strategy for their rational design. Here we show that most functional siRNAs have characteristic sequence features. We tested 601 siRNAs targeting one exogenous and three endogenous genes. The efficacy of the siRNAs was determined at the protein level. Using a decision tree algorithm in combination with information analysis, our analyses revealed four sets of rules with a mean knockdown efficacy ranging from 60% to 73%. (To distinguish between percentages used to describe the quality of an siRNA and the percentages used to describe parts of data sets we underlined the former throughout this paper.) The best rule comprises an A/U at positions 10 and 19, a G/C at position 1, and more than three A/Us between positions 13 and 19, in the sense strand of the siRNA sequence. Using these rules, there is a 99.9% chance of designing an effective siRNA in a set of three with more than 50% knockdown efficiency in a biological readout.
Keywords: siRNA design, gene silencing, RNA interference, decision tree algorithm, sequence analysis
INTRODUCTION
Fire et al. (1998) found that long double-stranded RNA (dsRNA) can specifically silence genes in Caenorhabditis elegans. An evolutionary conserved mechanism to protect against viral infections, RNA interference (RNAi) inhibits gene expression by degrading mRNA in a sequence-specific manner, upon introduction of double-stranded RNA (dsRNA). This long dsRNA is cut into 21–23-mer active intermediates, termed small interfering RNAs (siRNAs). The siRNAs are incorporated and unwound in the RNA-induced silencing complex (RISC) (Hammond et al. 2000). When loaded with a single-strand siRNA, RISC binds to a complementary sequence on the mRNA and cleaves it between nucleotides 10 and 11 of the siRNA (Elbashir et al. 2001a), thus initiating the degradation and inhibiting further gene expression.
RNAi was first used extensively in C. elegans, Drosophila melanogaster, and plants (Sharp 1999; Hannon 2002) as a gene knockdown technology. However, its use as a reverse genetic tool in mammalian systems remained limited because long dsRNAs elicit a strong interferon response (Stark et al. 1998). Elbashir et al. (2001a) and Caplen et al. (2001) demonstrated that, in contrast to long dsRNA, small interfering RNAs (21–23mers) generally do not induce an interferon response.
Different ways of producing these small interfering molecules, including chemical synthesis (Elbashir et al. 2002), in vitro transcription (Donze and Picard 2002), or vector-based delivery (Miyagishi et al. 2004) into mammalian cell lines, have been successfully developed over the last few years (Mittal 2004). Regardless of the transfection method, the efficiency of the siRNAs is mostly dependent on the successful rational design of the 21-mer sequences. Advances in the understanding of the biochemical mechanism of RNA interference (Schwarz et al. 2003) and statistical analyses of experimentally verified siRNAs (Gong and Ferrell 2004; Mittal 2004) have highlighted new biochemical and biophysical properties of the siRNA molecules.
In our studies, we have analyzed 601 chemically synthesized 21mer RNA duplexes with two 3′ nucleotide overhang (Elbashir et al. 2002). We used a subset of the highly active (>80% activity, POS) and nonactive siRNA sequences (<20% activity, NEG) for an information analysis and training a decision tree algorithm. (To distinguish between percentages used to describe the quality of an siRNA and the percentages used to describe parts of data sets we underlined the first ones.) The results from the information analysis were used to introduce ranked rules. Those rules were validated by analyzing an independent data set using siRNAs targeting unrelated genes and growing the data set with sequences of ambiguous quality (20%–80%). Here, we describe statistical evidence for certain biases of functional siRNAs and four sets of rules derived by a decision tree algorithm and information analysis. We applied our best rule to all human genes (ENSEMBL 19) and showed that 99.2% of the genome has at least three siRNA target sites, with a predicted average effectiveness of 73%.
RESULTS
To identify specific sequence features that are common among effective siRNAs, we designed and chemically synthesized siRNAs against three human endogenous genes (Rab6A, Tnfrsf1a, and p65/RelA) and one exogenous gene (LacZ). The sequences were chosen across a range of properties such as G/C ratio, melting point, secondary structure predictions, sequence variations, and codon usage on the mRNA. We measured the effectiveness of the siRNAs at the protein level using functional assays. The siRNA sequences together with their efficiency were then analyzed for their information content. We divided the data set into a training set and a test set and generated rules to predict the efficiency of the siRNAs. The best rule set increased the mean efficiency of a given siRNA from 49% (all 601 tested sequences) to 73%, giving a 99.9% chance of having one effective (>50%) siRNA in a set of three.
Result acquisition
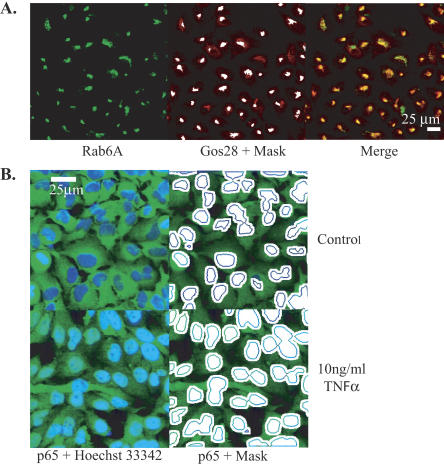
Knockdown efficiency of the siRNAs was determined by measuring either protein activity or level after siRNA transfection. We monitored the efficiency of LacZ siRNAs with a β-galactosidase enzymatic assay (data not shown). To allow for the accumulation of the protein before knockdown the plasmid encoding LacZ cDNA was transfected 24 h prior to siRNA transfection. To examine the relative level of endogenous protein two different methods were used. In the first method we coimmunolocalized the siRNA target with a marker protein for the same subcellular compartment and acquired the results on an automated confocal microscope (IN Cell 3000, GE Healthcare). The fluorescence of the target was determined in an object defined by the marker and extracted from the Object Intensity Analysis Module of the IN Cell Analyzer 3000 software. The readout was expressed as a percentage of fluorescence, standardized on mock transfected fluorescence levels. We used this method to measure the efficiency of Rab6A knockdown in the Golgi apparatus of HeLa cells (as defined by Gos28 localization; Fig. 1A). Briefly, the objects characterized by Gos28 expression (in red, right panel) were analyzed for Rab6A intensity (Fig. 1A, left panel, in green; middle panel, highlighted by a white mask). We used a similar method to measure the p65/RelA subunit of NFκB transcription factor knockdown efficiency. For this purpose, the level of p65 present in the nucleus (identified as marker object by Hoechst 33342 nucleic acid staining) of Tumor Necrosis Factor α stimulated cells was measured (data not shown). The second method tested the efficacy of the siRNA in a functional assay: the ability of an upstream gene knockdown to block a signal transduction cascade. For this purpose, we designed siRNAs against the Tumor Necrosis Factor α Receptor 1 gene (Tnfrsf1a) to block NFκB nuclear translocation upon TNFα stimulation (Leong and Karsan 2000; Aggarwal et al. 2004). The readout (see Materials and Methods) is illustrated in Figure 1B. Briefly, the mean fluorescence intensity of p65 (Fig. 1B, left panels, in green) was measured in two masks (Fig. 1B, right panels) defining the cytoplasm sampling zone (ring) and the nuclear region (plain circle). The results were then expressed as the nuclear to cytoplasmic fluorescence ratio for both TNFα stimulated and not stimulated cells. The knockdown efficiencies were in part confirmed by Western blot using anti-TNFR1 antibodies (data not shown).
FIGURE 1.
Measurements of expression level through immunostaining and high-throughput confocal microscopy. (A) Coimmunostaining of Rab6A (in green, left and right panels) and Gos28 (red, middle). The merged image appears in the right panel. The middle panel shows the mask (white) defined as the Gos28 subcellular expression compartment where the Rab6A expression level will be measured. (B) p65 translocation assay. p65 subunit localization (green) was monitored by immunostaining without (upper panels) or after addition of 10 ng/mL TNFα to monitor its translocation to the nucleus (lower left). The nuclei were counterstained with Hoechst 33342 (blue, left panels). The right panel shows the masks used to define the cytoplasmic and nuclear area where the mean intensities of p65 were measured.
Statistical analysis of results
We limited our studies to the 19mer duplex region of the siRNA and excluded the 3′ overhangs. Modifications of these two nucleotides have previously been shown not to alter efficiency (Martinez et al. 2002). Next, to rule out any biases we looked at the base-pair composition and melting temperatures. We did so since base-pair composition and information content of the combined positive and negative data sets can indicate whether a particular bias is present in the sequence. Any such bias would limit the usability of predictors derived from them.
As the training set we used the POS and NEG data (see Materials and Methods) derived from Rab6A and β-galactosidase experiments. To establish the predictive quality of the method, we gradually increased the data set in a stepwise manner. First, we included the remaining sequences of LacZ and Rab6A that were neither POS nor NEG (activity between 20% and 80%). Then we included the second set of experiments that included all sequences from Tnfrsf1α and p65. This procedure allows for a more realistic assessment of the prediction quality. In addition to that we give the results of a completely independent test data set: the POS, NEG sequences of Tnfrsf1α and p65 (Table 1).
TABLE 1.
Prediction accuracy of rules
| >70% | >50% | >20% | 1 out of 3 working (>70%) | 1 out of 3 working (>50%) | 1 out of 4 working (>50%) | |
| Rule 1 | 60% | 91% | 100% | 93.6% | 99.9% | >99.9% |
| Rule 2 | 59% | 77% | 96% | 93.1% | 98.8% | 99.7% |
| Rule 3 | 45% | 68% | 100% | 83.4% | 96.7% | 99.0% |
| Rule 4 | 43% | 68% | 93% | 81.5% | 96.7% | 99.0% |
| All data | 30% | 52% | 81% | 65.7% | 88.9% | 94.7% |
The prediction accuracy of the four rule sets (lines 2–5) and the complete data set (line 6) are shown. Columns 2–4 show the percentage of tested siRNA sequences that show more than 70%, 50%, or 20% activity for Rules 1–4 and for the whole data set. Columns 5–7 give the chance of getting at least one active siRNA in a set of three siRNAs (columns 5, 6) and in a set of four siRNAs (column 7), where active siRNA is defined by the percent activity: >70% (column 5) and >50% (columns 6, 7).
In measuring the information content in the combined data sets of POS and NEG we did not find any biases. The maximum information content, calculated with WebLogo (Schneider and Stephens 1990; Crooks et al. 2004), at any position in the 19mer siRNA sequences of the combined data set (POS, NEG) was lower than 0.02 bit (data not shown).
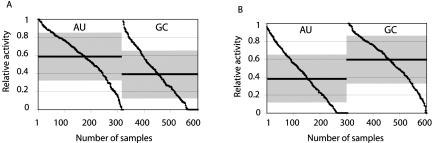
We only saw differences that were related to the A/U and G/C base-pair composition in the individual data sets (POS vs. NEG). When comparing the mean values and standard deviations of the base-pair compositions at different positions in the siRNA sequences, we observed the largest differences at positions 19 (Fig. 2A) and 1 (Fig. 2B). However, because of the large standard deviations and overlapping of the distributions, these properties can only be regarded as tendencies.
FIGURE 2.
Relative effectiveness for all tested sequences. The values are ordered according to their relative effectiveness. The mean values are indicated with a black bar and the gray area depicts the standard deviation. (A) On the left side, sequences having an A or U at position 19 (sense strand) are shown, whereas on the right side sequences with a G or C at position 19 are shown. Means and standard deviations: A/U 0.59 ± 0.26; GC 0.38 ± 0.28. (B) Efficiency for having an A/U (left) or a G/C (right) at position 1 of the sense strand. Means and standard deviations: A/U 0.40 ± 0.29; GC 0.58 ± 0.25.
We then calculated the information content for the sequences of the POS and NEG subsets individually using the WebLogo application (Schneider and Stephens 1990; Crooks et al. 2004). This application visualizes which nucleotides are overrepresented at a specific position by the letter size and order. The nucleotides are ordered by relative importance, having the most abundant nucleotide on top. The size coincides with the information content these nucleotides contain at a given position. As shown in Figure 3A,B, there are differences between the POS and the NEG siRNAs. The greatest differences were again found at positions 19 and 1 of the sense strand. At position 10 A/U is slightly dominating in the POS and G/C in the NEG siRNAs. These differences are complementary in the sense that A/U is in the POS and G/C is the NEG set dominant. Other minor differences that can be seen are not complementary. For example, at position 6 the most prominent bases for POS were A or U, which for the NEG were C or A. Since A is found in both the POS and NEG, the patterns do not complement, and therefore it is more likely that the feature at position 6 is an artifact.
FIGURE 3.
Graphic output from the WebLogo program for the POS (A) and NEG (B) sequences in the training set. Numbers on the x-axis represent the sequence positions in the siRNA (sense strand). The y-axis represents the information content measured in bits.
Decision tree algorithms are based on a principle of divide and conquer. They are robust and allow for an easy interpretation of the resulting rules because the relevant properties are specifically selected and weighted. Here we used the C4.5 decision tree algorithm to generate predictive rules (Quinlan 1993). We first estimated the optimal number of samples per two branches (parameter − m of the C4.5 program) by plotting the number of nodes of the resulting tree and the estimated error for a range of 1 to 30 samples (− m= 1...30). Both lines intersect at a minimum sample number of six (data not shown). After pruning, using standard parameters, C4.5 generated two rules that could be used for predicting functional siRNAs. The pruned version of the tree balances specificity of the rules with predictive accuracy. Rule 1 consisted of an A/U at position 19 and more than three A/U in the region of position 13 to position 19. This rather broad rule described 58 samples of the training set, misclassifying 10 NEG sequences. Then, we applied our knowledge from the information analysis and divided this data set further by distinguishing between A/U and G/C at position 1. Out of the 31 sequences with a G/C at position 1, 28 belonged to POS. Thus, we increased the selection accuracy of the training set from 83% to 90%. By including position 10 we selected 16 sequences with an A/U that were all active and 15 sequences with G/C of which 12 were highly active. We defined these rules as Rule 1 and Rule 2.
When looking at sequences with an A/C at position 1, we could not increase the accuracy by more than 4% when taking into account position 10. Thus, we did not divide this group and defined it as Rule 4.
The second of the original rules, defined as Rule 3, described only seven sequences and could not be improved by creating subgroups.
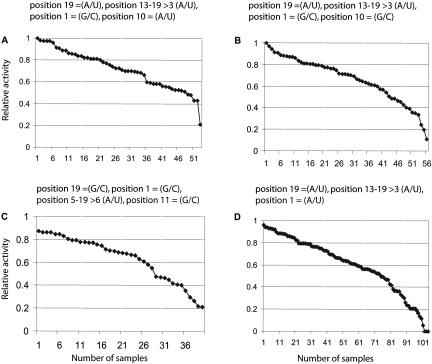
The distributions of the effectiveness of all tested sequences for these four rules are shown in Figure 4A–D.
FIGURE 4.
Sorted effectiveness of all tested sequences that comply with Rules 1–4 (A–D, respectively) are shown. The rules themselves are described above the graphs. The x-axis shows the number of siRNAs. Averages and standard deviations: (A) 0.72 ± 0.18; (B) 0.68 ± 0.22; (C) 0.62 ± 0.21; (D) 0.60 ± 0.25.
In the following, we describe the four rules in the context of the different data sets and the human genome: We applied Rule 1 to all the tested LacZ and Rab6A sequences (including the sequences with 20%–80% activity) and calculated the average activity to 70% (43 sequences). The test data set (POS and NEG of Tnfrsf1α and p65) contained four sequences with this motif, of which all were highly active. When applying this rule to all experimentally tested sequences, the average activity increases from 49% to 73% (53 sequences; Fig. 4A). Furthermore, there is only one sequence with less than 30% activity. Ninety-one percent of the sequences falling under Rule 1 are more than 50% active and 60% show an activity greater than 70%. There is a 93.6% chance of predicting at least one siRNA with more than 70% activity in a set of three and a 99.9% chance when using 50% activity as a cutoff (Table 1). When applying this rule to all 23,422 genes in the ENSEMBL database version 34 (Birney et al. 2004a,b), 99.2% (23,241) of the genes have at least three sequences complying with Rule 1. Interestingly, out of the theoretically possible 22,548,578,3045 sequences having this motif only 5,262,097 (0.23‰) are realized in the human genome.
Rule 2 differs from Rule 1 at position 10 where G/C replaces A/U. The average activity for Rab6A and LacZ is 53% (44 samples). The test set contained five sequences with this motif and all are active. When taking all tested sequences, the average activity increased to 68% (56 samples). Only three experimentally tested siRNAs with this motif were less than 30% active. Surprisingly, 9,155,723 sequences in the human genome have this motif, which is almost twice as many as for Rule 1. There is no easy explanation for this discrepancy because the overall chance of finding a G/C in the coding region is only 10% higher than finding an A/U. With this rule set, the chance of designing an effective siRNA is reduced to 59%, giving a 93.1% chance of having a least one working siRNA in a set of three. When applied to the human genome 23,305 (99.5%) genes have three or more sequences with this motif and 23,369 (99.8%) genes have three or more sequences following Rule 1 and/or Rule 2.
Rule 3 has a G/C at positions 1, 11, and 19 and more than six A/U in the region between positions 5 and 19. Only 40 siRNAs with this motif have been tested out of a possible 17,179,869,184, and 2,515,109 (0.15‰) of such sequences can be found in the human genome. The test set contained one sequence with this motif, which is active. The average activity for this rule is 62% and 45% of these sequences have more than 70% activity. This is again an improvement by comparison with the 30% siRNAs present in our full data set that are more than 70% active.
Rule 4 has an A/U at positions 19 and 1 and more than three A/U in the region between positions 13 and 19. The test set contains two sequences, of which one is active. The average activity of all 104 tested sequences with this motif is 60%, which is approximately a 10% increase over the overall average response of 49%. Only 43% of the sequences following this rule set show more than 70% activity, but 68% show at least 50% activity.
DISCUSSION
In this study, we developed four sets of rules that can be used to design siRNA sequences covering more than 99% of the human genome with high silencing efficiency. The rules we found are based on experimentally verified siRNAs targeting four genes (LacZ, Rab6A, Tnfrsf1a, and p65/RelA). The siRNAs were chosen to have not more than 15 consecutive nucleotides in common with any unrelated gene (see Materials and Methods). In contrast to Jackson et al. (2003), this was sufficient for us to minimize any potential cross-hybridization, off-target effect, as shown by micro array analysis (A. Volchuk et al., in prep. data not shown). Every siRNA sequence was also checked against the ENSEMBL SNPs database (Birney et al. 2004a,b) to avoid any loss of functionality due to polymorphism.
To assess the knockdown efficiency of the siRNAs, we measured the protein level after transfection using different methods. Assessing the interference efficacy at the protein level has been shown to highly correlate with the messenger RNA level remaining in the cells (Hsieh et al. 2004). We normalized the efficiencies on a per gene basis for comparing the results. The sequences were described by a total of 55 variables: All single sequence positions encoded to A/U= 1 and G/C= 0; melting temperature. Additionally we used the G/C ratio for all contiguous subsequences that include either the first or last nucleotide of siRNA sequence; these are: position 1–2, ..., 1–19, and 2–19, ..., 18–19 (see Materials and Methods).
We performed an information content analysis and showed that there are no specific biases in the selected sequences. We then used a decision tree algorithm to derive rules that classify siRNAs as being effective or not. This algorithm uses predefined features describing the sequences, and combines and ranks them. Compared to other classification algorithms like artificial neural networks, these rules can be directly interpreted. When training the decision tree we used only working (POS) and nonworking (NEG) siRNAs; siRNAs with ambiguous activity (20%–80%) were not used for training purposes. Then we used our knowledge from the information analysis to optimize the resulting decision tree and distinguished between different quality levels of siRNAs. We used siRNAs from one exogenous (LacZ) and one endogenous gene (Rab6A) to train the decision tree algorithm. The rules were then applied to a gradually increasing data set to point out tendencies of prediction accuracy. First, we used the complete set of sequence from LacZ and Rab6A and later all 601 sequences. For the first two rules, the average activity increased when applied to our larger data set (adding siRNA against Tnfrsf1a and p65/RelA to the ones against Rab6A and LacZ), thus demonstrating that these rules have a very good predictive value.
The four sets of rules we describe here are a mutually exclusive collection of properties. We only describe the rules to identify working siRNAs. The properties connected to these rules are either related to the 3′ region of the sense strand or are specific positions in the siRNA sequence.
We see an A/U enrichment toward the 3′ end of the siRNA and a dominating role of position 19 (Rules 1, 2, and 4 vs. Rule 3). The boundaries of this region are not consistently defined throughout all the rules, indicating that more data is needed to define these variables. Another possibility is that different correlations between positions and nucleotides would more accurately describe the matter; unfortunately we need more data to investigate this further. The enrichment in A/U at the 3′ end of the siRNA and specifically at position 19 has been demonstrated biochemically in the Zamore laboratory to allow better incorporation in the RISC complex (Schwarz et al. 2003). This has been confirmed by statistical analysis of the thermodynamical profiles of existing miRNA sequences (Khvorova et al. 2003) and several other statistical studies (Amarzguioui and Prydz 2004; Hsieh et al. 2004; Reynolds et al. 2004; Takasaki et al. 2004; Ui-Tei et al. 2004). A recent study by Chalk et al. (2004) also suggests that the two regions from positions 15 to 19 and from positions 5 to 9 are statistically important regions. The 3′ end of the siRNA is thought to be the first to interact with the RISC complex (Mittal 2004), thus explaining why this site has to be looser (less hydrogen bonds), to allow better interaction, unwinding of the duplex, and incorporation of the antisense strand. This property is further supported by the fact that the insertion of one to four mismatches at the 3′ end of the sense strand improves the efficiency by presumably destabilizing this end of the duplex (Hohjoh 2004).
Our findings show that A/U at position 10 (Rule 1 vs. Rule 2) and G/C at position 11 (Rule 3) increases the efficacy. Interestingly this corresponds to the nucleotides surrounding the cleavage site on the target mRNA (Elbashir et al. 2001b). It is therefore not surprising that these positions contain information.
At the 5′ end of the siRNA position 1 favors a G/C. This is in agreement with the results of others (Khvorova et al. 2003) asserting that the 5′ end of the siRNA consists of a tighter structure.
In our study, we did not confirm some rules described in other studies like an A at position 6 (Reynolds et al. 2004) or a G at position 16 (Hsieh et al. 2004). Although the information analysis revealed that these positions contain some information (Fig. 3) they were neither complementary for the POS and NEG sequences nor were they detected by the decision tree algorithm.
Reynolds et al. (2004) showed in a set of 180 siRNAs that positions 3, 10, 13, and 19 are biased toward certain nucleotides in functional siRNAs. Our results confirm these results at positions 10 and 19. Similar to their findings, we also see a trend toward a higher A/U ratio at the 3′ end of the sense strand, but our A/U-rich region from position 13 to 19 is 2 nt longer. The differences we see are most likely due to the limited data sets in both cases. Further analyses with a greater number of sequences have to be conducted to conclude a final rule set.
In addition to the rules proposed here, it is critical to ascertain that the sequences are as target specific as possible and no known SNPs are in the region of the siRNAs. For that purpose, targeting the coding region has the advantage of possessing fewer mutations than UTRs. Several siRNA molecules containing four consecutive cytidines were tested in this study. Their activity ranged from very potent to inactive. Despite this fact we caution the usage of this motif because it might pose problems in the chemical synthesis. It has also been demonstrated that the siRNA activity can be enhanced by destabilization of the duplex, by introducing mutations in the 3′ end of the sense strand (Hohjoh 2004), or by chemical modifications of the molecule (Chiu and Rana 2003). Krol et al. (2004) suggested that designing the siRNAs so that their properties resemble those of putative miRNA intermediates would produce highly effective siRNAs.
Our most dominant rule allows us to predict with 93.2% chance at least one effective siRNA (more than 70% activity) in a set of three (99.9% with more than 50% activity). Using only this rule we can already target more than 99% of the human genome with at least three siRNAs. Future work with more sequence samples will allow for more unrestricted parameters searches, where we will not limit ourselves to including the 5′ or 3′ end of the sequence. These extended data sets are needed to refine the parameters and map more precisely efficiency to sequence.
MATERIALS AND METHODS
siRNA synthesis and purification
The siRNAs used in this study were chemically synthesized using 2′-TOM RNA phosphoramidates (the precise methodology will be published independently).
Cell culture and reagents
HeLa CCL2 cells (ATCC) were cultured in DMEM high glucose supplemented with 10% fetal calf serum, penicillin, and streptomycin (MSKCC media preparation core facility). HUVEC, human umbilical vein endothelial cells, were grown in EGM-2 media supplemented with SingleQuot kit (Cambrex CC-3162). Cells were grown at 37°C in an atmosphere containing 5% CO2.
We used pCMV-Sport βgal (Invitrogen) as a reporter plasmid for the β-Galactosidase Assay (Invitrogen). The BCA Protein Assay (Pierce) was used to determine total cellular protein concentration.
We used the following antibodies for immunofluorescence and/or immunoblotting: mouse anti-p65 (Zymed #33–9900), mouse anti-Rab6A (Schiedel et al. 1995), rabbit anti-Gos28 (Nagahama et al. 1996), and goat anti-TNFR1 (R&D systems). Oregon Green 488 goat anti-mouse IgG (Molecular Probes), Cy-5 conjugated Donkey anti-mouse, or anti-rabbit IgG (Jackson Immunoresearch Labs) were used in immunostaining assays and horse radish peroxidase conjugated anti-mouse and anti-goat IgGs (Biorad) for Western blotting.
siRNA transfection
A day prior to transfection, 5000 cells per well were seeded in 96-well plates for immunostaining or β-galactosidase activity measurement. The transfection was performed as follows: 100 nM final concentration of siRNA was added to 15 μL Opti-MEM (Invitrogen). The mixture was then incubated for 5 min at room temperature. In the meantime, 0.5 μL of Oligofectamine (Invitrogen) were added to 2.5 μL of Opti-MEM. The two solutions were subsequently combined, gently mixed, and incubated at room temperature for 15–20 min. Cells were washed with PBS and supplemented with 80 μL of Opti-MEM. Following the incubation, the transfection complexes were added to the cells. After 4 h, media was supplemented with 100 μL of DMEM+ 20% FBS (for HeLa cells) or 150 μL EGM-2 media (for HUVEC cells). Cells were incubated for an additional 48 h at 37°C until assayed.
Quantification of knockdown: β-galactosidase activity
For the β-galactosidase assay, 5000 cells per well were seeded in 96-well plates (Nunc) and incubated overnight. One hundred nanograms of pCMV-Sport βgal were transfected into the cells using FuGene6 transfection reagent (Roche), using the standard manufacturer recommendations (reagent to plasmid ratio of 3:1). After 24 h, the cells were transfected with siRNAs as described. After an additional 48 h, β-galactosidase activity was measured using the β-Gal Assay Kit (Invitrogen). Activity was normalized to the total protein content of each of the samples (measured with the BCA Protein Assay Reagent Kit from Pierce, as advised by the manufacturer).
Quantification of knockdown: immunofluorescence
Protein abundance based on immunofluorescence was quantified using the IN Cell Analyzer 3000 high-throughput confocal microscope (GE Healthcare). One hundred nanomolar siRNAs were transfected in flat-bottom 96-well view plates (Packard) as described. Forty-eight hours after transfection the cells were fixed in 4% paraformaldehyde, permeabilized for 10 min (0.1% Triton X-100, 0.5% BSA in PBS), blocked for 1 h (2% BSA, 2% nonfat dry milk in PBS), and immunostained. The primary antibodies were incubated for 1 h at room temperature in blocking solution. After three washes, the adequate fluorophore-coupled secondary antibodies were added for another hour. When necessary, the nuclei were detected by Hoechst 33342 (Molecular Probes). The 96-well plates were then read and analyzed with the “Object Intensity Module” of the IN Cell Analyzer 3000 software. At least 1000 cells per data point were analyzed in four independent repeats. Briefly, the module uses two color channels: one for the object “marker” and another for the “signal”. First, the algorithm identifies the potential objects in the marker channel. It then applies size or intensity filters and generates an object mask that corresponds to the qualifying objects in the marker channel. Next, the algorithm establishes a measurement region by dilating the object masks by a user-specified number of pixels and generates a signal region mask that will be used to quantify the object intensities in the signal channel image (as recommended by the manufacturer in the IN Cell Analyzer 3000 user’s guide).
Quantification of knockdown: functional assay (NFκB translocation)
The last assay used tested the efficacy of the siRNA in a functional assay: the ability of a siRNA targeting the TNFα receptor gene 1 (Tnfrsf1a) to block the trafficking of the p65 subunit of NFκB transcription factor into the nucleus upon stimulation with TNFα. Cells were seeded and siRNA transfected as previously described in flat-bottom 96-well view plates (Packard). Forty-eight hours after siRNA transfection, cells were washed with PBS and replaced with media (either DMEM or EGM-2) containing 10 ng/mL TNFα (R&D Systems) or no cytokine. Plates were then incubated at 37°C for 20 min. Following stimulation, cells were immunostained for p65 (as described above). The nuclei were counterstained with Hoechst 33342 (Molecular Probes). The 96-well plates were subsequently read in the IN Cell Analyzer 3000 and analyzed using the Nuclear Trafficking Analysis module of the built-in software. At least 1000 cells per data point were analyzed in four independent repeats. The module provides a method to quantify the movement of a target molecule between the cytoplasm and the nucleus. To do this, the quantitative image analysis routine measures the fluorescence intensity of a target molecule in discrete nuclear and cytoplasmic regions and then calculates the ratio of the sampled intensities as an index of translocation. Briefly, the algorithm relies on the information captured in two color channels. The nucleus channel contains an image of the fluorescently stained cell nuclei (Hoescht 33342) and the cytoplasm channel an image of the fluorescently labeled translocating molecules of interest (p65). First, the algorithm identifies the potential objects in the marker channel image by applying a series of set-up parameters. It then applies any selected size or intensity filter and generates an object mask that corresponds to the qualifying objects in the marker channel image. Next, the algorithm uses the object mask to generate the sampling masks that are used to calculate the translocation index. To generate a nuclear sampling mask, the algorithm erodes the object mask by a user-specified number of pixels. Eroding the mask ensures that the sampling takes place well within the boundaries of the nucleus and helps to avoid edge-associated artifacts. To generate a cytoplasmic sampling mask, the algorithm expands (dilates) the object mask outward by a user-specified number of pixels. The algorithm then subtracts the original object mask from the dilated mask to obtain a ring-shaped mask that identifies the exclusively cytoplasmic sampling area. The algorithm then uses the nuclear and cytoplasmic masks to analyze the image stored in the cytoplasmic channel. It calculates the ratio of the average pixel intensities in the two sampling regions. The resulting nucleus/cytoplasm ratio provides an indication of the degree of translocation (as recommended by the manufacturer in the IN Cell Analyzer 3000 user’s guide).
Design of siRNAs
Gene sequences and SNP information are based on ENSEMBL 19 and the human genome version 34 therein (Birney et al. 2004a,b). The BLAST algorithm for small nearly identical matches (parameters used: −F F −I T −v 50 −b 50 −g T −W 7 −e 100 −M PAM30) (Lipman et al. 1989) was used to identify siRNAs that have less than 15 contiguous nucleotides aligning with a nonrelated gene. Additionally, siRNAs were checked not to overlap with any SNP annotated in the ENSEMBL database. Sequences were chosen to cover the greatest possible range of properties and values, not all of which are discussed here, but included G/C ratio, melting temperatures (Oligo 6.0; Molecular Biology Insights, Inc.), secondary structure (Hofacker 2003), MFold (Zuker 2003), and codon usage (Rice et al. 2000).
We used 601 siRNA sequences targeting the following genes: Tnfrsf1a, 67; LacZ, 344; Rab6A, 171; p65/RelA, 19.
Definition of response
The effectiveness of a siRNA was normalized against all siRNAs for that gene, where the siRNA with the highest activity was assigned 1 (or 100%) and the one with the lowest activity 0 (or 0%). For training, the sequences were divided into those clearly working well, having more than 80% activity (POS) and those not working, having less than 20% activity (NEG).
Decision tree classification
C4.5 is an implementation of a decision tree algorithm that generates classification rules based on a training data set (Quinlan 1993). We used the POS and NEG sequences designed against Rab6A and LacZ to train a decision tree. The “minimal number of sequences of samples per two branches” parameter of thw C4.5 program was optimized.
To perform a reasonably reliable statistical analysis of the features of functional siRNAs, the number of potential variables has to be as small as possible. That is why we did not encode the nucleotides individually but combined the nucleotides A, U and G, C, respectively. In addition to these 19 variables (individual sequence positions on the siRNA), we used the melting temperature as calculated by Oligo 6.0 and the G/C content of different sequence windows within the siRNA. These windows covered all possible coherent subsequences that contained either the 5′ or 3′ end of the siRNA. These features explicitly describe a correlation between positions in the siRNA. In total, we used 55 different variables. All positions mentioned in this publication are relative to the sense strand of the siRNA. The whole data set, including siRNAs from all tested genes and with all activity values, was used to evaluate the algorithm (test).
WebLogo
WebLogo is a computer program to calculate the information content in aligned sequences and is based on entropic measurements. We used a locally installed version of the WebLogo program (Schneider and Stephens 1990; Crooks et al. 2004) to calculate the information content at specific positions on the siRNA sequence.
We are generally making three siRNAs per gene and needed to estimate the probability that none of these siRNAs is effective [p(n)] to be able to better evaluate negative results in our studies. This can be calculated by the following formula, where p is the ratio of working siRNAs in a pool of siRNAs and n is the number of siRNAs made: (p(n) [%]) = 100 × (1 − (1 − p/100)n) [%].
Acknowledgments
We thank the FPP members for their contributions: Earl Kim (Informatics), Danuta Zatorska, Hui Fang, and Minxue Liu (siRNA chemical synthesis), and Constantin Radu, Chunyan Cao, and Geralda Torchon (Precursor phosphoramidite synthesis). We thank Memorial Sloan-Kettering and GE Healthcare Biosciences, Discovery Systems, for generous support of these studies. We also thank the members of the Rothman laboratory for helpful discussions during the course of this study. We especially thank Ai Yamamoto for critical reading of the manuscript and all the investigators at The Sloan-Kettering Institute for Cancer Research for providing us with results to confirm our algorithm.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7275905.
Footnotes
We counted 2688 different sequences with at least two A/U in a string of length 6 (region 13–18). At three positions only 2 nt are allowed (1, 10, 19). The remaining 10 positions allow for all four possible nucleotides. Then 2688 × 23 × 410 is the theoretical number of sequences having this motif.
REFERENCES
- Aggarwal, B.B., Takada, Y., Shishodia, S., Gutierrez, A.M., Oommen, O.V., Ichikawa, H., Baba, Y., and Kumar, A. 2004. Nuclear transcription factor NF-κ B: Role in biology and medicine. Indian J. Exp. Biol. 42: 341–353. [PubMed] [Google Scholar]
- Amarzguioui, M. and Prydz, H. 2004. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 316: 1050–1058. [DOI] [PubMed] [Google Scholar]
- Birney, E., Andrews, D., Bevan, P., Caccamo, M., Cameron, G., Chen, Y., Clarke, L., Coates, G., Cox, T., Cuff, J., et al. 2004a. Ensembl 2004. Nucleic Acids Res. 32: D468–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Andrews, T.D., Bevan, P., Caccamo, M., Chen, Y., Clarke, L., Coates, G., Cuff, J., Curwen, V., Cutts, T., et al. 2004b. An overview of Ensembl. Genome Res. 14: 925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplen, N.J., Parrish, S., Imani, F., Fire, A., and Morgan, R.A. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. 98: 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalk, A.M., Wahlestedt, C., and Sonnhammer, E.L. 2004. Improved and automated prediction of effective siRNA. Biochem. Biophys. Res. Commun. 319: 264–274. [DOI] [PubMed] [Google Scholar]
- Chiu, Y.L. and Rana, T.M. 2003. siRNA function in RNAi: A chemical modification analysis. RNA 9: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.M., and Brenner, S.E. 2004. WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze, O. and Picard, D. 2002. RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res. 30: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001a. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. 2001b. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20: 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Weber, K., and Tuschl, T. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26: 199–213. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Gong, D. and Ferrell Jr., J.E. 2004. Picking a winner: New mechanistic insights into the design of effective siRNAs. Trends Biotechnol. 22: 451–454. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Hofacker, I.L. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 31: 3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh, H. 2004. Enhancement of RNAi activity by improved siRNA duplexes. FEBS Lett. 557: 193–198. [DOI] [PubMed] [Google Scholar]
- Hsieh, A.C., Bo, R., Manola, J., Vazquez, F., Bare, O., Khvorova, A., Scaringe, S., and Sellers, W.R. 2004. A library of siRNA duplexes targeting the phosphoinositide 3-kinase pathway: Determinants of gene silencing for use in cell-based screens. Nucleic Acids Res. 32: 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., and Linsley, P.S. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21: 635–637. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., and Jayasena, S.D. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216. [DOI] [PubMed] [Google Scholar]
- Krol, J., Sobczak, K., Wilczynska, U., Drath, M., Jasinska, A., Kaczynska, D., and Krzyzosiak, W.J. 2004. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J. Biol. Chem. 279: 42230–42239. [DOI] [PubMed] [Google Scholar]
- Leong, K.G. and Karsan, A. 2000. Signaling pathways mediated by tumor necrosis factor α. Histol. Histopathol. 15: 1303–1325. [DOI] [PubMed] [Google Scholar]
- Lipman, D.J., Altschul, S.F., and Kececioglu, J.D. 1989. A tool for multiple sequence alignment. Proc. Natl. Acad. Sci. 86: 4412–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Mittal, V. 2004. Improving the efficiency of RNA interference in mammals. Nat. Rev. Genet. 5: 355–365. [DOI] [PubMed] [Google Scholar]
- Miyagishi, M., Matsumoto, S., and Taira, K. 2004. Generation of an shRNAi expression library against the whole human transcripts. Virus Res. 102: 117–124. [DOI] [PubMed] [Google Scholar]
- Nagahama, M., Orci, L., Ravazzola, M., Amherdt, M., Lacomis, L., Tempst, P., Rothman, J.E., and Sollner, T.H. 1996. A v-SNARE implicated in intra-Golgi transport. J. Cell Biol. 133: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, J.R. 1993. C4.5: Programs for machine learning. Morgan Kaufmann, San Mateo, CA.
- Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W.S., and Khvorova, A. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22: 326–330. [DOI] [PubMed] [Google Scholar]
- Rice, P., Longden, I., and Bleasby, A. 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Schiedel, A.C., Barnekow, A., and Mayer, T. 1995. Nucleotide induced conformation determines posttranslational isoprenylation of the ras related rab6 protein in insect cells. FEBS Lett. 376: 113–119. [DOI] [PubMed] [Google Scholar]
- Schneider, T.D. and Stephens, R.M. 1990. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208. [DOI] [PubMed] [Google Scholar]
- Stark, G.R., Kerr, I.M., Williams, B.R., Silverman, R.H., and Schreiber, R.D. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67: 227–264. [DOI] [PubMed] [Google Scholar]
- Takasaki, S., Kotani, S, and Konagaya, A. 2004. An effective method for selecting siRNA target sequences in mammalian cells. Cell Cycle 3: 790–795. [PubMed] [Google Scholar]
- Ui-Tei, K., Naito, Y., Takahashi, F., Haraguchi, T., Ohki-Hamazaki, H., Juni, A., Ueda, R., and Saigo, K. 2004. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32: 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]