Abstract
RNase P is a ubiquitous endoribonuclease responsible for cleavage of the 5′ leader of precursor tRNAs (pre-tRNAs). Although the protein composition of RNase P holoenzymes varies significantly among Bacteria, Archaea, and Eukarya, the holoenzymes have essential RNA subunits with several sequences and structural features that are common to all three kingdoms of life. Additional structural elements of the RNA subunits have been found that are conserved in eukaryotes, but not in bacteria, and might have functions specifically required by the more complex eukaryotic holoenzymes. In this study, we have mutated four eukaryotic-specific conserved regions in Saccharomyces cerevisiae nuclear RNase P RNA and characterized the effects of the mutations on cell growth, enzyme function, and biogenesis of RNase P. RNase P with mutations in each of the four regions tested is sufficiently functional to support life although growth of the resulting yeast strains was compromised to varying extents. Further analysis revealed that mutations in three different regions cause differential defects in holoenzyme assembly, localization, and pre-tRNA processing in vivo and in vitro. These data suggest that most, but not all, eukaryotic-specific conserved regions of RNase P RNA are important for the maturation and function of the holoenzyme.
Keywords: nuclear RNase P, RPR1, tRNA maturation, ribonucleoprotein, RNA affinity tag, kinetics
INTRODUCTION
The best studied function of ribonuclease P (RNase P) is the removal of the 5′ leader of precursor tRNAs (pre-tRNAs), in which the enzyme catalyzes the hydrolysis of a specific phosphodiester bond, leaving a phosphate at the 5′ end of the mature tRNA and a hydroxyl group at the 3′ end of the leader. The RNase P activity is found in all living cells, as well as mitochondria and chloroplasts (Frank and Pace 1998; Xiao et al. 2002). Most forms of RNase P holoenzymes are ribonucleoprotein complexes, i.e., they consist of RNA and protein subunits (Frank and Pace 1998; Xiao et al. 2002). Bacterial RNase P has an essential RNA moiety and a small, basic protein subunit. The RNA subunit by itself is able to catalyze the pre-tRNA cleavage reaction in vitro, and is one of the first identified catalytic RNAs or “ribozymes” (Guerrier-Takada et al. 1983).
Although eukaryotic nuclear RNase P contains an essential RNA subunit, the protein content is far more complex compared to the bacterial enzyme. The yeast Saccharomyces cerevisiae and human nuclear enzymes have 9 and 10 tightly associated protein subunits, respectively (Chamberlain et al. 1998; Jarrous 2002). The nomenclature for the nine yeast protein subunits is Pop1p, Pop3p, Pop4p, Pop5p, Pop6p, Pop7p, Pop8p, Rpp1p, and Rpr2p, and the RNA subunit is encoded by the RPR1 gene (Chamberlain et al. 1998). The RPR1 RNA is synthesized as a precursor form, pre-RPR1 RNA, which contains an 84-nt 5′ leader, where an internal promoter element for RNA polymerase III resides, and a short 3′-trailing sequence (Lee et al. 1991a). In actively growing yeast culture, the pre-RPR1 RNA appears to be converted to the mature form by removal of the 5′-leader and 3′-trailing sequences after seven of the nine protein subunits have associated with the RNA (Srisawat et al. 2002), and the ratio of mature to precursor RPR1 RNA is ~5:1.
In eukaryotes, RNase P is structurally related to another ribonucleoprotein enzyme, RNase MRP, which is involved in the nucleolar maturation of ribosomal RNAs (Lindahl and Zengel 1995; Reilly and Schmitt 1995; Tollervey 1995) and the turnover of selected messenger RNAs (Gill et al. 2004). Yeast RNase P and RNase MRP share eight protein subunits (Pop1p, Pop3p, Pop4p, Pop5p, Pop6p, Pop7p, Pop8p, and Rpp1p), but each enzyme has at least one unique protein subunit (Schmitt and Clayton 1994; Chamberlain et al. 1998). The RNA subunits of RNases P and MRP conform to a similar fold although the RNA sequences retain only limited identity between the two enzymes (Frank et al. 2000; Li et al. 2002). Eukaryotic RNase P and RNase MRP seem to have evolved from a common ancestor, but have diverged to carry out their distinct cellular functions.
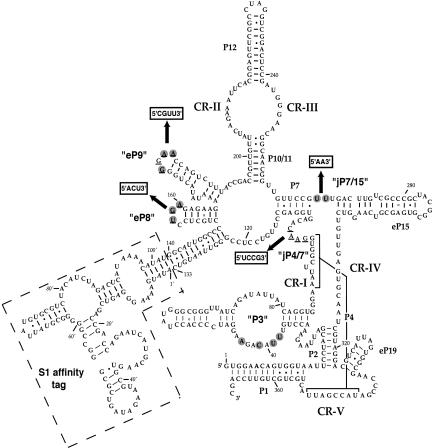
In spite of the varied protein compositions in RNase P from different organisms, the RNA subunit remains indispensable for activity. Phylogenetic-comparative studies have identified a core structure that contains five critical regions (CR I–V, Fig. 1) conserved in bacterial, archaeal, and eukaryotic nuclear RNase P RNAs (Chen and Pace 1997; Frank et al. 2000). The base pairing of CRI with CRV results in helix P4, which is postulated to be the catalytic core of the bacterial ribozyme (Nolan et al. 1993; Harris and Pace 1995; Kazantsev and Pace 1998; Christian et al. 2000; Crary et al. 2002). Structural elements have also been found that are conserved in eukaryotic RNase P RNA, but absent from the bacterial RNAs (Frank et al. 2000). One of the obvious eukaryotic-specific conserved regions is the internal loop of the P3 stem (Fig. 1). Nucleotide identities of the eukaryotic P3 loops are particularly well conserved between RNase P and RNase MRP RNA subunits in the same organism, suggesting that the sequences might have co-evolved with one or more protein subunits that bind to the P3 loop of both enzymes. Mutational and biochemical analyses have shown that this is the case. Mutation of the most conserved residues in the P3 internal loop of the yeast RPR1 RNA leads to a loss of specific binding by the largest protein subunit, Pop1p (Ziehler et al. 2001). In the case of human RNase P, the P3 region has also been shown to interact with an autoimmune To/Th protein antigen (Liu et al. 1994).
FIGURE 1.
Eukaryotic-specific conserved elements in the S1-RPR1 RNA. (P) Base-pair region. (eP) Stem structure in eukaryotic RNase P RNA that does not seem to have an obvious bacterial homolog (Frank et al. 2000). Loop regions between two stems are denoted with ‘jP’. Five critical regions (CR I–V) are conserved in bacterial, archaeal, and eukaryotic RNase P RNAs. Elements in quotation marks, P3, eP8, eP9, jP4/7, and jP7/15, are conserved features either unique to eukaryotic RNase P RNAs or with different sequence conservation in eukaryotic RNAs than in bacterial RNAs (Frank et al. 2000). Nucleotides in gray circles are most conserved in the corresponding eukaryotic conserved regions among yeast RNase P RNAs (Tranguch and Engelke 1993; Frank et al. 2000; Ziehler et al. 2001). Four conserved regions were mutated in this study where the underlined nucleotides were changed to the boxed sequences indicated with arrows. An S1 affinity tag with a short linker was inserted between nucleotides 133 and 140 of the wild-type RPR1 RNA as indicated within dashed lines.
Several other eukaryotic-specific conserved elements in RNase P RNAs have been found by comparison of the eukaryotic and bacterial consensus structures (Frank et al. 2000). These regions include the eP8 hairpin, eP9 stem-loops, and the junction between P4 and P7 (designated as jP4/7; Fig. 1). Although in bacterial RNase P RNAs, P8 and P9 stem-loops seem to occupy positions similar to those of eP8 and eP9, their functional equivalence remains to be confirmed. The fungal eP8 tetraloops (or pentaloops in some species) always have the sequence NUGA, while most of the fungal eP9 tetraloops have GNAA sequence (Frank et al. 2000). The jP4/7 loop, which is absent in bacterial RNase P, is found in fungal and human RNase P RNAs, although the loops are variable in size with low sequence conservation (Frank et al. 2000). In addition to the above conserved regions, two uridines are always found in an unpaired loop at the junction of the P7 stem and the eP15 stem in fungal RNase P RNAs (Fig. 1, jP7/15), whereas two adenosines occupy similar positions in bacterial RNAs (Tranguch and Engelke 1993; Frank et al. 2000).
Little is known about the roles of the eukaryote-specific sequences and structural features of RNase P RNA. Their possible functions include (1) providing binding sites for protein subunits of RNase P, as is the case for the P3 internal loop; (2) binding to other molecules in the nucleus to provide needed eukaryotic-specific functions, such as proper localization and transport of the enzyme; and (3) interacting with pre-tRNA substrates to form eukaryotespecific contacts, for example, recognition of the 3′ end of pre-tRNAs by eukaryotic RNase Ps is different than by the bacterial enzymes (Ziehler et al. 2000).
In this study, we have made mutations in the yeast RPR1 RNA at the loop regions of eP8 hairpin (eP8m), eP9 hairpin (eP9m), the junction of P4/P7 (jP4/7m), and the junction of P7/eP15 (jP7/15m). Mutations in these conserved regions have varying effects on RNase P assembly, localization, and activity. The mutation of jP7/15m does not affect cell growth, tRNA processing, or RNase P maturation. In contrast, mutants in eP8m, eP9m, and jP4/7m display pronounced growth defects and pre-tRNA processing defects in vivo. Assembly of the RNase P ribonucleoprotein complexes harboring each of these three mutations is affected, although each mutation has a distinctive effect. Mutations of eP9mand jP4/7m also result in significantly different subcellular distribution of RNase P, while mutation of eP8m has no obvious effect on RNase P localization. Steady-state kinetic studies of the pre-tRNA cleavage reaction reveal changes in KM and/or kcat for the reaction catalyzed by the three mutated enzymes.
RESULTS
Growth phenotypes of the S1-RPR1 mutants
The mutated sequences of the four RPR1 mutations, eP8m, eP9m, jP4/7m, and jP7/15m, are indicated in Figure 1. A streptavidin affinity tag, S1, was inserted between nucleotides 133 and 140 of the RPR1 RNA to facilitate characterization and purification of the enzymes (Srisawat and Engelke 2001). A secondary structure of the resulting S1-RPR1 RNA is shown in Figure 1.
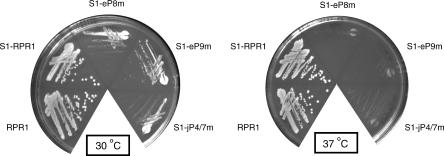
Strains carrying wild-type or various mutated RPR1 RNAs were tested for growth at 30°C, 37°C, and 16°C. The wildtype and the S1 tagged RPR1 strains show indistinguishable growth phenotypes at 30°C and 37°C, confirming that the S1 affinity tag does not interfere with growth. In contrast, mutant strains S1-eP8m, S1-eP9m, and S1-jP4/7m grow relatively slowly at 30°C, and the growth defect is more severe at 37°C (Fig. 2). Although several strains grow slowly at both 16°C (data not shown) and 30°C, no notable cold-sensitive growth defects are observed. The mutation of jP7/15m, which converted the conserved UU to AA, had no effect on growth, tRNA processing, or RNase P maturation (data not shown), and was not characterized further.
FIGURE 2.
Growth of strains containing mutated S1-RPR1 RNAs on YPD medium. The plates were incubated for 2 d at 30°C and 37°C, respectively.
Pre-tRNA and Pre-rRNA processing in the S1-RPR1 mutants
Effects of the RPR1 mutations on RNase P function in vivo were examined by analyzing the accumulation of precursors for two tRNA species, tRNALeu and tRNATyr. Processing of both of these pre-tRNAs by RNase P had been studied previously (Lee et al. 1997; Ziehler et al. 2000), and both were tested because pre-tRNATyr is more rapidly cleaved by RNase P than pre-tRNALeu.
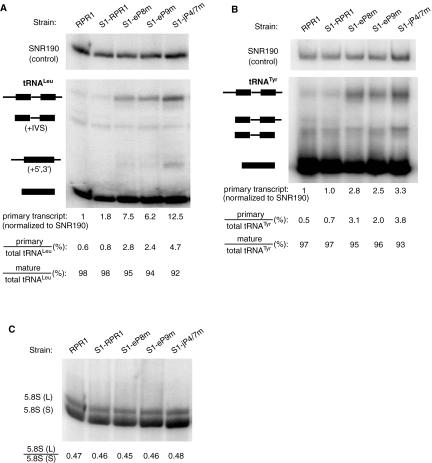
In the maturation of pre-tRNALeu and most other yeast pre-tRNAs, 5′ cleavage precedes 3′ processing, in part because the structure formed between the 5′ leader and 3′ trailer blocks access to cleavage of the 3′ trailer (Lee et al. 1997). Three major tRNALeu species are normally detected in Northern blots (Lee et al. 1991b)—i.e., the primary transcript, an intermediate with mature 5′ and 3′ ends containing the 32-nt intervening sequence (“+IVS” in Fig. 3), and the mature tRNA. In wild-type strains, the majority of the tRNALeu exists as the mature form, whereas the primary transcript and the +IVS species are low in abundance. This processing pattern is observed in the untagged wild-type strain FSY1/RPR1 and strain FSY1/S1-RPR1 (Fig. 3A). Although the abundance of pre-tRNALeu (but not pre-tRNATyr) increases slightly when the RNase P carries the S1 affinity tag, the defect is apparently not severe enough to slow growth significantly (Figs. 2, 3).
FIGURE 3.
Pre-tRNA and 5.8S rRNA processing in S1-RPR1 mutant strains. (A) Northern blot analysis on pre-tRNALeu. The four major tRNALeu species detected on a Northern blot are the primary transcript, an intermediate with the intervening sequence (+IVS), an intermediate with 5′ and 3′ extensions (+5, 3), and the mature tRNALeu (Lee et al. 1991b). (B) Northern blot analysis on pre-tRNATyr. Identities of the four major tRNATyr species on the blot are the primary transcript, an intermediate with the intron and a 3′ extension, the intermediate with an intron, and the mature tRNATyr (Kufel et al. 2002). Names and schematic diagrams of tRNA species are shown on the left of the blots. Black bars represent mature tRNA sequences whereas lines represent the 5′ leader, the intron, and the 3′ trailer sequences. Signal of the primary transcript was normalized to that of the loading control, SNR190 RNA. The amount of the primary transcript in the RPR1 wild-type strain was arbitrarily set at 1, and was used for comparison with data from the mutant strains. The relative amounts of primary transcript and mature tRNA are also listed as a percentage of the total tRNA species detected in the Northerns. (C) Northern blot analysis on the 5.8S rRNA processing. The same blot shown in A was reprobed for the 5.8S rRNA. Ratio of the long to short forms of 5.8S in each strain is listed.
In contrast, the amount of primary transcript increases significantly in mutant strains S1-eP8m (7.5-fold), S1-eP9m (6.2-fold), and S1-jP4/7m (12.5-fold) (Fig. 3A). Consistent with this, the percentage of the primary transcript relative to the total tRNALeu species also increases in the mutants, with 2.4–4.7% in different mutant strains versus 0.8% in the S1-RPR1 strain (Fig. 3A). However, the ratio of the mature to the total tRNALeu remains relatively constant in all the strains tested (Fig. 3A). An additional processing intermediate with the intron removed, but still containing the 5′ and 3′ extensions (denoted “+5, 3” in Fig. 3) normally appears when RNase P processing is severely slowed in vivo, allowing splicing to precede terminal processing (Lee et al. 1991b). This intermediate appears most prominently in the S1-jP4/7m strain (Fig. 3A), but also accumulates at a lower level in the S1-eP8m and S1-eP9m strains (darker exposure not shown).
To make sure that the tRNA processing defects observed were not specific to tRNALeu, we also examined the processing of tRNATyr in the S1-RPR1 mutant strains. The primary transcript of tRNATyr, which contains a 12-nt 5′ leader, an intron, and a 3′ trailing sequence, is accumulated modestly in all three mutant strains (2.5- to 3.3-fold accumulation; Fig. 3B). The ratio of the primary transcript to the total tRNATyr species is increased in the mutants, which is similar to what is observed for tRNALeu (Fig. 3A,B). These data suggest that the RPR1 RNA mutations cause a general defect in the 5′-end processing of pre-tRNAs.
Previously, it was observed that a severe mutation in the CR IV domain of RPR1 RNA (Fig. 1) mildly affected processing of 5.8S rRNA for unknown reasons, giving a 3′ extended form of about 7S rRNA in size (Chamberlain et al. 1996). We do not see the appearance of such aberrant bands in probing the 5.8S rRNA sequence in the RNase P mutants examined here (Fig. 3C). We also do not observe any change in the ratio of 5.8S (L) to 5.8S (S) forms, which would have indicated some perturbation in the function of RNase MRP (Fig. 3C). Although this lack of effect on 5.8S rRNA is not surprising, we tested this as a control to demonstrate that RNA processing in general is not slowed in these mutant strains.
Maturation of the S1-RPR1 RNA
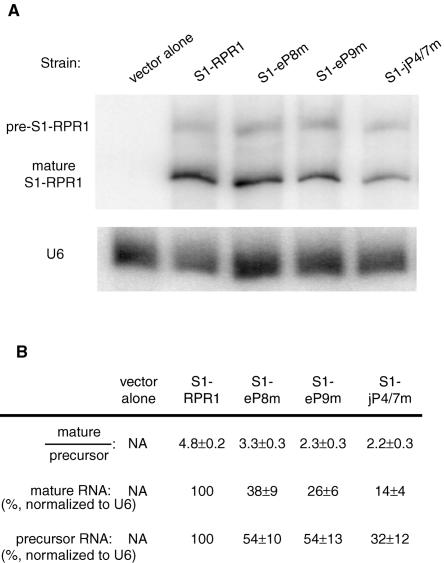
To further understand why the RPR1 mutations cause defective RNase P activity, we tested the effect of the mutations on maturation of the RPR1 RNA. We have previously found that mutations that either compromise the structure of the RPR1 RNA or prevent protein binding also block maturation of the precursor RPR1 RNA subunit (Pagan-Ramos et al. 1996a,b; Ziehler et al. 2001).
The maturation profiles of the mutated S1-RPR1 RNAs expressed from plasmids were examined in strains containing the untagged wild-type RPR1 RNA on the chromosome. The S1-RPR1 precursor and mature RNAs were detected by Northern blotting with a probe complementary to the S1 tag sequence. No bands are detected in the total RNA isolated from a control strain containing the vector alone, demonstrating that the probe does not hybridize to the endogenous untagged RPR1 RNA (Fig. 4, lane 1). The pre-RPR1 RNA generally migrates as a slightly diffuse band on a Northern blot because the 3′ end is heterogeneous (Lee et al. 1991b). When the S1-RPR1 strain is grown to mid-log phase, the ratio of mature to precursor RPR1RNAis~5 (Fig.4), which is normal for the maturation of wild-type RPR1 RNA. However, the ratio decreases twofold in strains S1-eP9m and S1-jP4/7m (ratio of 2.3 and 2.2, respectively), with an intermediate ratio in strain S1-eP8m (ratio of 3.3; Fig. 4). Moreover, the steady-state levels of the precursor and mature RPR1 RNAs in the mutant strains are consistently lower when normalized to the U6 snRNA (Fig. 4), suggesting that the mutated RPR1 RNAs might not be as stable.
FIGURE 4.
Processing of the S1-RPR1 RNA in the mutant strains. (A) Northern blotting of the S1-RPR1 RNA. Both the precursor (pre-S1-RPR1) and the mature S1-RPR1 RNA were detected on the Northern blot. The U6 RNA was probed as a loading control. (B) Quantitation of the S1-RPR1 RNA. Ratio of the mature to the precursor S1-RPR1 RNAs is listed with standard deviation from four Northern blots. Signals of the mature and precursor S1-RPR1 RNAs were also normalized to that of the U6 RNA. Amounts of the precursor and mature RNAs from the S1-RPR1 strain were set at 100% and were used for comparison with data from the mutant strains. Standard deviations obtained from four Northern blot analyses are provided.
The relative accumulation of the RPR1 precursor implies a defect in the assembly or stability of RNase P holoenzyme. Since the pre-RPR1 RNA binds at least seven of nine protein subunits before maturation (Pop1p, Pop4p, Pop5p, Pop6p, Pop7p, Pop8p, and Rpp1p) (Srisawat et al. 2002), defects in the RNA structure could alter the rate of precursor processing either directly or indirectly through blocking some essential RNP formation. Depletion of any of the seven protein subunits in the precursor complex, or compromised RNA structure or protein binding, will block maturation (Lygerou et al. 1994; Pagan-Ramos et al. 1996a,b; Chu et al. 1997; Stolc and Altman 1997; Chamberlain et al. 1998; Ziehler et al. 2001). In contrast, point mutations at CR II and CR III residues, which are primarily involved in pre-tRNA substrate interactions, allow pre-RPR1 maturation (Pagan-Ramos et al. 1996a). The relative accumulation of pre-RPR1 RNA is therefore a likely indication that either the RNA structure is altered or protein association is blocked, or both, thus hampering maturation. It is also possible that the stability of the mature RNase P particles is impaired, making them more susceptible to degradation, which could alter the ratio of the two species.
Association of the mutated S1-RPR1 RNA with RNase P protein subunits
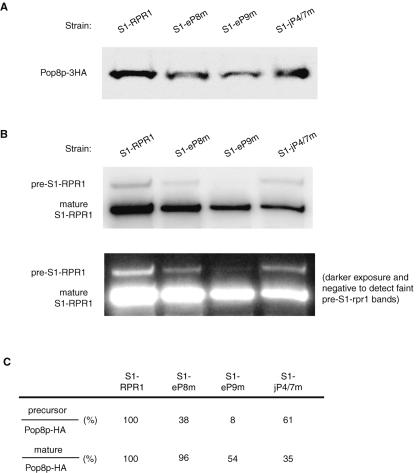
To address directly whether the mutations on the RPR1 RNA affect protein association, we performed co-immunoprecipitation of RNA with one of the early associating protein subunits. For unknown reasons, epitope tagging of most protein subunits is synthetically lethal in the presence of the S1 affinity tag on the RNA subunit. However, we were able to place three hemagglutinin (3HA) epitopes at the C terminus of Pop8p without negative effects on growth. Therefore, immunoprecipitation of Pop8p-3HA was performed, following which the S1-RPR1 RNA and Pop8p-3HA in the precipitate were examined by Northern blotting and Western blotting, respectively.
Pop8p-3HA is found in all the pull-downs in roughly equal quantities, as detected with anti-HA antibody in the Western blotting (Fig. 5A). Both the precursor and mature forms of all mutated S1-RPR1 RNAs are associated with Pop8p-3HA in the control and mutant strains, but the RNA-to-protein ratio changes significantly (Fig. 5B). Compared to the control, less precursor S1-eP8m RNA (38%) associates with the protein subunit (Fig. 5C). However, the binding of mature S1-eP8m RNA to Pop8p-3HA does not change significantly (Fig. 5C). This result suggests that the protein might be added more slowly to the precursor S1-eP8mRNA, but that the association is likely stable once the RNA is processed to the mature form because the mature RPR1-to-protein ratio is normal. The S1-eP9m mutation produces a more severe version of this, with a more than 10-fold decrease in the amount of bound precursor RNA, but only a twofold decrease in the amount of bound mature RPR1 RNA (Fig. 5C). The S1-jP4/7m RNA presents a different case. Although both the precursor and mature RPR1 RNAs associate less with Pop8p-3HA, the mutation affects the interaction with the mature RNA more significantly (Fig. 5C). The result suggests that all three of these regions contribute to some extent to the efficiency of pre-RPR1 association with proteins, whereas jP4/7 contributes to the stability of the mature RNase P complex once formed.
FIGURE 5.
Association of the mutated S1-RPR1 RNA with Pop8p-3HA in vivo. (A) Western blotting of Pop8p-3HA protein in the HA immunoprecipitates. (B) Northern blotting of the S1-RPR1 RNA copurified with Pop8p-3HA. Positions of the precursor and mature S1-RPR1 RNA on the blot are indicated. (C) Quantitation of the amount of Pop8p-3HA and S1-RPR1 in the immunoprecipitates. Signals of the precursor and mature S1-RPR1 RNA on the Northern blot were normalized to the signals of Pop8p-3HA on the Western blot to account for the difference in the immunoprecipitation efficiency among samples. The resulting ratio in the S1-RPR1 control strain was arbitrarily set at 100% and was used for comparison with ratios from the mutant strains.
Localization of the mutated S1-RPR1 RNA
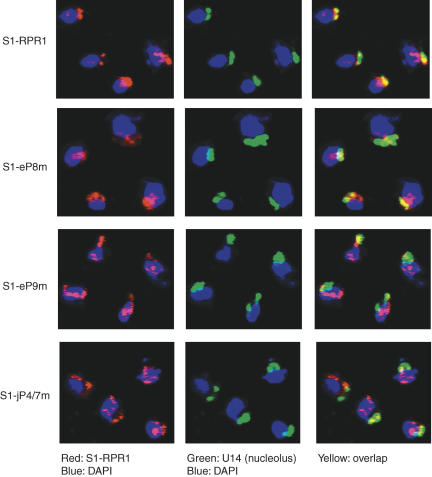
Early steps of the tRNA biogenesis pathway, including cleavage by RNase P, have been shown to occur in the nucleolus in yeast, and both the precursor and mature forms of RPR1 RNAs are primarily nucleolar (Bertrand et al. 1998; Srisawat et al. 2002). If the RNase P holoenzyme is assembled slowly or misassembled, then the nucleolar localization of the RPR1 RNA might be compromised. To test this hypothesis, we examined the subcellular distribution of the mutated S1-RPR1 RNA in strains FSY1/S1-eP8m, FSY1/S1-eP9m, and FSY1/S1-jP4/7m.
In the FSY1/S1-RPR1 control strain, the S1-RPR1RNAsignal substantially overlaps with the nucleolar U14 snoRNA marker (Fig. 6), although there is also nucleoplasmic signal as seen in earlier work (Bertrand et al. 1998). The signal for S1-eP8m RNA is not strikingly different from that in the S1-RPR1 strain, although examination of large numbers of cells could not rule out minor differences. This is consistent with the relatively small defect in RPR1 maturation in this strain (Fig. 5). In contrast, signals for S1-eP9m RNA and S1-jP4/7m RNA are reproducibly more diffuse, with considerably less overlap with the nucleolar U14 signal (Fig. 6). Thus, there is a correlation between holoenzyme assembly and localization, although it is not possible to determine causality between these events.
FIGURE 6.
Localization of the S1-RPR1 RNA in the mutant strains. The signal of S1-RPR1 RNA is shown in red and the signal of the nucleolar U14 RNA is shown in green. The blue DAPI staining indicates the nucleus. Right panels are the merged images of the corresponding left and middle pictures. Overlap of the red and green signals is shown in yellow. Images shown are representatives of large numbers of cells.
Effects of the S1-RPR1 mutations on RNase P activity in vitro
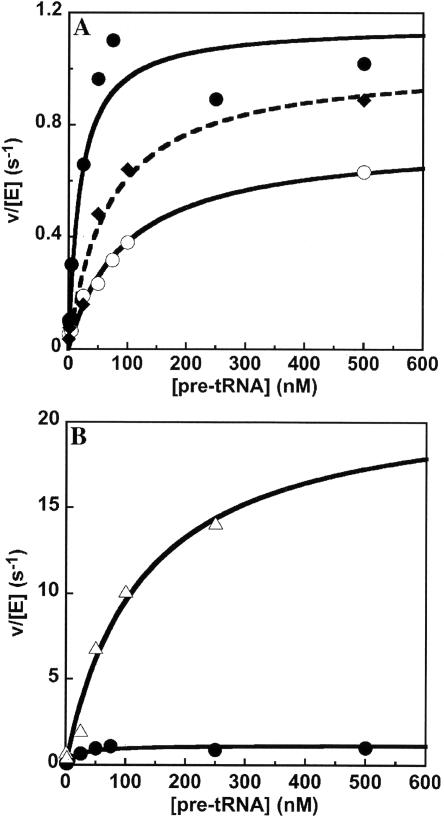
To determine the effects of the RPR1 mutations on RNase P catalytic activity, we measured the steady-state turnover for cleavage of S. cerevisiae precursor tRNATyr with a 12-nt 5′ leader catalyzed by RNase P mutants under initial velocity conditions. For each mutant the position of pre-tRNA cleavage appears normal (data not shown). We measured the activity of RNase P as a function of substrate concentration to determine the steady-state kinetic parameters, kcat, kcat/ KM and KM. The presence of the S1 affinity tag does not affect the value of kcat, as compared to the previously determined nontagged wild-type holoenzyme (1.2±0.1 and 1.3±0.1 sec−1, respectively; Fig. 7, Table 1) under identical conditions (Ziehler et al. 2000). The presence of the S1 tag decreases the observed KM from 55±10 nM in the nontagged enzyme to 20±8 nM in the S1-RPR1 enzyme. The S1 tag also increases the value of kcat/KM two- to threefold, moving it closer to the value of 1×108 M−1sec−1 observed for wildtype RNase P cleavage of a pre-tRNA substrate containing no trailer sequence (Ziehler et al. 2000). Previous data suggest that kcat most likely describes the rate constant for product release and kcat/KM nearly reflects the rate constant for substrate association under the conditions studied (Ziehler et al. 2000). For this kinetic mechanism, the value of KM cannot equal the substrate dissociation constant (Kd) but rather describes the apparent dissociation constant for the formation of all bound enzyme intermediates (Fersht 1985). These data suggest that the presence of the S1 affinity tag increases the apparent substrate association rate constant, likely by altering the partitioning between substrate dissociation and substrate cleavage.
FIGURE 7.
Processing of S. cerevisiae pre-tRNATyr in vitro. (A) The cleavage of a pre-tRNA substrate with a 12-nt leader catalyzed by either S1-RPR1 (•), S1-eP9m (♦) or S1-jP4/7m (○) in 10 mM HEPES (pH 8), 100 mM KCl, and 10 mM MgCl2 was determined under steady-state conditions where substrate concentration exceeds enzyme concentration. (B) The cleavage of pre-tRNATyr by S1-eP8m (▵) was measured under identical conditions as listed in A. The data for the S1-RPR1-catalyzed reaction (•) from A is included for comparison. The Michaelis–Menten equation is fit to the data to determine the values for kcat, kcat/KM, and KM (listed in Table 1).
TABLE 1.
Summary of the characterizations of the RPR1 mutations
| S1-RPR1 | S1-eP8m | S1-eP9m | S1-jP4/7m | |
| Mutated sequence | S1 insertion between nucleotides 133 and 140 | 158 ACUG 161 | 178 CGUU 181 | 99 UCCG 102 |
| Growth | Normal | Slow | Slow | Slow |
| Pre-tRNA processing | Normal | Deficient | Deficient | Deficient |
| Holoenzyme assembly | Normal | Mildly impaired precursor assembly | Severely impaired precursor assembly | Impaired assembly of precursor and mature complexes |
| Localization | Predominantly nucleolar | Nucleolar | Diffused, more nucleoplasmic | Diffused, more nucleoplasmic |
| Kinetics | ||||
| kcat (sec−1) | 1.2 ± 0.1 | 22 ± 3 | 1.0 ± 0.1 | 0.74 ± 0.03 |
| kM (nM) | 20 ± 8 | 130 ± 40 | 70 ± 20 | 127 ± 16 |
| kcat/kM (×107 M−1 sec−1) | 6 ± 2 | 17 ± 6 | 1.5 ± 0.4 | 0.58 ± 0.08 |
The steady-state kinetic data for the reactions catalyzed by the RPR1 mutants S1-eP8m, S1-eP9m, and S1-jP4/7m are depicted in Figure 7 and summarized in Table 1. Mutations in S1-eP9m and S1-jP4/7m decrease the observed kcat slightly (1.0±0.1 and 0.74±0.03 sec−1, respectively) as compared to wild type (1.2±0.1 sec−1) and increase the KM to 70±20 nM and 127±16 nM, respectively. Consequently, the specificity constant, kcat/KM, for the S1-eP9mcatalyzed reaction is 1.5±0.4×107 M−1sec−1, and for the S1-ejP4/7m is 0.58±0.08×107 M−1sec−1. These values are decreased fourfold and 10-fold, respectively, as compared to the S1-RNase P-catalyzed reaction (Table 1). For pre-tRNA cleavage catalyzed by S1-eP8m, both kcat (22±3 sec−1) and KM (130±40 nM) increase. Surprisingly, this leads to a threefold increase in the specificity constant (Table 1; Fig. 7) to a value that is slightly higher than the kcat/KM observed for wild-type RNase P cleavage of a pre-tRNA substrate containing no trailer sequence (Ziehler et al. 2000). In the case of the S1-eP8m mutant, the reaction is likely diffusion controlled since the specificity constant is 1.7×108 M−1sec−1. This increased value of kcat/KM indicates that every pre-tRNA that encounters RNase P is rapidly cleaved. This behavior may decrease the substrate selectivity of RNase P if cleavage of all pre-tRNA substrates is diffusion controlled.
DISCUSSION
Characterization of RPR1 mutations at four eukaryotic-specific conserved regions has revealed different roles of the conserved elements in eukaryotic RNase P activity and maturation. Replacing the UU at the jP7/15 region with AA does not notably affect cell growth, pre-tRNA processing, or the maturation of RNase P RNA. However, mutations at the other three conserved sequences—i.e., eP8, eP9, and jP4/7 regions—lead to pronounced slow growth phenotypes and pre-tRNA maturation defects, although none of the mutations is lethal. Taking advantage of an RNA affinity tag, we are able to further demonstrate that the defective RNase P enzymes have problems in holoenzyme assembly, localization, and in vitro pre-tRNA cleavage to varying degrees. The mutations of eP8m and eP9m reduce the binding of the protein subunit to pre-RPR1 RNA more significantly than to the mature RPR1 RNA, with eP9m showing more severe defective phenotypes (Fig. 5). In contrast, jP4/7m mutation appears to impair the association of the protein subunit with the mature RPR1 RNA more than the precursor RNA (Fig. 5). Consistent with the defects in holoenzyme assembly, the localization of eP9m and jP4/7m mutated RNAs is diffuse compared to the wildtype RNA, and is less associated with the nucleolus (Fig. 6). Kinetic analyses show that ep9m and jP4/7m mutations decrease the catalytic efficiency (kcat/KM) of the pre-tRNA cleavage reaction by fourfold and 10-fold, respectively, whereas mutation of eP8m increases the kcat/KM of the reaction in vitro (Table 1). The apparent discrepancy between the increased efficiency of the eP8m enzyme in vitro and the pre-tRNA processing defect caused by the eP8m mutation in vivo will be discussed below. A summary of the phenotypes of the individual mutation is listed in Table 1. It is worth noting that the presence of the S1 affinity tag in the RPR1 RNA constructs in this study might contribute to the observed phenotypes, for example, by weakening the stability of RNA structures. However, since the S1-tagged wild-type RPR1 RNA is used as a control, the defective phenotypes must also reflect a contribution from the mutated conserved sequences.
Region jP4/7 is conserved in the RNase P RNA from fungi and vertebrates, but is not found in bacterial or archaeal RNase P RNA (Frank et al. 2000). Mutation at jP4/7 impairs the association between protein subunits and the RPR1 RNA, especially in the mature form, suggesting defects in holoenzyme assembly and, possibly, the stability of the RNP complexes (Fig. 5). The observation is consistent with previous structure-sensitive footprinting results indicating that jP4/7 is protected from nuclease and chemical attacks in the holoenzyme (Tranguch et al. 1994). It is not known, however, whether the jP4/7 loop contacts protein subunits directly.
Mutation at the jP4/7 region causes the most severe defect in pre-tRNA processing in vivo (Fig. 3). Moreover, this is accompanied by a significant decrease (10-fold decrease) in the enzyme efficiency (kcat/KM) in vitro (Fig. 7; Table 1). Deficient RNase P cleavage in vivo might be mainly attributed to destabilization of either the RNA subunit or the holoenzyme. However, it is also plausible that jP4/7 could participate more directly in pre-tRNA binding or catalysis since this region is right next to P4, the postulated catalytic core of RNase P. A recent study on the yeast RNase MRP, a sibling enzyme of yeast RNase P, suggested that a bulged-loop structure equivalent to jP4/7might exist in the RNA subunit (Walker and Avis 2004). It will be intriguing to see whether the loop region plays similar roles in assembly and catalysis of RNase MRP.
Most fungal eP9 hairpins contain tetraloops of the GNRA type (Fig. 1). Presence of a tetraloop structure could increase the stability of RNA duplexes or participate in RNA–RNA interactions or RNA–protein interactions (Cheong et al. 1990; Gluck et al. 1992; Jaeger et al. 1994). Structure footprinting analysis of the yeast RNase P has indicated that the eP9 tetraloop is protected in the presence of protein subunits (Tranguch et al. 1994). In this study, the tetraloop of eP9 appears to be crucial for the assembly of precursor RNase P complex, since mutation at this region drastically reduces the association of pre-RPR1 RNA with protein subunits (Fig. 5). A recently refined consensus secondary structure of eukaryotic RNase P RNA has suggested that eP9 might be structurally homologous to the bacterial P9 stem-loop (Frank et al. 2000). The P9 tetraloop in both Escherichia coli and Bacillus subtilis RNase P RNAs was shown to be adjacent to the P1 stem of the RNA by crosslinking experiments (Harris et al. 1997; Chen et al. 1998). Whether eukaryotic eP9 is similarly positioned in the holoenzyme awaits further investigations.
Crystal structures of the specificity domains of two bacterial RNase P RNAs have shown that the P9, P10, and P11 stems form an opening structure that contributes to pre-tRNA recognition (Krasilnikov et al. 2003, 2004). A single nucleotide change in the tetraloop of the bacterial P9 mildly increases the KM and kcat of the pre-tRNA cleavage reaction catalyzed by the holoenzyme (Pomeranz Krummel and Altman 1999). Consistent with the studies on the bacterial enzyme, mutation in the tetraloop of eP9 in the yeast RPR1 RNA increases the KM of pre-tRNA processing in vitro without obviously changing the kcat of the reaction (Table 1). It is possible that eP9, like the bacterial P9, contributes to substrate recognition by eukaryotic RNase P, probably via formation of the substrate-binding site with P10/11 and potentially with protein subunits. However, with the increased protein content in eukaryotic RNase P and the diverged structural elements in the RNA subunit, eP9 might have different functions than the bacterial P9.
Mutation in the eP8 tetraloop only mildly reduces the association of protein subunits with precursor RPR1 RNA, and has little effect on the stability of the mature holoenzyme (Fig. 5). Consistent with this, there is not a major perturbation in the association of the RPR1 RNA with the nucleolus (Fig. 6). This mutation, however, causes a significant increase in KM and kcat/KM in the in vitro pre-tRNA cleavage reaction (Table 1). Therefore, this mutation leads to enhanced catalytic efficiency in vitro either by enhancing the association rate constant or changing the partitioning ratio between substrate dissociation and cleavage. If the rate constant for substrate association/dissociation is normally limited by a conformational change in wild-type RNase P, a less stable holoenzyme or one lacking one or more protein components could increase the rate of this conformational change.
The overall increase in catalytic efficiency (kcat/KM) caused by the eP8m mutation seems to be contradictory to the accumulation of pre-tRNA species with a 5′ extension in vivo (Fig. 3). While there might be a non obvious problem in nuclear localization or protein association, it is possible that the key change in the eP8m mutation is that it has become less specific for binding pre-tRNA substrates, since the reaction catalyzed by this mutated enzyme in vitro is diffusion controlled. In this case, the mutated enzyme might be more susceptible to competitive inhibition by other RNAs within the nucleoplasm or nucleolus. Consistent with this hypothesis, titration with total yeast RNAs as competitors shows that the apparent KI for the pre-tRNA cleavage reaction catalyzed by eP8m is 0.02 μg/μL, ~50- fold less than that for the reaction catalyzed by the wild type enzyme (1.0 μg/μL; data not shown).
Phylogenetic studies have demonstrated that the eukaryotic eP8 hairpin and the bacterial P8 might be homologs because they occupy similar positions in the four-way junction of RNase P RNA (Frank et al. 2000). Intriguingly, in the crystal structure of the specificity domain of an A-type RNase P RNA, P8 is involved in tertiary structural interactions to stabilize the conformation of the pre-tRNA recognition region (Krasilnikov et al. 2004). It is possible that eukaryotic eP8 might also stabilize the substrate recognition site, and that the mutated tetraloop could broaden the substrate specificity of the enzyme.
The studies presented here identify contributions of the eukaryotic-specific RPR1 sequences to holoenzyme assembly and pre-tRNA cleavage, but it is entirely possible that there are additional consequences of mutations at these sites. For example, it is possible that, like the bacterial RNase P, the nuclear enzyme has additional substrates that have not yet been identified and that require some contribution from these sequences. Alternatively, the RNase P holoenzyme might need to interact with some proteins that are associated with substrates in vivo, for instance, the Lsm (Sm-like) or La proteins bound to the 3′ ends of pre-tRNA transcripts (Kufel et al. 2002), or some other cellular component that facilitates substrate acquisition.
MATERIALS AND METHODS
Plasmids and mutagenesis
Mutation at eP8, eP9, or the junction of P4/P7 was individually introduced into the low-copy plasmid construct pRS315-S1-RPR1, which contained the RPR1 gene under its own promoter and a LEU2 selectable marker. PCR-based mutagenesis was performed as described (Srisawat and Engelke 2001). Each of the mutated RPR1 sequences, including a S1-RPR1 “wild-type” control, contained an “S1” RNA affinity tag that allowed the RNA to bind tightly to streptavidin, but to be released in the presence of biotin (Srisawat and Engelke 2001). A strain containing YCp50-RPR1 (Lee et al. 1991b), with the untagged wild-type gene on a low-copy plasmid, was used as a control for consequences of the S1 tag insertion.
Strains
Effects of the mutations on maturation of the RPR1 RNA were examined in the haploid S. cerevisiae strain W3031A (MATa ade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100) with mutated S1-RPR1 RNAs expressed on pRS315 plasmids. The resulting strains were W3031A/S1-RPR1, W3031A/S1-eP8m, W3031A/S1-eP9m, and W3031A/S1-jP4-7m. These strains also had a chromosomal wild-type RPR1 gene intact.
Mutated S1-RPR1 RNAs on pRS315 plasmids were transformed into strain FSY1 (MATa ade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 RPR1::kan) containing a chromosomal deletion of the RPR1 gene, but a copy of the wild-type gene on a counterselectable plasmid, YCp50-RPR1. Transformants containing the mutated RPR1 genes were selected for the loss of YCp50-RPR1 on medium with 5-fluoroorotic acid, resulting in strains FSY1/S1-RPR1, FSY1/ S1-eP8m, FSY1/S1-eP9m, and FSY1/jP4–7m. These strains, which contain only the mutated RPR1 RNA, were used to test growth phenotypes, pre-tRNA and pre-rRNA processing profiles, and localization of S1-RPR1 RNA.
To address whether the mutations on the RPR1 RNA cause assembly defects of the holoenzyme, a strain containing the chromosomal POP8 gene tagged with 3xHA sequences at the 3′ end was used (Srisawat et al. 2002). The pRS315 plasmids with mutated S1-RPR1 RNA genes were the only source of RPR1 RNA in the strain. HA-immunoprecipitation of RNase P was performed to examine the association of Pop8p-3HA with mutated RPR1 RNAs.
Pre-tRNA and pre-rRNA processing
Strains with pRS315-S1-RPR1 or its mutant derivatives in the FSY1 strain background were grown in YPD medium at 30°C to an OD600 of 1. Total cellular RNA was extracted with hot acidic phenol and precipitated with ethanol as described (Kohrer and Domdey 1991). Concentration of total RNA was determined by absorbance at 260 nm. Ten micrograms of isolated RNA were electrophoresed on an 8% denaturing polyacrylamide gel and transferred onto a nytran membrane. The blot was then probed with 32P-labeled DNA oligonucleotides complementary to tRNALeu, tRNATyr, and to 5.8S rRNA to assess the in vivo activities of RNase P and MRP. The tRNALeu probe was 5′-TGCTAAGAGATTCGAACTCTTGCA-3′, the tRNATyr probe was 5′-AGTCGAACGCCCGATCTCAAGATT-3′, and the 5.8S rRNA probe was 5′-CGCATTTCGCTGCGTTCTTCATCG-3′. The SNR190 RNA, a small nucleolar RNA that is not affected by RNase P mutations, was probed on the same blot as an internal normalization control (probe: 5′-ATGGTCGAATCGGACGAG-3′). Signals were detected using a PhosphorImager (Molecular Dynamics 445 SI), and quantified with IPlab Gel software (Signal Analytics). Quantitation shown in Figure 3 is the average of three independent experiments with consistent results.
Maturation of the RPR1 RNA
W3031A strains containing plasmid-borne S1-RPR1, S1-eP8m, S1-eP9m, S1-jP4/7m RNAs, or the empty vector were grown in SDC-Leu medium at 30°C to an OD600 of ~0.7. Ten micrograms of total RNA were separated on a denaturing 6% polyacrylamide gel and transferred onto a nytran membrane. The blot was probed with radiolabeled DNA oligonucleotides complementary to the S1 affinity tag that detected the mutated RPR1 RNA from the plasmid, but not the endogenous wild-type RPR1 RNA. Sequence of the S1 probe was 5′-GCATGATTCTGGTCGGTCGACTCCC-3′. As an internal loading control, U6 RNA was probed on the same blot with radiolabeled DNA oligonucleotide with sequence 5′-CTGATCATCTCTGTATTG-3′. The hybridized RNAs were detected and quantitated as above. The ratio of mature to precursor S1-RPR1 RNA listed in Figure 4 is the average of four Northern blotting analyses with standard deviation less than 0.3.
HA-immunoprecipitations and Western blotting
Strains containing Pop8p-3HA and pRS315 expressing S1-RPR1 or the mutated RNAs were grown in SDC-Leu medium at 30°C to an OD600 of ~1. Cells were washed with water and Buffer A (50mM Tris-HCl at pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% Triton-X-100, 10% glycerol, 1 mM DTT, 1 mM PMSF). About 2×109 cells were resuspended in 400 μL of Buffer A and were broken by vortexing with glass beads (425–600 μm diameter). Total cell extracts were centrifuged at 16,000 g for 2 min at 4°C. The supernatant was centrifuged again at 16,000 g for 15 min at 4°C. Protein content of the extract was determined with Micro Bicinchoninic Acid Assays (Pierce). FSY1 strain with wild-type Pop8p and S1-RPR1 RNA was included as a negative control for HA-dependent immunoprecipitation.
Protein A-agarose (BioRad Affi-Gel) was incubated with rabbit anti-mouse IgG (2 μL per 200 μL of packed beads, AffiniPure Rabbit Anti-Mouse IgG (H+L); Jackson ImmunoResearch) at 4°C for 1 h, with gentle rotation. Anti-HA antibody 12CA5 (10 μg per 200 μL packed beads; Roche) was added to the coupled protein A-agarose-IgG complex, and the mixture was incubated at 4°C for 2 h. The conjugates were washed six times with Buffer A.
Total yeast extract with 3 mg of protein was bound to 200 μL protein A–agarose–IgG–antiHA complex at 4°C for 1 h, and then washed with 1 mL of Buffer A five times. The volume of the bead slurry was brought to 300 μL with Buffer A. Portions of the slurry were saved for Western blotting of Pop8p-3HA (80 μL) and Northern blotting of S1-RPR1 RNA (220 μL).
Eighty microliters of the bead slurry were spun down at 1500 rpm for 2 min. Proteins bound to the beads were extracted with 40 μL of sample buffer with 6 M urea (60 mM Tris-HCl at pH 6.8, 2% SDS, 15% glycerol, 0.025% bromophenol blue, 5% β-mercaptoethanol, 6 M urea) at 95°C for 5 min. The supernatant sample was electrophoresed on a 12% polyacrylamide-Tris-HCl gel in SDS running buffer (BioRad) and transferred to a PVDF membrane. The blot was probed with anti-HA (12CA5; Roche) and goat anti-mouse HRP antibodies (Chemicon AP308) and was subjected to chemiluminescent substrate (ECL Plus Western Blotting Detection System; Amersham Biosciences). To detect the signals, the blot was exposed to an X-ray film (Kodak BIOMAX MR), and was also scanned with a Typhoon scanner (model 9400) with fluorescence detection settings (Amersham application note: Fluorescent Western Blotting).
Northern blotting of S1-RPR1 RNA in anti-HA immunoprecipitation
RNA bound to the 220 μL of Pop8p-3HA bead slurry was extracted with an equal volume of 5% SDS, 10% β-mercaptoethanol at 95°C for 5 min. Eluted RNAs were extracted with phenolchloroform, and ethanol precipitated with glycogen carrier. The RNA pellet was washed with 70% ethanol and resuspended in 2× formamide/EDTA/SDS loading buffer (95% formamide, 20 mM EDTA, 0.1% SDS, 0.05% bromophenol blue, 0.05% xylene cyanol). RNA samples were separated on a denaturing 6% polyacrylamide gel and transferred to a nytran membrane. The blot was probed with radiolabeled DNA oligonucleotide complementary to the RPR1 RNA (5′-GCTGGAACAGCAGCAGTAATCGGTA-3′). The signals were detected and quantitated as above.
To examine the association of the mutated S1-RPR1 RNAs with Pop8p-3HA, the amount of the S1-RPR1 RNA in the precipitates was then normalized to that of Pop8p-3HA to account for the difference in immunoprecipitation efficiency among samples. Data displayed in Figure 5 are representations of Western and Northern analyses of three independent experiments, which gave quantitatively similar results within expected error.
RNase P enzyme assays
Strains containing wild-type or the mutated S1-RPR1 RNA expressed on the pRS315 plasmid in the FSY1 strain background were grown in YPD medium at 30°C to an OD600 of ~1. Yeast extracts from 1.5×1010 cells were prepared as described by Srisawat and Engelke (2001). RNase P complexes associated with the S1-RPR1 RNA were purified by streptavidin affinity chromatography from yeast extract containing 50 mg of total protein as described (Srisawat and Engelke 2001). Concentrations of the eluted RNase P enzymes were determined by Northern blotting of the S1-RPR1 RNA in the biotin eluate in comparison to known amounts of in vitro transcribed S1-RPR1 RNA.
The eluted RNase P enzymes were tested for catalysis of pre-tRNA cleavage in vitro under steady-state conditions as described (Ziehler et al. 2000) in a reaction buffer containing 10 mM HEPES (pH 7.9) 100 mM KCl, and 10 mM MgCl2. The 5′ radiolabeled pre-tRNATyr substrate containing a 12-nt leader and a 3′ polyuridine trailer was incubated in 10 mM Tris HCl (pH 8), 0.1 mM EDTA at 95°C for 3 min prior to the addition of 2× reaction buffer and subsequent incubation at 37°C for 15 min. The enzyme assay was initiated by the addition of RNase P (0.04– 16.3 pM). At various times, an aliquot of the reaction mixture was diluted into an equal volume of stop solution (200 mM EDTA at pH 8.0, 8 M urea, 0.05% bromophenol blue, and 0.05% xylene cyanol). Reaction times were chosen so that <10% of substrate was cleaved (initial velocity conditions). The substrate and 5′ leader product were separated on a 6% polyacrylamide 7 M urea gel. Quantitation was performed using a PhosphorImager with ImageQuant software (Molecular Dynamics). The Michaelis–Menten equation was fit to the data using KaleidaGraph software (Synergy Software) to determine the kinetic parameters kcat, KM, and kcat/KM.
Localization of S1-RPR1 RNA in the mutants
Subcellular locations of the RNase P RNA in FSY1/S1-RPR1, FSY1/ S1-eP8m, FSY1/S1-eP9m, and FSY1/S1-jP4-7m were detected by fluorescent in situ hybridization (FISH). Yeast strains were grown in SDC-Leu medium at 30°C to anOD600 of ~0.4. Cells were fixed, hybridized with fluorescently labeled oligodeoxynucleotides, then stained with DAPI as described previously (Bertrand et al. 1998; Thompson et al. 2003), except that cells were spheroplasted with 0.2 mg/mL Zymolyase at 37°C for 40 min. The S1-RPR1 RNA was probed with two fluorescent oligonucleotides, the anti-S1 fluorescent probe (anti-S1-f) 5′-GACT*ATCTTACGCACT*TGCAT GATTCT*GGTCGGTCGACT*CCC-3′ and anti-RPR1 fluorescent probe (anti-RPR1-f) 5′-GACCT*AGGCCGAACTCCGT*GAATTT CTGAT*AACAACGGTCGGT*A-3′ with Cy3 (Amersham) attached to the dT positions denoted with asterisks. The U14 snoRNA was probed as a nucleolar marker with oligonucleotides 5′-CGAT* GGGTTCGTAAGCGT*ACTCCTACCGT*GG-3′, with asterisks indicating dTs labeled with Oregon Green 488 (Molecular Probes).
Acknowledgments
We are grateful to F. Houser-Scott, P. Good, S. Walker, D. Coughlin, and members of the Engelke laboratory for helpful discussions. We thank J. Hsieh and M. Tsoi for technical assistance. This work was supported by National Institutes of Health (NIH) Grant GM34869 (D.R.E.) and GM55387 (C.A.F.). Partial support was provided by NIH Training Grant GM08270 (J.J.D.), and the H.H. Rackham Graduate School at the University of Michigan (S.X.).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7282205.
REFERENCES
- Bertrand, E., Houser-Scott, F., Kendall, A., Singer, R.H., and Engelke, D.R. 1998. Nucleolar localization of early tRNA processing. Genes & Dev. 12: 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain, J.R., Pagan, R., Kindelberger, D.W., and Engelke, D.R. 1996. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 24: 3158–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain, J.R., Lee, Y., Lane, W.S., and Engelke, D.R. 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes & Dev. 12: 1678–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.L. and Pace, N.R. 1997. Identification of the universally conserved core of ribonuclease P RNA. RNA 3: 557–560. [PMC free article] [PubMed] [Google Scholar]
- Chen, J.L., Nolan, J.M., Harris, M.E., and Pace, N.R. 1998. Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. EMBO J. 17: 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, C., Varani, G., and Tinoco Jr., I. 1990. Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature 346: 680–682. [DOI] [PubMed] [Google Scholar]
- Christian, E.L., Kaye, N.M., and Harris, M.E. 2000. Helix P4 is a divalent metal ion binding site in the conserved core of the ribonuclease P ribozyme. RNA 6: 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, S., Zengel, J.M., and Lindahl, L. 1997. A novel protein shared by RNase MRP and RNase P. RNA 3: 382–391. [PMC free article] [PubMed] [Google Scholar]
- Crary, S.M., Kurz, J.C., and Fierke, C.A. 2002. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis ribonuclease P. RNA 8: 933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht, A. 1985. Enzyme structure and mechanism. W.H. Freeman, New York.
- Frank, D.N. and Pace, N.R. 1998. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67: 153–180. [DOI] [PubMed] [Google Scholar]
- Frank, D.N., Adamidi, C., Ehringer, M.A., Pitulle, C., and Pace, N.R. 2000. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA 6: 1895–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, T., Cai, T., Aulds, J., Wierzbicki, S., and Schmitt, M.E. 2004. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: Novel method of mRNA degradation. Mol. Cell. Biol. 24: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck, A., Endo, Y., and Wool, I.G. 1992. Ribosomal RNA identity elements for ricin A-chain recognition and catalysis. Analysis with tetraloop mutants. J. Mol. Biol. 226: 411–424. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N., and Altman, S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. [DOI] [PubMed] [Google Scholar]
- Harris, M.E. and Pace, N.R. 1995. Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA 1: 210–218. [PMC free article] [PubMed] [Google Scholar]
- Harris, M.E., Kazantsev, A.V., Chen, J.L., and Pace, N.R. 1997. Analysis of the tertiary structure of the ribonuclease P ribozyme-substrate complex by site-specific photoaffinity crosslinking. RNA 3: 561–576. [PMC free article] [PubMed] [Google Scholar]
- Jaeger, L., Michel, F., and Westhof, E. 1994. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol. 236: 1271–1276. [DOI] [PubMed] [Google Scholar]
- Jarrous, N. 2002. Human ribonuclease P: Subunits, function, and intranuclear localization. RNA 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev, A.V. and Pace, N.R. 1998. Identification by modification-interference of purine N-7 and ribose 2′-OH groups critical for catalysis by bacterial ribonuclease P. RNA 4: 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer, K. and Domdey, H. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194: 398–405. [DOI] [PubMed] [Google Scholar]
- Krasilnikov, A.S., Yang, X., Pan, T., and Mondragon, A. 2003. Crystal structure of the specificity domain of ribonuclease P. Nature 421: 760–764. [DOI] [PubMed] [Google Scholar]
- Krasilnikov, A.S., Xiao, Y., Pan, T., and Mondragon, A. 2004. Basis for structural diversity in homologous RNAs. Science 306: 104–107. [DOI] [PubMed] [Google Scholar]
- Kufel, J., Allmang, C., Verdone, L., Beggs, J.D., and Tollervey, D. 2002. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol. Cell. Biol. 22: 5248–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y., Evans, C.F., and Engelke, D.R., 1991a. Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc. Natl. Acad. Sci. 88: 6986–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y., Rohlman, C.E., Molony, L.A., and Engelke, D.R. 1991b. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell. Biol. 11: 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Kindelberger, D.W., Lee, J.Y., McClennen, S., Chamberlain, J., and Engelke, D.R. 1997. Nuclear pre-tRNA terminal structure and RNase P recognition. RNA 3: 175–185. [PMC free article] [PubMed] [Google Scholar]
- Li, X., Frank, D.N., Pace, N., Zengel, J.M., and Lindahl, L. 2002. Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA 8: 740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, L. and Zengel, J.M. 1995. RNase MRP and rRNA processing. Mol. Biol. Rep. 22: 69–73. [DOI] [PubMed] [Google Scholar]
- Liu, M.H., Yuan, Y., and Reddy, R. 1994. Human RNaseP RNA and nucleolar 7–2 RNA share conserved ‘To’ antigen-binding domains. Mol. Cell. Biochem. 130: 75–82. [DOI] [PubMed] [Google Scholar]
- Lygerou, Z., Mitchell, P., Petfalski, E., Seraphin, B., and Tollervey, D. 1994. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes & Dev. 8: 1423–1433. [DOI] [PubMed] [Google Scholar]
- Nolan, J.M., Burke, D.H., and Pace, N.R. 1993. Circularly permuted tRNAs as specific photoaffinity probes of ribonuclease P RNA structure. Science 261: 762–765. [DOI] [PubMed] [Google Scholar]
- Pagan-Ramos, E., Lee, Y., and Engelke, D.R. 1996a. A conserved RNA motif involved in divalent cation utilization by nuclear RNase P. RNA 2: 1100–1109. [PMC free article] [PubMed] [Google Scholar]
- ———. 1996b. Mutational analysis of Saccharomyces cerevisiae nuclear RNase P: Randomization of universally conserved positions in the RNA subunit. RNA 2: 441–451. [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel, D.A., and Altman, S. 1999. Verification of phylogenetic predictions in vivo and the importance of the tetraloop motif in a catalytic RNA. Proc. Natl. Acad. Sci. 96: 11200–11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly, T.H. and Schmitt, M.E. 1995. The yeast, Saccharomyces cerevisiae, RNase P/MRP ribonucleoprotein endoribonuclease family. Mol. Biol. Rep. 22: 87–93. [DOI] [PubMed] [Google Scholar]
- Schmitt, M.E. and Clayton, D.A., 1994. Characterization of a unique protein component of yeast RNase MRP: An RNA-binding protein with a zinc-cluster domain. Genes & Dev. 8: 2617–2628. [DOI] [PubMed] [Google Scholar]
- Srisawat, C. and Engelke, D.R. 2001. Streptavidin aptamers: Affinity tags for the study of RNAs and ribonucleoproteins. RNA 7: 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat, C., Houser-Scott, F., Bertrand, E., Xiao, S., Singer, R.H., and Engelke, D.R. 2002. An active precursor in assembly of yeast nuclear ribonuclease P. RNA 8: 1348–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc, V. and Altman, S. 1997. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae.Genes & Dev. 11: 2414–2425. Erratum 11: 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, M., Haeusler, R.A., Good, P.D., and Engelke, D.R. 2003. Nucleolar clustering of dispersed tRNA genes. Science 302: 1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D. 1995. Genetic and biochemical analyses of yeast RNase MRP. Mol. Biol. Rep. 22: 75–79. [DOI] [PubMed] [Google Scholar]
- Tranguch, A.J. and Engelke, D.R. 1993. Comparative structural analysis of nuclear RNase P RNAs from yeast. J. Biol. Chem. 268: 14045–14055. [PubMed] [Google Scholar]
- Tranguch, A.J., Kindelberger, D.W., Rohlman, C.E., Lee, J.Y., and Engelke, D.R. 1994. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry 33: 1778–1787. [DOI] [PubMed] [Google Scholar]
- Walker, S.C. and Avis, J.M. 2004. A conserved element in the yeast RNase MRP RNA subunit can participate in a long-range basepairing interaction. J. Mol. Biol. 341: 375–388. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Scott, F., Fierke, C.A., and Engelke, D.R. 2002. Eukaryotic ribonuclease P: A plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 71: 165–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehler, W.A., Day, J.J., Fierke, C.A., and Engelke, D.R. 2000. Effects of 5′ leader and 3′ trailer structures on pre-tRNA processing by nuclear RNase P. Biochemistry 39: 9909–9916. [DOI] [PubMed] [Google Scholar]
- Ziehler, W.A., Morris, J., Scott, F.H., Millikin, C., and Engelke, D.R. 2001. An essential protein-binding domain of nuclear RNase P RNA. RNA 7: 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]