Abstract
Increasing evidence suggests that low-abundant transcripts may play fundamental roles in biological processes. In an attempt to estimate the prevalence of low-abundant transcripts in eukaryotic genomes, we performed a transcriptome analysis in Drosophila using the SAGE technique. We collected 244,313 SAGE tags from transcripts expressed in Drosophila embryonic, larval, pupae, adult, and testicular tissue. From these SAGE tags, we identified 40,823 unique SAGE tags. Our analysis showed that 55% of the 40,823 unique SAGE tags are novel without matches in currently known Drosophila transcripts, and most of the novel SAGE tags have low copy numbers. Further analysis indicated that these novel SAGE tags represent novel low-abundant transcripts expressed from loci outside of currently annotated exons including the intergenic and intronic regions, and antisense of the currently annotated exons in the Drosophila genome. Our study reveals the presence of a significant number of novel low-abundant transcripts in Drosophila, and highlights the need to isolate these novel low-abundant transcripts for further biological studies.
Keywords: low abundant, transcript, SAGE, EST, Drosophila, genome
INTRODUCTION
RNA reassociation experiment indicates that the majority of transcripts expressed in eukaryotic genomes are present at low levels (Bishop et al. 1974). While low-abundant transcripts were traditionally considered unimportant “noise” transcripts, recent studies suggest that they may be biologically significant, with roles in cellular differentiation, metabolism, and phenotypic alternation (Reanney et al. 1983; Elowitz et al. 2002; Kuznetsov et al. 2002; Ozbudak et al. 2002; Blake et al. 2003; Paulsson 2004).
Low-abundant transcripts may also be a driving force in the evolutionary process (Alvarez 2001). Answers to some fundamental biological questions may emerge from the systematic study of low-abundant transcripts. Low-abundant transcripts must be isolated before their biological roles can be determined. Despite intensive efforts on transcript identification, however, little is known about the prevalence of low-abundant transcripts, primarily because of isolation difficulties due to their small mass and high heterogeneity (Bishop et al. 1974; Holland 2002; Czechowski et al. 2004).
SAGE is a method for genome-level transcript analysis (Velculescu et al. 1995). Through isolating a short tag from a transcript and concatemerizing multiple tags for a single sequencing reaction, SAGE provides high sensitivity for transcript detection. SAGE can detect both known and novel transcripts, and provides quantitative information about the detected transcripts (Zhou et al. 2001; Saha et al. 2002). Drosophila is a well-established eukaryotic animal model. The Drosophila genome has been well sequenced and annotated, and its transcriptome has been extensively characterized by the large-scale EST approach (Adams et al. 2000; Rubin et al. 2000; Stapleton et al. 2002). Compared with higher eukaryotic genomes, the smaller size of the Drosophila genome enables Drosophila SAGE tags to represent their original transcripts and to map in the Drosophila genome with high specificity (Jasper et al. 2001, 2002; Fujii and Amrein 2002; Pleasance et al. 2003).
Taking advantage of the vast information known about the Drosophila genome and the high sensitivity of the SAGE technique for transcript detection, we performed a thorough Drosophila transcriptome analysis using the SAGE method to investigate the prevalence of low-abundant transcripts. We expected to detect low-abundant transcripts as long as a significant quantity of low-abundant transcripts exists and a sufficient number of SAGE tags could be collected. Here we report the results from this study.
RESULTS
General procedures of the study
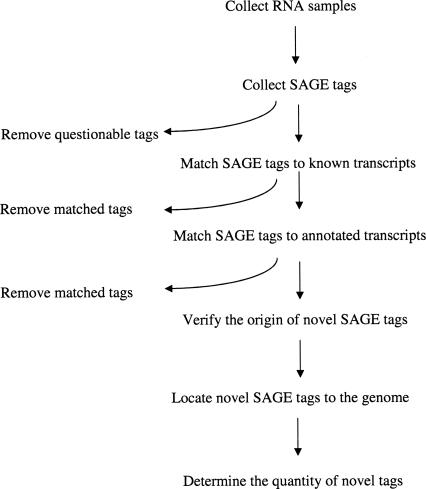
Figure 1 outlines the overall procedures of the study. RNA was collected from different samples, SAGE libraries were constructed, SAGE tags were collected, and questionable SAGE tags were eliminated. SAGE tags were then matched to known transcripts and annotated transcripts, the origins of novel SAGE tags were verified, the loci of novel SAGE tags in the Drosophila genome were located, and the quantities of novel SAGE tags were determined.
FIGURE 1.
Schematic of the procedures used in the study. See text for details.
Data collection and processing
We collected 359,139 SAGE tags from normal and radiated embryos, larvae, pupae, male and female adults, and testes from male adults. To ensure a high confidence in the downstream analysis, we excluded 114,826 uncertain SAGE tags, which included tags not mapped in the Drosophila genome, tags from potentially contaminated transcripts from the yeast that were used for feeding Drosophila in the laboratory environment, tags of SAGE linkers, and tags from the Drosophila mitochondrial genome. We obtained 244,313 final SAGE tags. From these SAGE tags, we identified a total of 40,823 unique SAGE tags. Each unique SAGE tag has quantitative information according to its copy number and maps to the Drosophila genome (Table 1; [http://www.ncbi.nlm.nih.gov/geo; accession no. GSE2347]; Supplementary Table 1 and Supplementary Table 2 [http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm]).
TABLE 1.
Summary of SAGE tag collection
| A. Preprocessed SAGE tags | ||
| Classification | Copy number (%) | Unique SAGE tags (%) |
| Total collected SAGE tags | 359,139 (100) | 56,974 (100) |
| Total removed SAGE tags | 114,826 (32) | 16,151 (28) |
| Unmapped tags | 38,509 | 14,641 |
| Yeast tags | 7,502 | 1,336 |
| SAGE linker tags | 111 | 2 |
| RT primer tags | 1,271 | 121 |
| Mitochondrial tags | 67,433 | 51 |
| Final SAGE tags | 244,313 (68) | 40,823 (72) |
| B. Quantitative distribution of SAGE tags | ||
| Copy number | Number of SAGE tags | Percentage (%)a |
| aThe percentage of SAGE tags from each class among the total unique SAGE tags. | ||
| >100 | 286 | 1 |
| 100–10 | 3,456 | 8 |
| 9–5 | 3,812 | 9 |
| 4–2 | 10,326 | 25 |
| 1 | 22,943 | 56 |
| Total | 40,823 | 100 |
Comparison of SAGE tags with known Drosophila transcripts
We compared the 40,823 unique SAGE tags with known Drosophila transcripts physically isolated from the Drosophila genome, including full-length cDNAs, 3′ ESTs, and 5′ ESTs. First, we matched the SAGE tags to the 10 bases adjacent to the last CATG in the full-length cDNAs and 3′ ESTs. Because SAGE tags are adjacent to the last CATG of the detected transcripts, a SAGE tag matched to such a location is an indication that the SAGE tag originates from the matched transcript. Second, we matched the SAGE tags to the 10 bases adjacent to all CATG sites in full-length cDNA, 3′ ESTs, and 5′ ESTs (except those adjacent to the last CATG in full-length cDNAs and 3′ ESTs). A SAGE tag matched to those locations suggests the presence of an alternatively spliced or polyadenylated transcript in which the sequences around the last CATG were removed and an upstream CATG was exposed for SAGE tag releasing. From these two comparisons, we observed that 45% of the 40,823 unique SAGE tags match the known expressed sequences. The remaining 55% do not have matches to the known Drosophila transcripts (Table 2; Supplementary Tables 3, 4 [http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm]).
TABLE 2.
Matching of 40,823 SAGE tags to known transcripts
| Class | mRNA | 3′ EST | 5′ EST | Total (%) |
| Matched SAGE tags | 16,378 | 6950 | 8,770 | 18,246 (45) |
| Adjacent to last CATG | 9,104 | 5,500 | ||
| Adjacent to an upstream CATG | 7,274 | 1,450 | ||
| Unmatched SAGE tags | 24,445 | 33,873 | 32.053 | 22,577 (55) |
We also compared the 40,823 unique SAGE tags with the annotated Drosophila transcripts. SAGE tags are located toward the 3′ part of the transcripts. Because the majority of known Drosophila transcripts are ESTs and most of the ESTs are 5′ ESTs, the comparison between SAGE tags and physically isolated transcripts may be biased due to the unbalanced 3′ EST and 5′ EST collection. The Drosophila genome project provides a full set of annotated transcripts. These annotated transcripts were generated using current knowledge of gene structure, computational gene prediction, and experimental evidence —including, but not necessarily restricted by, the full-length cDNAs, 5′ ESTs, and 3′ ESTs. The biased distribution of 5′ ESTs and 3′ ESTs has been normalized in the annotated transcripts. This comparison shows that 41% of the 40,823 unique SAGE tags match the annotated transcripts, whereas 59% do not match.
Taken together, the results from these two comparisons show that over half of the SAGE tags do not match the known Drosophila transcripts.
Verification of the origins of unmatched SAGE tags
We performed four types of experiments to verify the origins of the unmatched SAGE tags.
We converted the unmatched SAGE tags into 3′ cDNAs using the GLGI (generation of longer 3′ cDNA from SAGE tag for gene identification) method (Chen et al. 2002). We analyzed 90 3′ cDNAs converted from unmatched SAGE tags. The results show that all are mapped to the Drosophila genome, and 88 are novel transcripts. Of these 88 novel transcripts, 81 contain a polyA tail, 41 contain a polyA signal, and 32 are antisense of the known transcripts (Table 3; Supplementary Table 5 [http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm]).
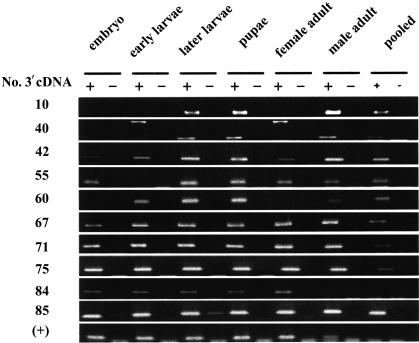
We performed RT-PCR to confirm the transcripts detected by unmatched SAGE tags. A set of 3′ cDNAs generated in 1 was used to design sense and antisense primers. The SAGE tag itself was used as the sense primer; the downstream sequences in 3′ cDNA were used to design antisense primers. Total RNA samples from embryo, early larvae, later larvae, pupae, female and male adults, and pooled samples were used for the detection. Of the 11 targeted transcripts, 10 were detected in either different samples or in all samples. No genomic DNA signal was detected in these reactions (Fig. 2; Supplementary Table 6 [http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm]).
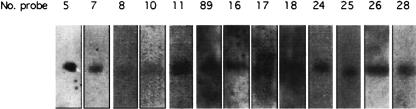
We performed Northern hybridization to verify the transcripts detected by unmatched SAGE tags. A set of 3′ cDNAs generated in 1 was used as the probes. RNA samples from embryos and whole adults were used for the detection. Of the 16 targeted transcripts, 13 were detected (Fig. 3; Supplementary Table 7 [http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm]).
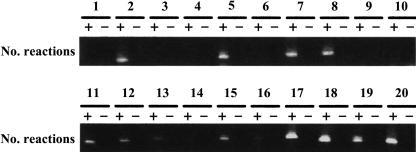
We performed RT-PCR to detect transcripts expressed from genomic regions mapped by unmatched SAGE tags. We considered that if a genomic locus mapped by a SAGE tag is transcriptionally active, we should be able to detect directly the transcripts expressed from this locus. A group of genomic segments was selected, starting from SAGE tag-mapped locations until the polyA signal sequence AATAAA or ATTAAA was reached downstream. RT-PCR was used to detect these potential transcripts in the pooled total RNA sample. Sense primers were designed based on the SAGE tags, and antisense primers were designed based on the genomic sequences upstream of the polyA signal sequence. Of the 20 targeted reactions, 14 were positive. No signals were present in RNase-digested RNA samples, indicating that the detected signals were indeed from the transcripts expressed from the genomic loci mapped by the unmatched SAGE tags (Fig. 4; Supplementary Table 8 [http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm]).
TABLE 3.
Features of 88 3′ cDNAs converted from the unmatched SAGE tags
| Feature | Number (%) |
| Map to genome | 88 (100) |
| Mapped genomic sequences with intron | 12 (14) |
| PolyA tail | 81 (92) |
| PolyA signal | 41 (47) |
| Antisense of known transcripts | 32 (36) |
| Average length (bp) | 110 |
FIGURE 2.
RT-PCR confirmation of the novel transcripts identified by novel SAGE tags. RT-PCR was performed using multiple tissue RNA samples. The novel 3′ cDNAs converted from novel SAGE tags were used for designing sense and antisense primers. “+” refers to RT-PCR; “−” refers to the controls with RNase A-digested RNA sample. See details in Supplementary Tables 5 and 6 (http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm).
FIGURE 3.
Northern blot confirmation of the novel transcripts identified by novel SAGE tags. Northern blots were performed using embryo or whole adult RNA. The cDNA probes were the 3′ cDNAs converted from novel SAGE tags. The figure shows the 13 positive signals from the 16 total reactions. See details in Supplementary Tables 5 and 7 (http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm).
FIGURE 4.
RT-PCR detection of transcripts expressed from novel SAGE tag-mapped, unannotated genomic regions. Sense primers were designed based on mapped SAGE tags, and antisense primers were designed based on the mapped genomic sequences upstream of the polyA signal AATAAA or ATTAAA of the mapped regions. The pooled RNA samples were used as the templates for the detection. “+” refers to RT-PCR; “−” refers to the control with RNase A-digested RNA for monitoring genomic DNA contamination. See details in Supplementary Table 8 (http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm).
In summary, these experimental results indicate that most of the unmatched SAGE tags are novel SAGE tags representing currently unidentified novel transcripts.
Location of novel SAGE tags in the Drosophila genome
To investigate the correlation between the novel transcripts detected from novel SAGE tags and known genes, we mapped the novel SAGE tags in the Drosophila genome. To provide high mapping specificity, we focused on the 18,913 unique SAGE tags that map only to a single locus in the Drosophila genome. These tags include 7,106 matched SAGE tags and 11,807 novel SAGE tags. Of the 7,106 matched SAGE tags, 88% are located in the annotated exons, and 12% are mapped in the unannotated loci. In contrast, of the 11,807 novel SAGE tags, only 1% are mapped within the annotated exons, while 99% are mapped in the unannotated loci (Table 4A). Further analysis revealed that 48% of those mapped in the unannotated loci are located in the intergenic regions, 16% are located in the intragenic regions (most of which are intronic), and 36% are antisense of the intragenic regions (two-thirds of which are exonic). Since some annotated genes may end at a translational stop codon without 3′ UTR sequences, we further mapped the 11,807 novel SAGE tags to the genomic sequences 1,000 bp downstream of the annotated genes. Only 7.5% of these novel tags mapped within the region. The 11,807 novel SAGE tags were rather uniformly distributed among different chromosomes (Table 4B), although many tags mapped in particular chromosomes tend to be clustered. In conclusion, most novel transcripts detected by novel SAGE tags were expressed outside the annotated exons or genes, or were antisense of annotated exons or genes in the Drosophila genome.
TABLE 4.
Mapping SAGE tags to the Drosophila genome
| A. Distribution of mapped SAGE tags | |||
| Location of mapped tags (%) | |||
| SAGE taga | Number (%) | Annotated exon | Unannotated loci |
| aEach SAGE tag has only one mapped location in the Drosophila genome. | |||
| Matched tags | 7,106 (100) | 6,234 (88) | 872 (12) |
| Novel tags | 11,807 (100) | 155 (1) | 11,631 (99) |
| Intergenic | 5,607 (48) | ||
| Intragenic | 1,903 (16) | 155 | 1,748 (intronic) |
| Antisense | 4,297 (36) | 2,842 | 1,455 (intronic) |
| B. Distribution of novel SAGE tags in euchromatin of different chromosomes | |||
| Location | Size (kb) | Mapped SAGE tags | Tags/kb |
| 2L | 22,200 | 2,193 | 0.099 |
| 2R | 20,300 | 2,044 | 0.101 |
| 3L | 23,400 | 2,286 | 0.098 |
| 3R | 27,900 | 2,746 | 0.098 |
| 4 | 1,200 | 132 | 0.110 |
| X | 21,800 | 2,072 | 0.095 |
| Total | 116,800 | 11,473 | 0.100 |
Quantitative distribution of novel SAGE tags
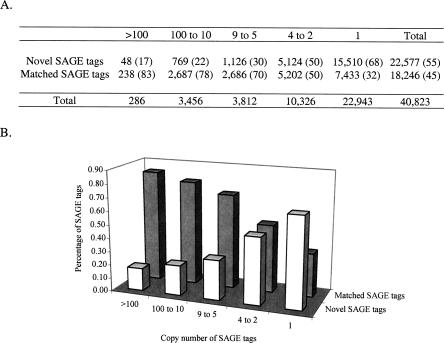
We measured the quantitative distribution of the novel SAGE tags and compared it with that of the matched SAGE tags. The matched SAGE tags have proportionally higher copy numbers, while the novel SAGE tags have proportionally lower copy numbers (Fig. 5). For example, the novel SAGE tags account for 68% (15,510) of the 22,943 single copy SAGE tags. The pattern of novel SAGE tag distribution implies that most of the novel transcripts detected by SAGE are low-abundant transcripts.
FIGURE 5.
Quantitative distribution of SAGE tags. (A) SAGE tags were grouped based on their copy numbers. Within the same group, the number and percentage of novel SAGE tags and matched SAGE tags were further determined. (B) The quantitative distribution of the matched SAGE tags and the novel SAGE tags. The percentages shown in A are used for the plot.
DISCUSSION
The total number of Drosophila SAGE tags collected in this study approximates the number of Drosophila EST sequences. However, over half of the transcripts detected by SAGE tags are not present in the EST collection. The discrepancy between the SAGE data and the EST data may be explained by the following:
Sequencing regular cDNA libraries detects fewer low-abundant transcripts. Of the nine EST libraries used in the Drosophila EST project, seven were regular cDNA libraries that contributed over half of the ESTs (http://www.fruitfly.org/EST/EST.shtml). The redundant nature of transcripts prevents the detection of low-abundant copies (Bishop et al. 1974). The redundancy problem is largely overcome when the highly sensitive SAGE method is applied, which detects far more transcripts present at lower levels (Sun et al. 2004).
cDNA libraries processed by normalization may lose low-abundant copies. Of the nine EST libraries used in the Drosophila EST project, two were constructed through the normalization process and contributed nearly half of the ESTs (http://www.fruitfly.org/EST/EST.shtml). While normalization can indeed remove many abundant templates (Bonaldo et al. 1996), this treatment can also lead to the loss of templates due to cross-hybridization between templates with partial similarities or between different templates through the formation of polydA/polydT tail hybrids (Wang et al. 2000). The loss may affect particularly the low-abundant copies. Because many low-abundant copies may be missing from the libraries, increasing sequencing collection may not increase the detection of low-abundant copies. In contrast, normalization is not used for SAGE library construction. The content of original transcripts, particularly the low-abundant copies, is well preserved in SAGE libraries. Low-abundant copies hiding in the high-abundant copies in the SAGE library are detected by collecting multiple tags per sequencing reaction.
Other factors, such as cDNA size selection in EST library construction and poor replication of clones with long cDNA inserts, may also prevent detection of certain low-abundant copies. These limitations do not exist in SAGE library construction.
We consider it unlikely that the following possibilities could be the major source of novel SAGE tags detected in this study.
Genetic background of Drosophila strains. The Drosophila strain used in our SAGE tag collection is the same as that used in the Drosophila genome and EST projects. Therefore, there should not be sequence variation affecting data comparison.
Experimental errors. Strict conditions set in this study excluded potential error SAGE tags collected in this study.
Novel alternatively spliced or polyadenylated transcripts from known transcripts. All SAGE tags were classified as matched tags if they matched to locations adjacent to all CATG sites in the known transcripts. These matched SAGE tags cover the potential novel alternatively spliced or polyadenylated isoforms of the known transcripts. However, this does not include the situation where an alternate 3′ exon exists that is downstream of the known 3′ exon and has a CATG site(s).
Identification of low-abundant transcripts is more difficult than identification of high-abundant transcripts due to the redundancy and complexity of transcripts. In the last decade, new technologies with increased sensitivity, such as EST, subtraction/normalization EST, SAGE, and MPSS, have been developed and applied in transcriptome studies (Adams et al. 1992; Velculescu et al. 1995; Bonaldo et al. 1996; Brenner et al. 2000). When techniques with higher sensitivity are used, greater numbers of less abundant novel transcripts are identified. However, identification of all low-abundant transcripts remains a challenge, as evidenced by our current study. Although mathematic calculations can be used to estimate the scope of transcript collection for identification of full-set transcripts (Stern et al. 2003; Reverter et al. 2005), the final answer will likely come from experimental data showing that few or no novel transcripts could be identified. Thus far, this stage of transcript identification has not been achieved in most of the genomes studied (Kapranov et al. 2002; Okazaki et al. 2002; Seki et al. 2002; Bertone et al. 2004; Imanishi et al. 2004; Schadt et al. 2004; Scheetz et al. 2004).
The genomic locations of the novel SAGE tags are interesting. Among the novel SAGE tags, nearly half are located intergenically, implying that more novel transcribed regions than current annotated ones are present in the Drosophila genome. Using a tilling array technique, a recent study also detected transcriptional activities in 41% of the intergenic region and 43% of the intronic region in the Drosophila genome (Stolc et al. 2004). Furthermore, a third of the novel SAGE tags are antisense transcripts of the annotated genes, most of which are located in the known exons. The wide presence of antisense novel transcripts for known genes revealed in this study supports the concept that antisense transcript is one of the major means for gene expression regulation (Yelin et al. 2003).
In conclusion, our study demonstrates the presence of a large quantity of low-abundant transcripts in Drosophila, which may also occur in other species (Bertone et al. 2004). Systematic identification of low-abundant transcripts in model species is an important step toward the elucidation of the biological roles of low-abundant transcripts.
MATERIALS AND METHODS
Collection of SAGE tags
The Drosophila melanogaster strain (y; cn bw sp) used in the Drosophila genome and EST projects was used for the study. RNA was extracted from six types of samples, including embryo (0–24 h), embryo with radiation (0- to 24-h embryos, 30 min after treatment with 40 Gy γ radiation), larvae (second and third day), pupae (third day), adults (10 males and 10 females, up to 10 d), and testis from testicular tissue of adult males. Four SAGE libraries were constructed: (1) pooled sample that included an equal amount of total RNA from embryo, larvae, pupae, and young and aged adults; (2) embryo; (3) irradiated embryo; and (4) testis. SAGE libraries were constructed following the procedures (Lee et al. 2001). Large-scale SAGE tag sequence collection was performed using the DYEnamic ET Terminator Cycle Sequencing kit in Megabase1000 DNA sequencers (Amersham) with Phred20 as the cutoff. SAGE tags were extracted using SAGE- 300 software. Questionable SAGE tags were removed from the collected SAGE tags, including yeast SAGE tags (http://www.sagenet.org/SAGEData/sagedata.htm), the SAGE tag linkers TCCCTATTAA and TCCCCGTACA, the primer tag AAAGCGGCCG and its derivatives, and the mitochondrial tags (extracted from the Drosophila melanogaster mitochondrial genome, NC_001709, http://www.ncbi.nlm.nih.gov/).
Construction of SAGE reference databases
The Drosophila genome sequences used were Drosophila Release 3.1 (http://www.fruitfly.org/cgi-bin/seq_tools/fasta_download.cgi). The genomic tag reference database was generated by extracting 10-base tags from all CATG sites in the genomic sequences, including the tags from the sense strand immediately adjacent to the CATGs and the tags from the antisense before CATG with reverse/complementary sequences. The SAGE tag reference database from the physically isolated known transcripts was constructed by extracting 10 bases adjacent to CATGs in the full-length cDNA, 3′ ESTs, and 5′ ESTs (UniGene Drosophila melanogaster database release 17, http://www.ncbi.nlm.nih.gov/). Of these sequences, 94% of mRNA, 78% of 3′ ESTs, and 80% of 5′ ESTs contain CATG and are therefore detectable by SAGE. The SAGE tag reference database from the Drosophila annotated transcripts was constructed by extracting 10 bases adjacent to all CATGs in each annotated “transcript” sequence in Release 3.1 (http://flybase.net/annot/download_sequences.html).
Conversion of SAGE tags into 3′ cDNAs
A set of unmatched SAGE tags was randomly selected from the total unmatched SAGE tag list and converted into 3′ cDNAs using the GLGI method (generation of longer 3′ cDNA from SAGE tags for gene identification) (Chen et al. 2002; Supplementary Table 5 [http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm]). The 3′ cDNA sequences were deposited in GenBank with accession numbers CB305186–CB305318.
RT-PCR confirmation of novel transcripts detected by novel SAGE tags
RT-PCR was used to confirm novel transcripts detected by novel SAGE tags. Sense primers and antisense primers were designed based on the 3′ cDNAs converted from novel SAGE tags. Total RNA samples from embryonic, early larval, later larval, pupal, male and female adult, and pooled tissues were used as the templates for the analysis (Supplementary Table 6 [http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm]). RNase A-treated RNA samples were used as negative control.
Northern blot confirmation of novel transcripts detected by novel SAGE tags
Northern blot was used to confirm novel transcripts detected by novel SAGE tags. RNA samples from whole adults were used for the detection. The 3′ cDNAs converted from novel SAGE tags were used as probes. Probe labeling, hybridization, and signal detection were performed using the Bright Star Bio-Detection system (Ambion) following the protocol.
RT-PCR detection of novel transcripts expressed from novel SAGE tag-mapped, unannotated genomic regions
Genomic segments mapped by novel SAGE tags were used for the test. Each segment starts at the novel SAGE tag-mapped location and moves downstream to the polyA signal sequences AATAAA or ATTAAA. Sense primers were designed based on the mapped novel SAGE tag; antisense primers were designed based on the genomic sequences upstream of AATAAA or ATTAAA (Supplementary Table 8 [http://www.biochem.northwestern.edu/ibis/faculty/smwang.htm]). The pooled total RNA samples were used as the templates for the detection. RNase A-treated RNA samples were used as control for monitoring genomic DNA contamination.
Acknowledgments
This study was funded by a Howard Hughes Fellowship (J.S.), a Department of Defense MURI program (C.T., S.M.W.), the Ludwig Fund for Cancer Research (W.D., also a Leukemia and Lymphoma Society Scholar), National Institutes of Health (C-I.W.), Natural Science Foundation of China and Chinese Academy of Sciences (H.Y.), the G. Harold and Lelia Y. Mathers Charitable Foundation (S.M.W.), the Daniel F. and Ada L. Rice Foundation (S.M.W.), and National Institutes of Health (S.M.W.).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7239605.
REFERENCES
- Adams, M.D., Dubnick, M., Kerlavage, A.R., Moreno, R., Kelley, J.M., Utterback, T.R., Nagle, J.W., Fields, C., and Venter, J.C. 1992. Sequence identification of 2,375 human brain genes. Nature 355: 632–634. [DOI] [PubMed] [Google Scholar]
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Alvarez, L.H. 2001. Does increased stochasticity speed up extinction? J. Math. Biol. 43: 534–544. [DOI] [PubMed] [Google Scholar]
- Bertone, P., Stolc, V., Royce, T.E., Rozowsky, J.S., Urban, A.E., Zhu, X., Rinn, J.L., Tongprasit, W., Samanta M., Weissman, S., et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246. [DOI] [PubMed] [Google Scholar]
- Bishop, J.O., Morton, J.G., Rosbach, M., and Richardson, M. 1974. Three abundance classes in HeLa cell messenger RNA. Nature 250: 199–204. [DOI] [PubMed] [Google Scholar]
- Blake, W.J., Kaern, M., Cantor, C.R., and Collins, J.J. 2003. Noise in eukaryotic gene expression. Nature 422: 633–637. [DOI] [PubMed] [Google Scholar]
- Bonaldo, M.F., Lennon, G., and Soares, M.B. 1996. Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res. 6: 791–806. [DOI] [PubMed] [Google Scholar]
- Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D.H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. 2000. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18: 630–634. [DOI] [PubMed] [Google Scholar]
- Chen, J., Lee, S., Zhou, G., and Wang, S.M. 2002. High-throughput GLGI procedure for converting a large number of serial analysis of gene expression tag sequences into 3′ complementary DNAs. Genes Chromosomes Cancer 33: 252–261. [DOI] [PubMed] [Google Scholar]
- Czechowski, T., Bari, R.P., Stitt, M., Scheible, W.R., and Udvardi, M.K. 2004. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: Unprecedented sensitivity reveals novel rootand shoot-specific genes. Plant J. 38: 366–379. [DOI] [PubMed] [Google Scholar]
- Elowitz, M.B., Levine, A.J., Siggia, E.D., and Swain, P.S. 2002. Stochastic gene expression in a single cell. Science 297: 1183–1186. [DOI] [PubMed] [Google Scholar]
- Fujii, S. and Amrein, H. 2002. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 21: 5353–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, M.J. 2002. Transcript abundance in yeast varies over six orders of magnitude. J. Biol. Chem. 277: 14363–14366. [DOI] [PubMed] [Google Scholar]
- Imanishi, T., Itoh, T., Suzuki, Y., O’Donovan, C., Fukuchi, S., Koyanagi, K.O., Barrero, R.A., Tamura, T., Yamaguchi-Kabata, Y., Tanino, M., et al. 2004. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2: 856–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper, H., Benes, V., Schwager, C., Sauer, S., Clauder-Munster, S., Ansorge, W., and Bohmann, D. 2001. The genomic response of the Drosophila embryo to JNK signaling. Dev. Cell 1: 579–586. [DOI] [PubMed] [Google Scholar]
- Jasper, H., Benes, V., Atzberger, A., Sauer, S., Ansorge, W., and Bohmann, D. 2002. A genomic switch at the transition from cell proliferation to terminal differentiation in the Drosophila eye. Dev. Cell 3: 511–521. [DOI] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919. [DOI] [PubMed] [Google Scholar]
- Kuznetsov, V.A., Knott, G.D., and Bonner, R.F. 2002. General statistics of stochastic process of gene expression in eukaryotic cells. Genetics 161: 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Chen, J., Zhou, G., and Wang, S.M. 2001. Generation of highquantity and quality tag/ditag cDNAs for SAGE analysis. Biotechniques 31: 348–354. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573. [DOI] [PubMed] [Google Scholar]
- Ozbudak, E.M., Thattai, M., Kurtser, I., Grossman, A.D., and van Oudenaarden, A. 2002. Regulation of noise in the expression of a single gene. Nat. Genet. 31: 69–73. [DOI] [PubMed] [Google Scholar]
- Paulsson, J. 2004. Summing up the noise in gene networks. Nature 427: 415–418. [DOI] [PubMed] [Google Scholar]
- Pleasance, E.D., Marra, M.A., and Jones, S.J. 2003. Assessment of SAGE in transcript identification. Genome Res. 13: 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reanney, D.C., MacPhee, D.G., and Pressing, J. 1983. Intrinsic noise and the design of the genetic machinery. Aust. J. Biol. Sci. 36: 77–90. [PubMed] [Google Scholar]
- Reverter, A., McWilliam, S.M., Barris, W., and Dalrymple, B.P. 2005. A rapid method for computationally inferring transcriptome coverage and microarray sensitivity. Bioinformatics 21: 80–89. [DOI] [PubMed] [Google Scholar]
- Rubin, G.M., Hong, L., Brokstein, P., Evans-Holm, M., Frise, E., Stapleton, M., and Harvey, D.A. 2000. A Drosophila complementary DNA resource. Science 287: 2222–2224. [DOI] [PubMed] [Google Scholar]
- Saha, S., Sparks, A.B., Rago, C., Akmaev, V., Wang, C.J., Vogelstein, B., Kinzler, K.W., and Velculescu, V.E. 2002. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20: 508–512. [DOI] [PubMed] [Google Scholar]
- Schadt, E.E., Edwards, S.W., GuhaThakurta, D., Holder, D., Ying, L., Svetnik, V., Leonardson, A., Hart, K.W., Russell, A., Li, G., et al. 2004. A comprehensive transcript index of the human genome generated using microarrays and computational approaches. Genome Biol. 5: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz, T.E., Laffin, J.J., Berger, B., Holte, S., Baumes, S.A., Brown, R., Chang, S., Coco, J., Conklin, J., Crouch, K., et al. 2004. Highthroughput gene discovery in the rat. Genome Res. 14: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A., Akiyama, K., Oono, Y., et al. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145. [DOI] [PubMed] [Google Scholar]
- Stapleton, M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, K., et al. 2002. The Drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12: 1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, M.D., Anisimov, S.V., and Boheler, K.R. 2003. Can transcriptome size be estimated from SAGE catalogs? Bioinformatics 19: 443–448. [DOI] [PubMed] [Google Scholar]
- Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M.F., Rifkin, S.A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P.E., et al. 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306: 655–660. [DOI] [PubMed] [Google Scholar]
- Sun, M., Zhou, G., Lee, S., Chen, J., Shi, R.Z., and Wang, S.M. 2004. SAGE is far more sensitive than EST for detecting low-abundance transcripts. BMC Genomics 5: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Vogelstein, B., and Kinzler, K.W. 1995. Serial analysis of gene expression. Science 270: 484–487. [DOI] [PubMed] [Google Scholar]
- Wang, S.M., Fears, S.C., Zhang, L., Chen, J.J., and Rowley, J.D. 2000. Screening poly(dA/dT)-cDNAs for gene identification. Proc. Natl. Acad. Sci. 97: 4162–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21: 379–386. [DOI] [PubMed] [Google Scholar]
- Zhou, G., Chen, J., Lee, S., Clark, T., Rowley, J.D., and Wang, S.M. 2001. The pattern of gene expression in human CD34+ hematopoietic stem/progenitor cells. Proc. Natl. Acad. Sci. 98: 13966–13971. [DOI] [PMC free article] [PubMed] [Google Scholar]