Abstract
Small interfering RNAs (siRNAs) are being used to induce sequence-specific gene silencing in cultured cells to study mammalian gene function. Libraries of siRNAs targeting entire human gene classes can be used to identify genes with specific cellular functions. Here we describe high-throughput siRNA delivery methods to facilitate siRNA library screening experiments with both immortalized and primary cells. We adapted chemical reverse transfection for immortalized adherent cell lines in a 96-well format. The method is fast, robust, and exceptionally effective for many cell types. For primary cells and immortalized cells that are recalcitrant to lipofection-based methods, we developed electropermeabilization (electroporation) conditions that facilitate siRNA delivery to a broad range of cell types, including primary human T-cells, hMSC, NHA, NDHF-Neo, HUVEC, DI TNC1, RPTEC, PC12, and K562 cells. To enable high-throughput electropermeabilization of primary cells, we developed a novel 96-well electroporation device that provides highly efficient and reproducible delivery of siRNAs. The combination of high-throughput chemical reverse transfection and electroporation makes it possible to deliver libraries of siRNAs to virtually any cell type, enabling gene function analysis and discovery on a genome scale.
Keywords: RNAi screening, gene expression, siRNA, reverse transfection, electroporation
INTRODUCTION
RNA interference (RNAi), a form of post-transcriptional gene silencing induced by introduction of double-stranded RNA (dsRNA), has become a powerful experimental tool for studying gene function. The RNAi phenomenon was first discovered in Caenorhabditis elegans and is characterized by sequence-specific gene silencing elicited by introduction of dsRNA (Fire et al. 1998; Elbashir et al. 2001) complementary to a target mRNA. In the endogenous RNAi pathway, long dsRNA is cleaved by the RNase III type endonuclease, Dicer, to produce 21–23 base pair (bp) short interfering RNAs. The siRNAs are in turn unwound and incorporated into a multiprotein complex known as the RNA-induced silencing complex (RISC), generating a sequence-specific nuclease that guides the cleavage of specific complementary mRNAs. In mammalian cells, direct introduction of siRNAs is used to experimentally initiate RNAi, because introduction of long dsRNA induces a potent antiviral response in addition to RNAi (Hammond et al. 2000).
Because of its ease of use, RNAi has been rapidly adopted for functional genomics, pathway analysis, and drug target validation experiments, and is now being used in high-throughput experiments with large numbers of siRNAs, or siRNA libraries (Sachse and Echeverri 2004; Ren et al. 2004; Silva et al. 2004; Xin et al. 2004). A key to all successful siRNA experiments is efficient delivery of the siRNA into cells and subsequent uptake of the siRNA by the RISC. RNAi can be successfully elicited in mammalian cells using exogenously derived siRNA only when the correct method and matrix of delivery conditions are employed for the cell type being used (Caplen et al. 2001; Elbashir et al. 2001; Harborth et al. 2001).
High-throughput delivery of siRNAs remains a challenge for many laboratories due to the variability of cell culture and transfection efficiency in 96- and 384-well plate formats. To overcome this variability, we adapted a procedure termed reverse chemical transfection, or transfection in suspension (Amarzguioui 2004), to facilitate the transfection of hundreds or thousands of siRNAs comprising an siRNA library. We found chemical transfection to be restricted primarily to immortalized cell lines like HeLa, HepG2, A549, and others.
Efficient transfer of siRNAs into primary cells by chemical transfection has so far been restricted to only a few cell types (Olivera and Goodell 2001; Laderach et al. 2003). Nevertheless, primary cells are often desired for gene silencing experiments because they better model their in vivo counterparts than immortalized cells. An alternative to chemical transfection-mediated nucleic acid delivery, electroporation, typically provides higher transfection efficiencies in primary cell types (Olivera and Goodell 2001; Walters and Jelinek 2002; Weil et al. 2002; Dunne et al. 2003; Jiang et al. 2003; Wilson et al. 2003). Electroporation involves applying an electric field pulse to induce transient cell membrane permeability via the formation of microscopic pores (electropores) in the cell membrane. These electropores allow molecules, ions, water, and nucleic acids to traverse the membrane. If the electric field pulse has the proper characteristics, the “electroporated” cells recover and continue to function normally. Although advances have been made in electroporation equipment and protocols (Chu et al. 1987; Yan et al. 1998; Gehl 2003; Herweijer and Wolff 2003), it largely remains a process for single-sample RNA and DNA delivery. We have created a device and associated protocols that enable the delivery of siRNAs to as many as 96 samples at a time. The 96-sample electroporator was extensively characterized to ensure its applicability to siRNA library screening.
RESULTS
As a precursor to performing gene function screens using libraries of siRNAs, we tested a variety of methods for delivering active siRNAs to immortalized and primary cells. We found that the best approach for primary cells is quite different than for immortalized adherent cells. Methods for both cell types are presented below.
High-throughput delivery for immortalized cells chemical reverse transfection
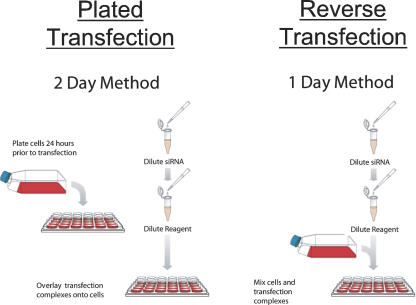
Small interfering RNAs can be transiently transfected into immortalized cell lines using chemical transfection reagents such as cationic liposomal and polyamine-based agents. Most published siRNA studies have involved the analysis of no more than a few genes with transfections in six-well and 24-well formats. We adapted the reverse transfection method (Amarzguioui 2004) to facilitate the rapid and robust delivery of siRNAs in 96-well and 384-well plates (Fig. 1). The reverse transfection method involves simultaneous mixing of siRNAs with transfection reagent in the wells of 96-well or 384-well plates. Adherent immortalized cells that have been detached by trypsinization and suspended in medium are added to the wells with the transfection complexes. Tissue culture plate(s) with cells, transfection complexes, and media are placed in an incubator to allow the cells to adhere to the surface of the plate. SiRNAs enter the cells during the first 24 h of incubation and significantly reduce target mRNA expression. If the viability of the cells being transfected is significantly affected at 24 h post-transfection, the level of cytotoxicity can be decreased by changing the growth medium and eliminating redundant exposure of cells to transfectant (Table 1).
FIGURE 1.
Schematic diagram of reverse transfection procedure.
TABLE 1.
Time course of siRNA-induced gene silencing and cell viability HeLa cells were reverse-transfected with 30 nM of siRNA targeting GAPDH mRNA
| Time of media replacement | Percent mRNA knockdown | Percent cell viability |
| None | 90%–95% | 60%–65% |
| 4 h post-transfection | 60%–65% | 90%–95% |
| 24 h post-transfection | 90%–95% | 80%–85% |
There are three important components of a successful siRNA library screen that are directly related to delivery: transfection efficiency, reproducibility, and cell viability. High transfection efficiency maximizes the reduction in target gene expression and improves the likelihood that a significant phenotype will be observed. High reproducibility of the siRNA delivery between wells and between plates typically reduces the variability of the quantitative phenotypic results and improves the resolution of the screening data. High cell viability increases the specificity of the siRNA effect, further improving the likelihood that significant phenotypic differences will be observed in the screen. Cytotoxicity induced by the delivery method will alter the biochemistry of the cells and might influence the identities of the siRNAs that produce distinct phenotypes. We characterized the transfection efficiency, reproducibility, and cell viability of the high-throughput reverse transfection procedure.
High-throughput reverse transfection efficiency
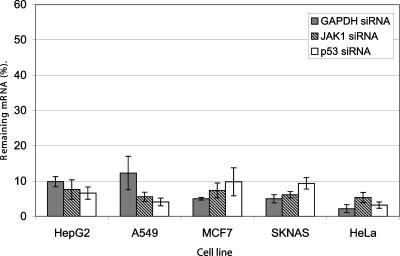
To measure siRNA delivery, we mixed well characterized siRNAs targeting GAPDH, p53, and JAK1 with transfection reagent in the wells of a 96-well plate. Five different cell lines were harvested and added to the siRNA-lipid complexes. Forty-eight hours post-transfection, we monitored GAPDH mRNA levels by real-time RT-PCR. As shown in Figure 2, the reverse transfection method provided significant reduction in GAPDH, JAK1, and p53 mRNA in all five cell types when using only 10 nM siRNA.
FIGURE 2.
Efficiency of reverse transfection in multiple cell types. Indicated cell lines were plated in 96-well format and simultaneously transfected (8000 cells/well) with addition of transfection complexes, prepared in Opti-mem serum-free medium, by mixing 0.3 μL siPORT NeoFX Transfection Reagent and 1 pmol siRNA targeting GAPDH, JAK1, p53, or nonsilencing control. Forty-eight hours post-transfection cells were harvested and analyzed by real-time RT-PCR for both target mRNA and 18S rRNA levels. Remaining gene expression was calculated as a percentage of target mRNA in cells transfected with siRNA targeting GAPDH, JAK1, p53, and cells transfected with the nonsilencing control siRNA. Data were normalized against the 18S rRNA signal. Transfections were performed in duplicate. Data are presented as means ± SD.
In general, 30 nM siRNA was sufficient to achieve maximal target gene reduction of genes: GAPDH, MMP-2, JAK1, P53, Cyclin E1, STAT1, PKCα, Cyclin D1, CDK2 and RAF 1, while at least 100 nM siRNA was required to induce the same reduction when using a standard transfection method (cell plating followed 24 h later with the addition of transfection complexes). This observation suggests that cells that have adhered to a surface are less compatible with lipid-induced siRNA delivery, perhaps because the cell membrane is less exposed to the siRNA-lipid transfection complexes in the surrounding medium (data not shown).
Another notable benefit of chemical reverse transfection is the broad range of cell numbers that could be reverse transfected. While standard transfection is limited to transfecting 5000–10,000 cells per well on a 96-well plate with high transfection efficiency (>70% reduction in GAPDH mRNA expression level by real-time RT-PCR), reverse transfection exhibited similar reductions in target GAPDH mRNA expression when transfecting 5000–25,000 cells per well (data not shown).
Reproducibility of chemical reverse transfection
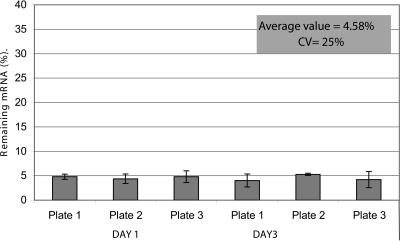
While testing the transfection efficiency of the reverse transfection procedure in 96-well plates, we noted that the reductions in GAPDH mRNA in wells having the same amount of siRNA were extremely consistent. To confirm this reproducibility, we reverse transfected cells in eight wells of each of three different 96-well plates with 10 nM GAPDH siRNA. Two days later, we repeated the experiment with three more 96-well plates. Analysis of GAPDH mRNA expression 48 h post-transfection revealed that the reverse transfection procedure is indeed highly reproducible (Fig. 3). We also tested the reproducibility of reverse transfection in the context of an siRNA library screen by measuring the reduction in target mRNA expression in positive control siRNA wells present on each plate. As above, the reduction in gene expression across seven 96-well plates comprising a medium-scale siRNA screen consistently exceeded 95% (data not shown).
FIGURE 3.
Well-to-well reproducibility of the reverse transfection method. A GAPDH and nonsilencing siRNA (10 nM) were reverse transfected in triplicate into HeLa cells in a 96-well format. Forty-eight hours post-transfection, cells were harvested and analyzed by real-time RT-PCR for both target mRNA and 18S rRNA levels. Remaining gene expression was calculated as a percentage of target mRNA in cells transfected with siRNA targeting GAPDH compared to cells transfected with the nonsilencing control siRNA. Data were normalized against the 18S rRNA signal and are presented as means ± SD.
Cell viability associated with reverse transfection
To ensure that reverse transfection does not cause undue stress on the treated cells, we measured the viability of cells reverse transfected under a broad range of conditions. Although slight reduction in cell viability was observed in reverse-transfected cells at the higher siRNA concentrations, there was no distinguishable difference in the health of the cells when we used siRNA concentrations of 30 nM or less. We suspect that the increased cytotoxicity at higher siRNA concentrations relates to the increased transfection efficiency of the method; we observe the same level of cytotoxicity when using standard transfection to deliver more than 100 nM siRNA (data not shown).
High-throughput delivery for primary cells—96-well electroporation
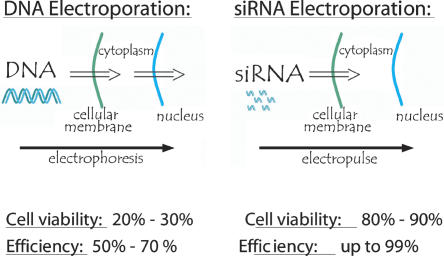
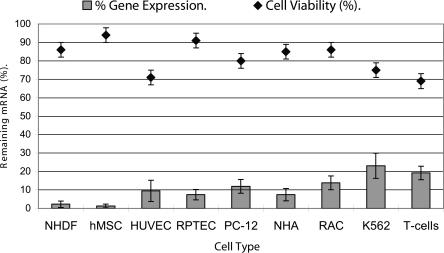
For primary cells, suspension cells, and difficult to transfect immortalized cells, we have found electroporation to be an extremely efficient alternative to chemical transfection for siRNA delivery. To date, mammalian cell electroporation has been used primarily to transfer plasmid DNA into cell nuclei to facilitate exogenous gene expression. The long electrical pulses required to transfer DNA through the cellular and nuclear membrane induce significant cell mortality; thus electroporation is often associated with massive cell death (Fig. 4). Because siRNAs are smaller and only need to be delivered into the cellular cytoplasm to be active, a higher applied external field and shorter electropulse duration can be used to ensure efficient siRNA delivery. These pulse conditions induce significantly less cell toxicity than typical plasmid electroporation conditions. We used electroporation to deliver siRNAs to nine different cell types, and we found the procedure to consistently provide >70% reduction in target mRNA expression and >70% cell viability (Fig. 5). For several cell types, the transfection efficiency and cell viability exceed 90%. It is worth noting that the optimal electroporation conditions are specific for every cell type (Table 2). The three key parameters that influence square wave electroporation efficiency are pulse-length, voltage, and number of pulses. Peak voltage and pulse length are cell type-specific parameters, while number of pulses can be modulated to achieve the desired ratio of electroporation efficiency to cell viability (see Supplementary Data at http://www.ambion.com/techlib/data/RNA.pdf).
FIGURE 4.
Schematic illustrating electroporation of plasmid DNA vs. electroporation of siRNA.
FIGURE 5.
Reduction of GAPDH gene expression and maintenance of cell viability after electroporation across several cell types. The electroporation parameters listed in Table 2 were used to deliver siRNA into seven primary cell types: human primary T-cells (T-cells), human mesenchymal stem cells (hMSC), normal human astrocytes (NHA), normal human dermal fibroblasts-neonatal (NHDF-Neo), rat astrocytes (DI TNC1), normal human umbilical vein endothelial cells (HUVEC), normal human renal proximal tubule cells (RPTEC), and two immortalized cell lines—K562 (human erythroleukemia cells) and PC12 (rat pheochromocytoma) cells. An siRNA targeting GAPDH or nonsilencing sequence siRNA (1.5 μg) was electroporated into each cell type. Forty-eight hours post-electroporation, the cells were harvested and analyzed by real-time RT-PCR for GAPDH mRNA and 18S rRNA levels. 18S rRNA levels were used to normalize GAPDH expression. “Remaining mRNA (%)” was calculated as a percentage of GAPDH mRNA levels in cells electroporated with GAPDH siRNA relative to cells electroporated with nonsilencing siRNA. Electroporations were performed in duplicate. Data are presented as means ± SD.
TABLE 2.
Electroporation parameters applied to different cell types
| Cell type | Voltage (Volts) | Pulse length (μsec) | Number of pulses | Time between pulses (sec) | Number of cells |
| HMSC | 700 V | 90 μsec | 2 | 5 sec | 75,000 |
| HUVEC | 250 V | 150 μsec | 1 | — | 75,000 |
| RPTEC | 300 V | 300 μsec | 1 | — | 75,000 |
| Human T-cells | 300 V | 400 μsec | 1 | — | 200,000 |
| NHDF-neo | 900 V | 70 μsec | 2 | 5 sec | 75,000 |
| PC-12 | 450 V | 200 μsec | 1 | — | 75,000 |
| Rat astrocytes | 300 V | 90 μsec | 3 | 0.1 sec | 75,000 |
| NHA | 400 V | 120 μsec | 2 | 0.1 sec | 75,000 |
| K562 | 350 V | 130 μsec | 2 | 0.1 sec | 150,000 |
Time-course studies using electroporation followed by real-time PCR revealed that target mRNA levels are reduced as soon as 6 h after electroporation; maximum reduction of mRNA levels is achieved 24–48 h after electroporation, depending on the cell type and expression level of the target gene. Analysis of target protein expression revealed that protein reduction slightly lagged mRNA reduction and that maximal protein reduction occurred 48–72 h post-electroporation (data not shown). To determine expression levels of active siRNA in electroporated cells, and to correlate knockdown of target mRNA with a reduction in the corresponding protein levels, GAPDH knockdown was triggered by electroporating primary HUVEC cells with siRNA targeting GAPDH (see Supplementary Data at http://www.ambion.com/techlib/data/RNA.pdf). A reduction of GAPDH expression was observed at both the mRNA and protein levels in these cells, but not in HUVECs electroporated with nonsilencing siRNA. No variation in expression level was detected in a nontarget mRNA (β-actin), small RNAs (mir-16, 5S rRNA, 5.8S rRNA), or the control protein (Ku).
To enable high-throughput delivery of siRNAs to primary cells, we developed an instrument with the capacity to delivery siRNAs to as many as 96 samples at a time. The 96-well electroporation device features two gold-plated electrode plates, the lower with 96 concave sample wells and the upper with 96 convex protrusions. Cells, siRNAs, and a low-conductivity electroporation buffer are added to the lower, concave plate using a multichannel pipetter. The upper, convex plate is then snapped onto the bottom plate, connecting the lower plate and the upper plate via the sample. The siRNA/cell samples are constrained laterally by their own surface tension between the upper and lower electrodes. The 96-well electroporation device is powered by a Gene Pulser Xcell electroporator power supply. Since the electrode plates are made from large blocks of metal, there is no delay or decrease in the applied electrical pulse between samples; each sample receives an identical pulse. An internal resistor protects the power supply from high peak currents.
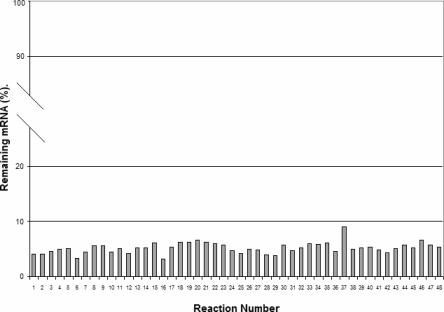
This system was used for high-throughput electroporation with delivery parameters identified previously (Table 2) for primary NHDF-neo cells. To demonstrate consistent functionality of electroporated siRNA and to assess well-to-well reproducibility, a GAPDH siRNA (48 samples) or nonsilencing siRNA (48 samples) was simultaneously electroporated. Forty-eight hours after electroporation, total RNA was isolated from each of the cell populations and assayed for 18S rRNA and GAPDH mRNA using real-time PCR. The GAPDH levels were normalized between samples using the 18S rRNA Ct values. As shown in Figure 6, GAPDH mRNA was reduced by 92%–96%, with an average reduction of 94.8%, CV=18.9%. In addition, cell viability remained above 85% across the plate (Fig. 6). These results indicate that the 96-well electroporator is highly reproducible. Unlike other electroporation protocols which often require a minimum of 200,000 cells to achieve acceptable delivery and enough viable cells for analysis, we found that the combination of the 96-well device and accompanying electroporation facilitates experiments with as few as 25,000 cells per reaction.
FIGURE 6.
Well-to-well variability across the 96-well plate after electroporation of siRNA into primary NHDF-neo cells. GAPDH or nonsilencing siRNA (1.0 μg) was electroporated into normal human dermal fibroblast cells using the 96-well electroporation device; samples 1–48 targeted GAPDH, and samples 49–96 were electroporated with nonsilencing siRNA sequence. Twenty-four hours post-electroporation, the cells were harvested and analyzed by real-time RT-PCR for GAPDH and 18S rRNA. “Remaining mRNA (%)” was calculated as a percentage of GAPDH mRNA in samples 1–48 relative to samples 49–96.
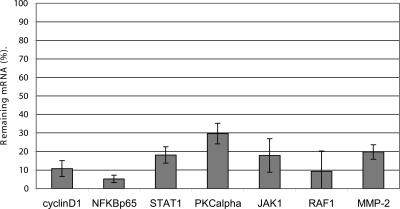
We used the 96-well electroporator with siRNAs to reduce target gene expression in eight cell types: primary human T-cells, normal human dermal fibroblasts-neonatal (NHDF-Neo), renal proximal tubule cells (RPTEC), human mesenchymal stem cells (hMSC), normal human umbilical vein endothelial cells (HUVEC), mouse embryonic fibroblasts (MEF), rhesus monkey stem cells (RMSC), and acute T-cell leukemia (Jurkat) cells. Consistent with single-cuvette electroporation, target gene expression is reduced within the population by >70% and cell viability exceeds 70% in all cell types (data not shown). Gene silencing was induced in primary NHDF-neo cells by several different siRNAs targeting Cyclin D1, NFKBp65, STAT1, PKCα, JAK1, RAF1, and MMP2 (Fig. 7).
FIGURE 7.
SiRNAs elicit RNAi when electroporated into NHDF-neo cells. Seven siRNAs (1.5 μg) targeting Cyclin D1, NFKBp65, STAT1, PKCα, JAK1, RAF 1, MMP-2 and a scrambled sequence were electroporated into NHDF-neo cells. At 48 h post-transfection the cells were harvested and analyzed by real-time RT-PCR for gene expression levels. 18S rRNA levels were used to normalize the Cyclin D1, NFKBp65, STAT1, PKCα, JAK1, RAF 1, MMP-2 data. Percent gene expression was calculated as a percentage of gene expression compared with the negative control siRNA. Electroporations were performed in duplicate. Data are means ± SD.
Application of high-throughput siRNA delivery
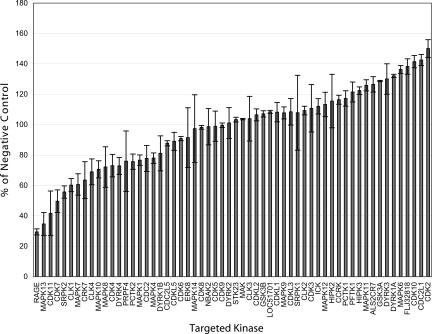
Using the high-throughput, chemical reverse transfection method, we delivered 174 siRNAs targeting 58 individual human kinases (three distinct siRNAs per target gene) plus positive and negative control siRNAs into HeLa cells. Transfections were done in triplicate. We assessed knockdown efficiencies across the multiplate screen using realtime RT-PCR. The positive control siRNAs on each plate provided at least 95% reduction in specific mRNA expression with a CV of 25%. Seventy-two hours after transfection, cell proliferation was measured using an alamar blue assay. Individual data points were compared to that generated by a nonsilencing siRNA (Fig. 8). The screen identified several kinases whose down-regulation either inhibited or enhanced cell proliferation (Table 3). Worth noting is that for several genes, the phenotypes induced by each of the three different siRNAs were subtly or even significantly different. We measured the efficacies of the siRNAs for these genes and found that target mRNAs for each gene were reduced to similar levels (data not shown). Given that siRNAs can target unintended genes (Jackson and Linsley 2004), we suspect that regarding the cases in which only a single siRNA caused an effect, the siRNA targeted a gene essential for cell proliferation in addition to the intended kinase. More interesting are the genes for which two or three siRNAs induced a cell proliferation defect. These genes represent kinases that are involved in cell proliferation. In fact, several genes, such as CDK7, CDK11, CDK2, CDC2L1, and RAGE have already been implicated in the cell cycle (Pagano et al. 1992; Liang et al. 2003; Shi et al. 2003; Brizzi et al. 2004; Feng et al. 2004).
FIGURE 8.
Screening for siRNAs that differentially affect cell proliferation. HeLa cells (5000 cells/well) were plated in 96-well plates simultaneously with addition of transfection complexes, prepared in Opti-mem serum-free medium, by mixing 0.3 μL siPORT NeoFX Transfection Reagent and 1 pmol siRNA (CMGC subset of kinase library). Three different siRNAs targeting each different kinase were transfected in triplicate. Data were normalized by a negative control—the average of three nonsilencing siRNAs (Silencer Negative Control #1, #2, #3 siRNA). Cells were incubated for 72 h, and cell proliferation was measured using an alamar blue cell proliferation assay. The average of the triplicate transfection for each of the three different siRNAs is plotted with error bars representing standard deviations of the nine wells per gene. The size of the error bars derives principally from the use of three different siRNAs per gene.
TABLE 3.
siRNAs that inhibit or enhance cell proliferation
| Inhibit | Enhance |
| SRPK2 | CDK2 |
| CDK7 | CDC2L1 |
| CDK11 | CDK10 |
| MAPK13 | FLJ32818 |
| RAGE | MAPK6 |
DISCUSSION
Double-stranded RNA library screening has enabled the classification of genes involved in apoptosis, phagocytosis, cell proliferation, cell cycle, p53-mediated senescence, cell morphogenesis, cytokinesis, and signaling (Sachse and Echeverri 2004; Somma et al. 2002; Kiger et al. 2003; Lum et al. 2003; Pearson et al. 2003; Berns et al. 2004; Ren et al. 2004; Silva et al. 2004; Xin et al. 2004). The availability of libraries of siRNAs has made it possible to evaluate the effects of sequentially reducing the expression of individual genes on cellular processes. Key to the effective use of siRNA libraries in screening studies are procedures that enable hundreds or thousands of active siRNAs to be delivered to cells with high efficiency while minimizing toxicity in the cells associated with the delivery method.
There are several existing approaches for screening large libraries of dsRNA in mammalian cells. First, individual siRNAs can be transfected in a multi-well format using standard transfection methods with preplated cells. Alternatively, an array-based platform has been described wherein siRNAs are spotted on a slide, to which cells are added and transfected (Ziauddin and Sabatini 2001; Chang et al. 2004; Redmond et al. 2004). Transfection arrays are well suited to high-throughput RNAi screening, except for experimental variability, caused by the small number of transfected cells and the limited number of cell lines that have been reported to have transfection efficiencies high enough to be used in this application.
We have found that chemical reverse transfection can be used to efficiently and reproducibly deliver siRNAs to immortalized, adherent cells in 96- and 384-well plates. In addition, we developed a 96-well electroporator to facilitate high-throughput delivery of siRNAs to primary cells and hard-to-transfect immortalized cell lines. While transfection efficiency and cell viability are cell type-specific for both high-throughput delivery methods, we find them both to be highly reproducible. Coefficients of variance, as measured from the percent of mRNA remaining after siRNA delivery, were similar between 96-well reverse transfection and 96-well electroporation, CV = 20%–27%.
The high reproducibility of the chemical reverse transfection method likely results from three characteristics of the technique: (1) Reverse transfection enables the use of master mixes for both cells and transfection reagent, reducing well-to-well reagent variability; (2) reverse transfection is more efficient than standard transfection, making it possible to achieve threshold target gene reduction at siRNA concentrations that are too low to induce cytotoxicity; and (3) there are fewer pipetting steps relative to standard transfection, reducing the variability of reagent concentration and volume that can occur with repeated pipetting and manipulation.
The high reproducibility of the 96-well electroporator relative to what can be achieved with a single cuvette electroporator is due to several factors: (1) The cells being electroporated are processed and electroporated together, reducing the variability that occurs in cells between the time that they are harvested and when they are ultimately electroporated; and (2) the voltage applied by an electroporator can vary by as much as 10% when using the same delivery parameters. Electropermeabilizing 96 cell samples at a time eliminates this variable.
A distinct advantage of electroporation over other methods of siRNA delivery is that electropermeabilization is independent of cell-cycle stage. This makes it possible to deliver siRNAs to even nondividing or slow-dividing cells. A disadvantage of electroporation approach is that an ~10-fold higher siRNA amount is required for successful gene silencing, compared to the chemical transfection method. Another interesting feature of electroporation is that gene silencing is apparent just a few hours after delivery. The observation that the 96-well electroporator can be used for siRNA studies with as few as 25,000 cells is extremely beneficial for primary cells that are difficult to obtain or culture. Large-scale RNAi screening in most biologically relevant primary cells, a setting where genomic manipulations have proven difficult, is a valuable research tool for studying gene function, target validation, and gene therapeutic approaches.
The combination of high-throughput chemical reverse transfection and 96-well electroporation will enable researchers to use siRNA libraries to identify genes in virtually any cellular process using virtually any cell type. The widespread application of this approach will make it possible to assign cellular functions to many or even most of the genes in the human, mouse, and rat genomes.
MATERIALS AND METHODS
Cells and cell culture
Primary human umbilical vein endothelial cells (HUVEC; Cambrex) were cultivated in modified CCMD 130 Endothelial Cell Medium (Cambrex) and split 1:3 at 90% confluence. Cells were electroporated between passages 3 and 8.
Human cervical adenocarcinoma cells (HeLa; ATCC) were cultivated in DMEM medium, 10% FBS (Gibco), and split 1:8 at 90% confluence. Cells were transfected between passages 12 and 16.
Human breast cancer cells (MCF7; ATCC) were cultivated in DMEM medium, 10% FBS (Gibco), and split 1:6 at 90% confluence. Cells were transfected between passages 22 and 26.
Human neuroblastoma cells (SKNAS; ATCC) were cultivated in DMEM medium, 10% FBS (Gibco), and split 1:6 at 90% confluence. Cells were transfected between passages 17 and 25.
Human lung epithelial cells (A549; ATCC) were cultivated in F12K medium, 10% FBS (Gibco), and split 1:8 at 90% confluence. Cells were transfected between passages 6 and 14.
Human hepatocarcinoma cells (HepG2; ATCC) were cultivated in DMEM medium, 10% FBS (Gibco), and split 1:6 at 90% confluence. Cells were transfected between passages 18 and 26.
Primary human mesenchymal stem cells (hMSC; Cambrex) were cultivated in MSCGM medium (Cambrex) and split 1:5 at 90% confluence. Cells were electroporated between passages 3 and 8.
Primary human renal proximal tubule cells (RPTEC; Cambrex) were cultivated in REGM medium (Cambrex) and split 1:6 at 90% confluence. Cells were electroporated between passages 2 and 5.
Primary normal human astrocyte cells (NHA; Cambrex) were cultivated in ABM medium (Cambrex) and split 1:4 at 90% confluence. Cells were electroporated between passages 3 and 6.
Rat astrocyte cells (DI TNC1; ATCC) were cultivated in DMEM/F12 medium, 10% FBS (Gibco) and split 1:4 at 90% confluence. Cells were electroporated between passages 4 and 7.
Human erythroleukemia cells (K562; ATCC) were cultivated in DMEM medium, 10% FBS (Gibco). Cultures were maintained by addition or replacement of fresh medium between 2 × 105 and 2 × 106 cells/mL. Cells were electroporated between passages 5 and 11.
Primary normal human dermal fibroblasts (NHDF-Neonatal; Cambrex) were cultivated in modified MCDB 202 Fibroblast Cell Medium (Cambrex) and split 1:4 at 90% confluence. Cells were electroporated between passages 3 and 8.
Rat pheochromocytoma cells (PC-12; ATCC) were cultivated in Ham’s F12K medium (Gibco) with 2 mM L-Glutamine and adjusted to contain 1.5 g/L sodium bicarbonate, 15% horse serum (Gibco), 2.5% fetal bovine serum (Gibco) and split 1:5 at 90% confluence. Cells were electroporated between passages 5 and 8.
Cells were maintained under a humidified atmosphere of 5% CO2 at 37°C.
Materials
The GAPDH siRNA (Silencer GAPDH siRNA), p53 siRNA (Silencer p53 siRNA), JAK1 siRNA (Silencer JAK1 siRNA), Cyclin D1 siRNA (Silencer Cyclin D1 siRNA), NFKBp65 (Silencer NFKBp65 siRNA), STAT1 siRNA (Silencer STAT1 siRNA), PKCα siRNA (Silencer PKCα siRNA), RAF1 siRNA (Silencer RAF1 siRNA), MMP-2 siRNA (Silencer MMP-2 siRNA), nonsilencing siRNA (Silencer Negative Control #1 siRNA), and low-conductivity electroporation pulse medium (siPORT siRNA Electroporation Buffer) were obtained from Ambion.
siRNA synthesis
Single-stranded RNA oligonucleotides were synthesized and column- purified (>90% purity as analyzed by mass spectrometry). The single-stranded RNA oligonucleotides were annealed to generate the double-stranded siRNAs that were used for reverse transfection and for electroporation (annealed siRNAs were analyzed by nondenaturing PAGE; Ambion).
Preparation of cells for chemical transfection
Cells were harvested by incubation with 0.05% trypsin-EDTA/PBS (Gibco) for 5 min at 37°C, and trypsin was inactivated by adding complete growth medium. Cell viability was assessed by Trypan Blue (Invitrogen) assay, and each cell suspension was incubated at 37°C in polypropylene tubes (1 h or less) until used in the chemical reverse transfection.
Chemical transfection
Transfection complexes were prepared in Opti-mem serum-free medium (Invitrogen) by mixing 0.3 μL of siPORT NeoFX Transfection Reagent (Ambion) and 10 nM of siRNA (individual siRNA members of the CMGC kinase library targeting CDK, MAPK, GSK3, and CLK gene families; Ambion). HeLa cells (5000 cells/well) were plated in 96-well format simultaneously with addition of transfection complexes. Cells were incubated for 72 h, and subsequently analyzed with an alamar blue cell proliferation assay (Biosource) according to the manufacturer’s recommended protocol.
The 384-well transfections were performed with an automatic liquid handling system (Beckman Biomek 2000) by complexing 30 nM of siRNA and 0.3 μL of siPORT NeoFX Transfection Reagent (Ambion) in Opti-mem serum-free medium (Invitrogen) for 15 min. HeLa cells (4000 cells/well) were plated in 384-well format. Plates were sealed with free-gas exchange lids (Axygen) to prevent water evaporation.
Preparation of cells for electroporation
Cells were harvested by incubation with 0.05% trypsin-EDTA/PBS (Gibco) for 5 min at 37°C, and trypsin was inactivated by adding growth medium. Cells were centrifuged at 200g for 7min, and the cell pellet was resuspended in an appropriate amount of pulse medium.
Single-cuvette electroporation
First, 75 μL of cell suspension containing 75,000 cells and 1.5 μg of siRNA were transferred into a 1-mm electroporation cuvette (BTX). An electrical field was applied to induce siRNA transfer into the cells. After electroporation, cells were incubated in the cuvette at 37°C for 10 min and then transferred into 1 mL of prewarmed (37°C) complete growth medium.
High-throughput electroporation
Here, 40 μL of cell suspension containing 25,000 cells and 1.0 μg of siRNA were used for each sample of a 96-well electroporation reaction. After electroporation, 30 μL of cell suspension was recovered and transferred into 160 μL of prewarmed (37°C) complete growth medium in 96-well tissue culture plates.
Electroporation
The Gene Pulser Xcell System (Bio-Rad) was used for single-cuvette electroporation. A 96-well electroporation device (siPORTer-96 Electroporation Chamber; Ambion) was used with the Gene Pulser Xcell power supply with CE Module (Bio-Rad) to enable high-throughput electroporation in a standard 96-well format. Electroporation reactions were performed in siPORT siRNA Electroporation Buffer (Ambion). Cell viability was measured by the Trypan Blue (Invitrogen) assay 24 h post-electroporation.
Real-time RT-PCR analysis
Total RNA from siRNA electroporated cells was isolated using the MagMAX96 Total RNA Isolation Kit (Ambion). The purified, DNase-treated RNA was reverse transcribed with random decamers using the RETROscript Kit (Ambion). Gene expression levels were determined by real-time PCR using SuperTaq reagents (Ambion) and SYBR Green (Molecular Probes) on the ABI Prism 7900 SDS (Applied Biosystems). The GAPDH data were collected using a human primer set (forward: gaaggtgaaggtcggagt, reverse: gaagatggtgatgggatttc) and rat primer set (forward: catcttcttgtg cagtgcca, reverse: gaggtcaatgaaggggtcgtt). 18S rRNA was amplified (forward: ttgactcaacacgggaaacct, reverse: agaaagagctatcaatctgt caatcct, probe: 5′-VIC-acccggcccggacacgga-TAMRA-3′) as an endogenous control to adjust for well-to-well variances in amount of starting template. The corrected values were normalized to a sample transfected with the nonsilencing siRNA.
Acknowledgments
We thank Ellen Prediger, Po-Tsan Ku, and Kathy Latham for helpful discussions and manuscript review.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7288405.
REFERENCES
- Amarzguioui, M. 2004. Improved siRNA-mediated silencing in refractory adherent cell lines by detachment and transfection in suspension. Biotechniques 36: 766–770. [DOI] [PubMed] [Google Scholar]
- Berns, K., Hijmans, E.M., Mullenders, J., Brummelkamp, T.R., Velds, A., Heimerikx, M., Kerkhoven, R.M., Madiredjo, M., Nijkamp, W., Weigelt, B., et al. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428: 431–437. [DOI] [PubMed] [Google Scholar]
- Brizzi, M.F., Dentelli, P., Rosso, A., Calvi, C., Gambino, R., Cassader, M., Salvidio, G., Deferrari, G., Camussi, G., and Pegoraro, L., et al. 2004. RAGE- and TGF-β receptor-mediated signals converge on STAT5 and p21waf to control cell-cycle progression of mesangial cells: A possible role in the development and progression of diabetic nephropathy. FASEB J. 18: 1249–1251. [DOI] [PubMed] [Google Scholar]
- Caplen, N.J., Parrish, S., Imani, F., Fire, A., and Morgan, R.A. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. 98: 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F.H., Lee, C.-H., Chen, M.-T., Kuo, C.-C., Chiang, Y.-L., Hang, C.-Y., and Roffler, S. 2004. Surfection: A new platform for transfected cell arrays. Nucleic Acids Res. 32: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=373424. [DOI] [PMC free article] [PubMed]
- Chu, G., Hayakawa, H., and Berg, P. 1987. Electroporation for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 15: 1311–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne, J., Drescher, B., Riehle, H., Young, B.D., Krauter, J., and Heidenreich, O. 2003. The apparent uptake of fluorescently labeled siRNAs by electroporated cells depends on the fluorochrome. Oligonucleotides 13: 375–380. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Feng, Y., Goulet, A.C., and Nelson, M.A. 2004. Identification and characterization of the human Cdc2l2 gene promoter. Gene 330: 75–84. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Gehl, J. 2003. Electroporation: Theory and methods, perspectives for drug delivery, gene therapy and research. Acta Physiol. Scand. 177: 437–447. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.L. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Bechert, T., Tuschl, T., and Weber, K. 2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114: 4557–4565. [DOI] [PubMed] [Google Scholar]
- Herweijer, H. and Wolff, J.A. 2003. Progress and prospects: Naked DNA gene transfer and therapy. Gene Ther. 10: 453–458. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L. and Linsley, P.S. 2004. Noise amidst the silence: Off-target effects of siRNAs? Trends Genet. 20: 521–524. [DOI] [PubMed] [Google Scholar]
- Jiang, Z.Y., Zhou, Q.L., Coleman, K.A., Chouinard, M., Boese, Q., and Czech MP. 2003. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc. Natl. Acad. Sci. 100: 7569–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger, A., Baum, B., Jones, S., Jones, M.R., Coulson, A., Echeverri, C., and Perrimon, N. 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laderach, D., Compagno D., Danos O., Vainchenker W., and Galy A. 2003. RNA interference shows critical requirement for NF-κ B p50 in the production of IL-12 by human dendritic cells. J. Immunol. 171: 1750–1757. [DOI] [PubMed] [Google Scholar]
- Liang, Y.C., Tsai, S.H., Chen, L., Lin-Shiau, S.Y., and Lin, J.K. 2003. Resveratrol-induced G2 arrest through the inhibition of CDK7 and p34CDC2 kinases in colon carcinoma HT29 cells. Biochem. Pharmacol. 65:1053–1060. [DOI] [PubMed] [Google Scholar]
- Lum, L., Yao, S., Mozer, B., Rovescalli, A., Von Kessler, D., Nirenberg, M., and Beachy, P.A. 2003. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299: 2039–2045. [DOI] [PubMed] [Google Scholar]
- Oliveira, D.M. and Goodell, M.A. 2003. Transient RNA interference in hematopoietic progenitors with functional consequences. Genesis 36: 203–208. [DOI] [PubMed] [Google Scholar]
- Ovcharenko, D., Jarvis, R., Kelnar, K., and Brown, D. 2004. Delivering siRNAs to difficult cell types. Ambion TechNotes 10: 15–16. [Google Scholar]
- Pagano, M., Draetta, G., and Jansen-Durr, P. 1992. Association of cdk2 kinase with the transcription factor E2F during S phase. Science 255: 1144–1147. [DOI] [PubMed] [Google Scholar]
- Pearson, A.M., Baksa, K., Ramet, M., Protas, M., McKee, M., Brown, D., and Ezekowitz, R.A. 2003. Identification of cytoskeletal regulatory proteins required for efficient phagocytosis in Drosophila. Microbes Infect. 5: 815–824. [DOI] [PubMed] [Google Scholar]
- Redmond, T.M., Ren, X., Kubish, G., Atkins, S., Low, S., and Uhler, M.D. 2004. Microarray transfection analysis of transcriptional regulation by cAMP-dependent protein kinase. Mol. Cell Proteomics 3: 770–779. [DOI] [PubMed] [Google Scholar]
- Ren, Y.G., Wagner, K.W., Knee, D.A., Aza-Blanc, P., Nasoff, M., and Deveraux, Q.L. 2004. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol. Biol. Cell. 15: 5064–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse, C. and Echeverri, C.J. 2004. Oncology studies using siRNA libraries: The dawn of RNAi-based genomics. Oncogene 23: 8384–8391. [DOI] [PubMed] [Google Scholar]
- Shi, J., Feng, Y., Goulet, A.C., Vaillancourt, R.R., Sachs, N.A., Hershey, J.W., and Nelson, M.A. 2003. The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. J. Biol. Chem. 278: 5062–5071. [DOI] [PubMed] [Google Scholar]
- Silva, J., Chang, K., Hannon, G.J., and Rivas, F.V. 2004. RNA-interference- based functional genomics in mammalian cells: Reverse genetics coming of age. Oncogene 23: 8401–8409. [DOI] [PubMed] [Google Scholar]
- Somma, M.P., Fasulo, B., Cenci, G., Cundari, E., and Gatti, M. 2002. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13: 2448–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, D.K. and Jelinek, D.F. 2002. The effectiveness of double-stranded short inhibitory RNAs (siRNAs) may depend on the method of transfection. Antisense Nucleic Acid Drug Dev. 12: 411–418. [DOI] [PubMed] [Google Scholar]
- Weil, D., Garcon, L., Harper, M., Dumenil, D., Dautry, F., and Kress. M. 2002. Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. Biotechniques 33: 1244–1248. [DOI] [PubMed] [Google Scholar]
- Wilson, J.A., Jayasena, S., Khvorova, A., Sabatinos, S., Rodrigue-Gervais, I.G., Arya, S., Sarangi, F., Harris-Brandts, M., Beaulieu, S., and Richardson, C.D. 2003. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. 100: 2783–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, H., Bernal, A., Amato, F.A., Pinhasov, A., Kauffman, J., Brenneman, D.E., Derian, C.K., Andrade-Gordon, P., Plata-Salamán, C.R., and Ilyin, S.E. 2004. High-throughput siRNA-based functional target validation. J. Biomol. Screen. 9: 286–293. [DOI] [PubMed] [Google Scholar]
- Yan, C., Li, F., Patterson, C., and Runge, M.S. 1998. High-voltage and high-salt buffer facilitates electroporation of human aortic smooth-muscle cells. Biotechniques 24: 590–592. [DOI] [PubMed] [Google Scholar]
- Ziauddin, J. and Sabatini, D.M. 2001. Microarrays of cells expressing defined cDNAs. Nature 411: 107–110. [DOI] [PubMed] [Google Scholar]