Abstract
The subcellular localization of mRNAs is a key step in the polarization of cells in organisms from yeast to man. Here, we use a transgenic fly/in situ hybridization assay system to define the positional, structural, and sequence requirements of the TLS, a stem loop RNA sequence element that mediates the subcellular localization of K10 and Orb transcripts in Drosophila oocytes. We find that the TLS is a highly robust and modular element. It mediates efficient RNA localization regardless of sequence context or position within the transcript. Site-specific mutagenesis experiments indicate that the size and shape of the stem and loop regions are critical determinants of TLS activity. Such experiments also identify specific base residues that are important for TLS activity. All such residues map to the stem portion of the structure. Significantly, mutations at these residues interfere with TLS activity only when they alter the stereochemistry of the stem’s minor groove. For example, mutation of the A:U base pair at position 3 of the TLS stem to G:C severely reduces TLS activity, while mutation of the same base pair to U:A has no effect. Extensive searches for TLS-like elements in other Drosophila mRNAs using sequence and structural parameters defined by our experiments indicate that the TLS is unique to K10 and Orb mRNAs. This unexpected finding raises important questions as to how the many hundreds of other mRNAs that are known or thought to exhibit K10 and Orb-like localization are localized.
Keywords: RNA localization, RNA recognition, stem–loop structure, Drosophila K10 gene, Drosophila Orb gene, minor groove recognition
INTRODUCTION
Many mRNAs are localized to specific subcellular sites prior to translation as a means of targeting the encoded protein to that region of the cell where it is needed and/or preventing it from accumulating in regions where it might do harm (for a recent review, see Kloc et al. 2002). The important role that mRNA localization plays in protein targeting is perhaps best illustrated in the Drosophila oocyte, where mutations that disrupt mRNA localization result in the production of misshaped eggs that cannot be fertilized or normal-shaped eggs that give rise to headless or other monstrous embryos that die before hatching (for reviews, see Bashirullah et al. 1998; Huynh and St. Johnston 2004).
Studies in Drosophila, Xenopus, yeast, and mammalian cell culture indicate that mRNAs are localized by one of three general mechanisms, namely, active transport on microtubule or actin tracks, diffusion to a localized trap, and region specific mRNA degradation and protection (Kloc et al. 2002). Each localized mRNA contains one or more cis-acting sequence elements (referred to here as RNA localization elements) that specify how and where the RNA will be localized through the recruitment of proteins that comprise a particular localization machinery.
Given the relatively small number of localization mechanisms and the large number of localized mRNAs—hundreds of mRNAs are estimated to be localized in the Drosophila oocyte alone (Dubowry and Macdonald 1998)—it is surprising that no one RNA localization element has been found in more than two or three different transcripts. One reason for this may simply be that no one RNA localization element is well enough defined to know what to look for when searching for it in other genes. The major difficulty here is that RNA localization elements are not defined solely by their primary sequences, but also by their secondary and, possibly, tertiary structures, which are difficult to search for given the large number of RNA sequences that can fold into a common structure (e.g., see Macdonald 1990).
In an attempt to define an RNA localization element with sufficient resolution to reliably detect it in other transcripts and to gain insights into the biophysical basis of its recognition by the localization machinery, we have focused on the Drosophila fs(1)K10 (K10) gene. K10 is representative of a large family of genes whose mRNAs are synthesized in ovarian nurse cells during early and middle stages (i.e., stages 1–7) of oogenesis and rapidly transported into the oocyte through a series of cytoplasmic bridges that connect these two cell types to each other (Cheung et al. 1992) (see Spradling 1993 for a complete description of the 14 stages of oocyte maturation). Such transport is microtubule-dependent and thought to be powered by a minus-end directed motor, most probably cytoplasmic dynein (Duncan and Warrior 2002; Januschke et al. 2002; Navarro et al. 2004). In response to programmed reorganization of the oocyte’s microtubule cyto-skeleton, K10 and many other transported mRNAs are directed to the oocyte’s anterior cortex during stage 8, where they may persist for many hours or days.
We have identified previously a 44 nucleotide RNA sequence element, called the TLS (Transport and Localization Sequence), that is both required and sufficient for the transport into and anterior localization within the oocyte of K10 transcripts (Serano and Cohen 1995a). More recent studies show that the TLS can also mediate the apical localization of K10 mRNA ectopically expressed in somatic follicle cells (Karlin-McGinness et al. 1996) or in syncytial embryos (Bullock and Ish-Horowicz 2001). While the transport/anterior localization activity of the TLS is thought to reflect its ability to recruit a dynein-containing motor complex to the RNA, no TLS binding proteins have been identified to date.
Our previous studies show that the active form of the TLS is a stem–loop secondary structure, consisting of an 8-nucleotide loop, and a 17-base pair stem, interrupted by two single nucleotide bulges (Serano and Cohen 1995a) (see Fig. 1A). Here, we use the same transgenic fly/in situ hybridization assay system used in our original study to define further the key features of the TLS. We find that TLS activity is not influenced by its position within the transcript. We further find that the base identities of the residues that comprise the loop and bulges are not important and conclude that any base-specific interactions between the TLS and the localization machinery are confined to the stem portion of the structure. Consistent with the idea that these interactions are directed toward the minor groove of the helix, base pair substitutions, e.g., A:U for G:C, that change the stereochemistries of the minor and major grooves of the helix, but not base pair substitutions, e.g., A:U for U:A, that change the stereochemistry of the major groove alone, severely reduce TLS activity. Extensive database searches with programs that probe RNA secondary structures indicate that the TLS is rare, occurring in K10 and Orb, but not in other Drosophila transcripts. Thus, while hundreds of mRNAs are known or thought to exhibit K10- and Orb-like localization, most, if not all, of them would appear to utilize other RNA localization elements.
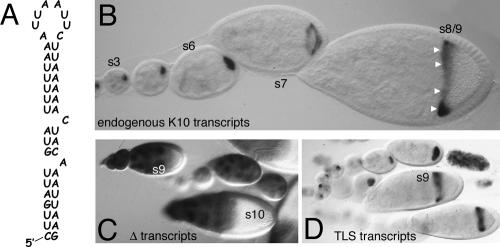
FIGURE 1.
The TLS is required and sufficient for the transport and anterior localization of K10-LacZ fusion transcripts. (A) Sequence and putative secondary structure of the 44 nucleotide TLS. (B) Whole-mount in situ hybridization to endogenous K10 transcripts in wild-type egg chambers, arranged Left to Right in order of increasing stage of development. Stages (s) of select egg chambers are indicated. The oocyte lies to the Right (posterior) of the nurse cell cluster in each of these and in all other displayed egg chambers. In young (s1–6) egg chambers, K10 mRNA fills the entire oocyte. Localization to the oocyte’s anterior cortex (arrowheads) is evident in the stage 8/9 egg chamber. In the stage 7 egg chamber, K10 mRNA is moving toward, but is not yet completely localized to, the oocyte’s anterior cortex. (C, D) Whole-mount in situ hybridization to K10-lacZ transcripts in egg chambers containing the Δ transgene (C), which lacks the TLS, or the wtTLS transgene (D), which contains the TLS. The probe in C and D and in all other experiments designed to detect K10-lacZ transcripts is complementary to the lacZ portion of the RNA and thus does not detect endogenous K10 transcripts.
RESULTS
The TLS is required and sufficient for the transport/anterior localization of a K10-lacZ reporter transcript
In this paper, we report the RNA transport/anterior localization activity of a series of engineered TLS elements using a transgenic fly/in situ hybridization assay system. Our starting point for these studies was a K10-lacZ-K10 (KZK) reporter construct that lacks the TLS due to the deletion of a 308 base pair sequence from the trailing (K10 3′UTR) portion of the construct (see Materials and Methods). This construct, called Δ, and all other constructs described in this paper contain the endogenous K10 promoter, which is active exclusively in nurse cells (Cheung et al. 1992; Serano et al. 1994). In situ hybridization experiments show that transcripts produced from Δ transgenes are completely defective for transport/anterior localization (Fig. 1C; Table 1). Thus, Δ transcripts never become enriched in the oocyte, but rather remain in nurse cells until late stages of oogeneis (after stage 10B), when nurse cells indiscriminately dump their entire cytoplasm into the oocyte (Spradling 1993). Moreover, the few Δ transcripts that enter the oocyte (presumably by diffusion from nurse cells) during earlier stages of oogenesis remain uniformly distributed throughout it, never becoming concentrated at the anterior cortex.
TABLE 1.
Summary of the structures and activities of the TLS elements used is this study
| Expression | |||||||
| In oocyte | |||||||
| TLS | −ΔG | Type of mutation | S1 | S6 | Ant. loc. | Overall activity | Rescue |
| Endogenous | 7.1 | none | + | + | + | wt | >99 |
| wtTLS | 7.1 | none | + | + | + | wt | |
| 5′TLS | 7.1 | position | + | + | + | wt | |
| intron TLS | 7.1 | position | + | + | + | wt | |
| Orb TLS | 11.3 | natural | + | + | + | wt | >99 |
| loop7 | 7.1 | structure | + | + | + | wt | |
| loop5 | 7.1 | structure | + | + | + | wt | |
| rev1 | 7.1 | major gr. | + | + | + | wt | |
| rev2 | 7.1 | major gr. | + | + | + | wt | |
| rev1,2 | 6.9 | major gr. | + | + | + | wt | >99 |
| rev8-17 | 7.1 | major gr. | + | + | + | wt | |
| rev17 | 7.1 | major gr. | + | + | + | wt | |
| 1AU→GC | 9.3 | minor gr. | + | + | + | wt | >99 |
| 8AU→GC | 9.0 | minor gr. | + | + | + | wt | |
| 15UA→CG | 9.0 | minor gr. | + | + | + | wt | |
| rev5 | 7.1 | major gr. | + | + | +/− | mod | >99 |
| rev3-7 | 6.9 | major gr. | + | + | +/− | mod | 92, >99 |
| 13AU→GC | 9.0 | minor gr. | + | + | +/− | mod | |
| Δbub | 16.2 | structure | + | + | +/− | mod | |
| loop10 | 7.1 | structure | − | + | − | weak | <1, 2 |
| stem14 | 6.6 | structure | − | + | − | weak | |
| 3UA→CG | 10.9 | minor gr. | − | + | − | weak | <1, 8 |
| 5UA→CG | 11.3 | minor gr. | − | + | − | weak | |
| 10GC→AU | 5.1 | minor gr. | − | + | − | weak | |
| 8&10 | 7.1 | minor gr. | − | + | − | weak | |
| 3&5 | 13.6 | minor gr. | − | +/− | − | weak/none | |
| Δ | na | deletion | − | − | − | none | <1, 2 |
| stem11 | 6.3 | structure | − | − | − | none |
The names of the TLS elements (and corresponding transgenes) are shown in the Left column. All of the TLS elements are predicted by mFOLD (Zuker 2003) to give the same basic stem–loop secondary structure (data not shown). The ΔG values (as calculated by the mFOLD program) give an indication of the stability of the predicted fold. The expression pattern of the endogenous TLS is based on analysis of endogenous K10 transcripts. The expression patterns of the other TLS elements is based on the analysis of K10-lacZ reporter transcripts in at least two different transgenic lines. Four different levels of activity (wild type, wt; moderate, mod; weak; and none) are evident by noting when the reporter transcripts become enriched in the oocyte (i.e., by stage 1 or by stage 6) and whether they show tight and persistent (+), weak and/or transient (+/−), or no (−) localization to the anterior cortex. The functional activity of select TLS elements was tested by determining the ability of the corresponding transgene to rescue the egg hatching defect of K10 mutants (see Materials and Methods). The numbers shown indicate the percent of eggs that hatched. Two numbers are given in some cases, reflecting two independent experiments, each utilizing a different copy (chromosomal insertion site) of the transgene.
In striking contrast to Δ transcripts, transcripts produced from a KZK transgene (called wtTLS) that contains a single copy of the TLS in place of the 308 base pair deletion become concentrated in the oocyte by stage 1 and show tight localization to the anterior cortex in all mid-stage (stages 7–10) oocytes (Table 1; Fig. 1D). At the onset of stage 11, wtTLS and endogenous K10 transcripts are released from the anterior and degraded (Cheung et al. 1992). We conclude from these findings that the transport/anterior localization activity of the TLS is fully recapitulated within the context of the KZK reporter construct used in this study.
TLS transport/anterior localization activity is not influenced by the position of the element within the transcript
The vast majority of known RNA localization elements map to the 3′UTRs of their respective transcripts (Kloc et al. 2002). To determine if the RNA transport/anterior localization activity of the TLS is dependent on its position within the transcript, we inserted a copy of it 49 nucleotides upstream of the ATG translation start codon of the Δ construct to create 5′TLS. The activity of the 5′TLS element was analyzed using the same transgenic fly/in situ hybridization assay system described above. We also monitored the translation of 5′TLS transcripts by immunostaining transgenic ovaries with an anti-β-galactosidase (β-gal) antibody (see Materials and Methods).
We found that 5′TLS transcripts exhibit wild-type transport and anterior localization (Table 1; Fig. 2A). The wtTLS transcripts become enriched in the oocyte by stage 1 and show strong and persistent localization to the oocyte’s anterior cortex in later stages. We also found that the 5′TLS transcripts are actively translated. Strong staining of the oocyte’s nucleus was observed upon immunostaining with the anti-β-gal antibody (Fig. 2A). The specific labeling of the nucleus was expected, since the K10 and β-gal portions of the encoded fusion protein both contain nuclear localization sequences (Serano et al. 1995). We conclude that the TLS retains its transport/anterior localization activity when placed in the 5′UTR of the transcript, and that such placement does not greatly interfere with translation.
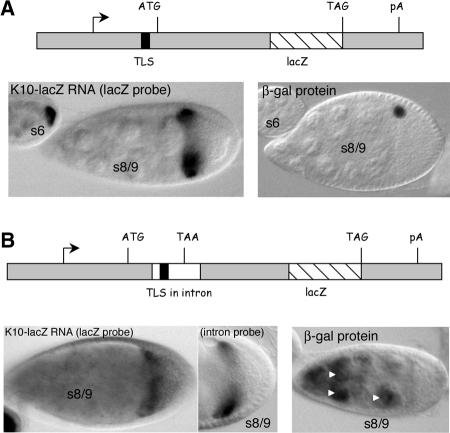
FIGURE 2.
The TLS directs transport and anterior localization when placed in the 5′UTR or intron portion of the K10-lacZ reporter transcript. (A) Structure and expression pattern of the 5′TLS transgene. (Top) Diagram to approximate scale of the 5′TLS transgene. The TLS is located in the 5′UTR of the gene, 49 nucleotides upstream of the ATG translation start codon. (Bottom Left) In situ hybridization for K10-lacZ transcripts in representative egg chambers containing the 5′TLS transgene. Wild-type transport and anterior localization is observed. Note, for example, the intense labeling of the stage 6 oocyte and the sharp localization of the K10-lacZ transcripts to the anterior cortex of the stage 8/9 oocyte. (Bottom Right) Immunodetection of β-galactosidase (β-gal) fusion protein. The single dot of staining in the stage 8/9 egg chamber corresponds to the oocyte nucleus. (B) Structure and expression pattern of the intronTLS transgene. (Top) Diagram to approximate scale of the intronTLS transgene. The TLS (black box) is located in the intron (white box), 15 nucleotides downstream of the intron/exon junction. The intron contains an in-frame TAA stop codon, such that translation of the lacZ portion of the transcript is dependent on removal of the intron by splicing. (Bottom Left) In situ hybridization for intronTLS transcripts in transgenic ovaries using the lacZ probe. The nurse cells and oocyte both stain, consistent with the presence of unspliced and spliced transcripts (see Results). (Bottom middle) In situ hybridization for intronTLS transcripts in transgenic ovaries using the intron-specific probe (see text). (Bottom Right) Immunodetection of β-gal fusion protein. Protein is detected in nurse cell nuclei (arrowheads) only, consistent with the idea that only the nontransported (nurse cell) transcripts are spliced (see Results).
The TLS interferes with splicing when placed close to the splice donor site
While the data presented above indicate that placement of the TLS close to the translational control elements does not interfere with TLS activity, they do not adequately address the reciprocal possibility that the TLS (or TLS activity) interferes with the activity of translation control elements. This uncertainty stems from the fact that the immunostain procedure used to monitor protein synthesis is not sensitive to small differences in expression levels. To address the possibility that the TLS may interfere with translation or other aspects of RNA metabolism when placed close to the corresponding regulatory elements, we sought to develop a more sensitive assay system where such interference would not only manifest itself as a change in the amount of protein or RNA products produced, but also in the cellular distribution of such products. To this end, we inserted a copy of the TLS into the intron (and close to the splice donor site) of an intron-containing version of the Δ construct to create intronTLS. No other TLS element was included in the construct such that intronTLS transcripts would accumulate in (be transported into) the oocyte and become localized to the anterior cortex if, and only if, there were a defect in splicing, i.e., if stable nonspliced transcripts were produced.
As seen in Figure 2B, readily detectable amounts of intronTLS reporter transcripts are transported into, and localized to the anterior cortex of, the oocyte. Two lines of evidence confirm that the transported/anteriorly localized transcripts are not spliced and thus contain the TLS. First, the transported/anteriorly localized transcripts were detected by in situ hybridization with an intron-specific probe as well as with the standard lacZ probe (Fig. 2B). Control experiments using the intron-specific probe against nontransgenic ovaries detected no cytoplasmic transcripts, indicating that the K10 intron is normaly efficiently spliced (data not shown). Second, no β-gal fusion protein was detected in the oocyte upon immunostaining with an anti-β-gal antibody (Fig. 2B); nonspliced transcripts cannot support synthesis of β-gal fusion protein as the intron contains multiple in-frame stop codons (see Materials and Methods). While the TLS strongly inhibited splicing, it did not eliminate it as evident by the presence of intronTLS transcripts in nurse cells (Fig. 2B). Consistent with the idea that these transcripts are spliced, immunostain experiments revealed high level accumulation of β-gal fusion protein in nurse cell nuclei. Importantly, the nurse cell transcripts were detected only when using the lacZ probe. Thus, all nonspliced (TLS-containing) transcripts were efficiently transported and anteriorly localized.
We draw two conclusions from the above findings: First, the TLS can inhibit splicing and, possibly, other aspects of RNA metabolism when placed close to the cis elements that control such processes. Second, the TLS mediates efficient RNA transport and anterior localization, even when located in the main body (protein coding portion) of a transcript providing that it is not removed by splicing.
A TLS-like sequence in the Drosophila Orb gene also mediates RNA transport and anterior localization
We previously identified with BLAST searches a TLS-like sequence in the Drosophila Orb gene (Serano and Cohen 1995a), whose transcripts are also transported into, and anteriorly localized within, the oocyte (Lantz et al. 1992; Lantz and Schedl 1994). To determine whether the Orb TLS-like sequence can mediate transport and anterior localization, we inserted a copy of it into the 3′UTR of the Δ construct to create OrbTLS. As seen in Figure 3, OrbTLS transcripts are efficiently transported and localized. We conclude that the Orb and K10 TLS elements are functionally equivalent.
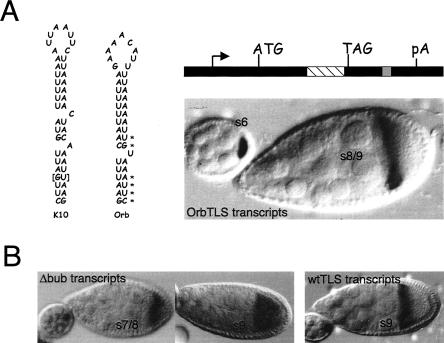
FIGURE 3.
A TLS-like sequence in Orb mediates RNA transport and anterior localization and highlights essential features of the TLS. (A) The nucleotide sequences and putative secondary structures of the TLS elements of the D. melanogaster K10 and Orb genes are shown at the Left. The overall structure of the two elements is very similar, except that the Orb TLS contains only one bulge, while the K10 TLS contains two. The asterisks highlight base pairs in the Orb stem whose orientation is reversed compared to the corresponding base pairs in the K10 stem. As described in the Results, differences in base pair orientation (e.g., 5′A:U vs. 5′U:A) alter the stereochemistry of H-bond donor and acceptor groups in the major groove, but not in the minor groove, of the double helix. The brackets highlight a base pair that is exclusive to the K10 TLS. The structure of the OrbTLS transgene is shown at the top Right. It is identical to the TLS construct of Figure 1, except that it contains a copy of the Orb TLS (gray box) in place of the K10 TLS. The lacZ portion of this construct is denoted with the striped box. In situ hybridization for K10-lacZ transcripts (bottom Right) shows that the Orb TLS mediates wild-type transport and anterior localization. (B) In situ hybridization experiments showing that maximal TLS localization activity requires at least one bulge. (Left) In situ hybridization for Δbub transcripts, which contain a TLS that lack both bulges. Transport into the oocyte is normal; however, localization to the anterior cortex is transient. Thus, while some localization to the anterior cortex is apparent in the stage 7/8 oocyte (s7/8), no localization is apparent in the stage 9 (s9) oocyte. (Right) Control in situ hybridization to a stage 9 oocyte showing persistent localization of transcripts that contain a wild-type copy of the TLS (wtTLS transcripts) to the anterior cortex.
A comparison of the nucleotide sequences and predicted secondary structures of the K10 and Orb TLS elements revealed several differences that presumably reflect nonessential features of the element. One such feature is the base sequence of the loop, 5′-AUUAAUUC for K10 and 5′-GAAAACAUU for Orb. The nonessential nature of the loop sequence was also borne out by our previous study, which showed that an engineered TLS with the loop sequence 5′-GCCGGCCU mediates wild-type or near wild-type transport and anterior localization (Serano and Cohen 1995a). Indeed, we discovered the TLS-like sequence in Orb only after learning that the sequence of the loop is not important and relaxing our BLAST search criteria accordingly.
A second nonessential feature of the TLS is the loop-proximal single-nucleotide bulge. No such bulge exists in the Orb TLS (Figs. 3, 7). A third nonessential feature of the TLS is the base identity of the loop-distal, single-nucleotide bulge, which can be A or U (Fig. 3). We reported previously that an engineered TLS (called TLS Δbub) that contains no bulges mediates transport and anterior localization (Serano and Cohen 1995a). However, reevaluation of this TLS with a more extensive criteria set reveals a slight loss of activity—transcripts lacking both single-nucleotide bulges become enriched in the oocyte during early stages of oogenesis, but localization to the anterior cortex is somewhat diffuse and does not persist beyond stage 8 (Fig. 3B; Table 1). We conclude from these findings that maximal TLS activity requires at least one bulge.
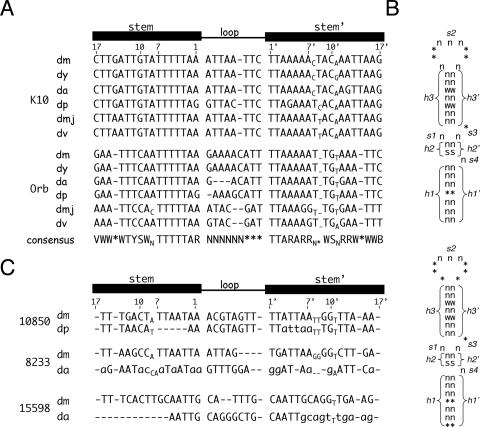
FIGURE 7.
The TLS element is highly conserved, but not found in other genes. (A) Sequence alignments of the D. melanogaster K10 and Orb TLS elements with their counterparts in other Drosophila species. A linear diagram of the TLS is shown above the alignments for reference, where the numbers indicate the positions of the nucleotides that comprise the 1st, 7th, 10th, and 17th base pairs. Abbreviations are as follows: dm, D. melanogaster; dy, D. yakuba; da, D. ananassae; dp, D. pseudoobscura; dmj, D. mojavensis; dv, D. virilis. Gaps in the sequence are denoted with dashes. Unpaired bases (bulges and bubbles) that lie outside of the loop are indicated in subscripts. Abbreviations for the deduced consensus sequences are as follows: V, not T; W, A or T; Y, C or T; S, C or G; N, any nucleotide; *, optional nucleotide of type N; R, A or G; B, not A. (B) Schematic representation of the TLS primary sequence and secondary structure as used for database searches with RNABOB. For these searches, the TLS is conceptually divided into three helical (h1/h1′, h2/h2′, and h3/h3′) and four single-stranded (s1–4) motifs as shown. The acceptable sequences for each motif is indicated, using the same code described in A. The top structure fits most closely with our data (see text), but identifies no TLS-like elements apart from those in the K10 and Orb genes. The bottom structure depicts the more relaxed search conditions (see text), that led to the identification of three candidate elements. (C) Attempted sequence alignments of the candidate TLS elements, which are found in the 3′UTRs of the CG10850 and CG8233 genes and in the 5′UTR of the CG111598 gene, respectively. Species abbreviations are as in A. Residues that are unable to make their normal base pairs (as defined by the K10 and Orb elements) are denoted with lowercase letters. The deletion at the beginning of the D. ananassae CG15598 sequence is based on more extensive sequence alignments (data not shown). The deletion is flanked on its 5′side by the sequence AAAGACACAG. When substituted for the deleted segment, this sequence only regenerates one base pair, i.e., less than the number expected for a random sequence.
Loop and stem (helix) length affect TLS activity
The lengths of the TLS loop (8 and 9 nucleotides, respectively) and stem (17 and 16 base pairs, respectively) are remarkably well-conserved between the K10 and Orb elements (Fig. 3). To determine if these features are critical determinants of TLS activity, we made a series of TLS elements, called loop5, loop7, loop10, stem11, and stem14, that contain slight alterations in stem or loop length (see Fig. 4); loop5 has a 5-nucleotide loop, stem11 an 11-base-pair stem, and so on. None of the mutations are predicted by mFOLD (Zuker 2003) to alter the base-pairing pattern of the element. Theoretical calculations of the free energies of folding (see ΔG values in Table 1) further indicate that the loop mutations do not alter the stability of the element, while the two stem mutations cause a slight reduction in stability (Table 1). The loop and stem variants were inserted into the 3UTR of the Δ construct, and their RNA localization activities analyzed using the same transgenic fly/in situ hybridization assay system as before.
FIGURE 4.
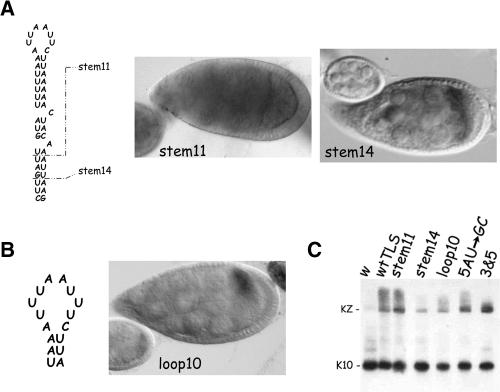
The lengths of the TLS stem and loop are critical for transport and anterior localization activity. (A) Structure and expression patterns of TLS stem truncation mutants. The wild-type TLS with its 17 base pair stem is shown at the Left. The breakpoints of the stem14 and stem11 mutants, which reduce the stem length by 3 and 6 base pairs, respectively, are indicated by the dashed lines. In situ hybridization for K10-lacZ transcripts in representative egg chambers of stem11 and stem14 transgenic lines are shown to the Right of the diagram. The stem11 TLS has no detectable activity; stem11 transcripts never become enriched in oocytes and no localization to the anterior cortex is ever observed. The stem14 TLS possesses only weak activity; stem14 transcripts do not become enriched in the oocyte until ~stage 6 and exhibit only transient and diffuse localization to the oocyte’s anterior cortex. (B) Structure and expression patterns of the loop10 mutant. The sequence of the mutated loop (and only a portion of the stem) is shown on the Left. The Right panel shows that this TLS possesses only weak transport and anterior localization activity; no accumulation is evident in the oocyte until stage 6 and localization to the anterior cortex is diffuse and transient. (C) Northern blot analysis of K10-lacZ (KZ) and endogenous K10 transcripts from transgenic ovaries. The KZ transcripts encoded by the stem11, stem14 and loop10 transcripts all migrate as a single major band of the expected size, ruling out the possibility that their poor transport/anterior localization is due to message instability. The Northern blot also rules out message instability as the reason for the poor transport/anterior localization of 5AU → GC and 3&5 transcripts (see text and Fig. 6 for further details). The w and wtTLS lanes are controls containing RNA from nontransgenic flies and flies harboring the wtTLS transgene, respectively.
We found that the loop5 and loop7 TLS elements possess wild-type transport/anterior localization activity. The loop5 and loop7 transcripts become enriched in the oocyte during very early stages of oogenesis and show tight and persistent localization to the anterior cortex (data not shown). We conclude from these findings that the loop can be reduced in size without adversely affecting TLS activity.
In marked contrast to the mutations that decrease the size of the loop, mutations that increase the size of the loop or that decrease the size of the stem clearly reduce the transport/anterior localization activity of the TLS (Fig. 4). Such reductions are most notable with stem11, where virtually no transport or anterior localization of the reporter transcript is apparent (Fig. 4). The stem14 and loop10 TLS elements support transport, but it is very much slowed—enrichment in the oocyte is not apparent until stage 6 or 7—and little or no anterior localization is evident (Fig. 4A,B).
It may be noted that stem11, stem14, loop10, and other transcripts that exhibit poor transport/anterior localization often give a less intense in situ hybridzation signal than do transcripts, e.g., wtTLS, that show wild-type transport/anterior localization. We believe the former transcripts give a weaker signal because they are distributed over a larger area (volume). Consistent with this interpretation, Northern blot analyses show that stem11, stem14, and loop10 transcripts accumulate to roughly the same steady state level as do wtTLS transcripts (Fig. 4C), even though the latter always gives the most robust signal by in situ hybridization.
We conclude from the above findings that the TLS stem must be at least 15 base pairs in length for wild-type transport/anterior activity and that the loop must be no larger than 9 nucleotides.
TLS activity is not influenced by base changes that specifically alter the stereochemistry of the major groove of the stem
The recognition of RNA control elements by their cognate binding proteins invariably involve the formation of one or more H-bonds between amino acid side chains and specific nucleotide bases (for review, see Hall 2002). Given our findings above that the identity of the bases that comprise the loop and bulge regions of the TLS is not critical for TLS activity, it is likely that any H-bonding between the TLS and the localization machinery is confined to the stem region of the RNA. The major and minor grooves of RNA helices both contain a number of functional groups that could potentially form hydrogen bonds with the localization machinery (Saenger 1984). In the major groove, the stereochemistry of these groups varies for each Watson-Crick (and G:U) base pair. Accordingly, H-bond formation in the major groove is sensitive to all possible base pair substitutions, even those that simply reverse the orientation of the bases, e.g., A:U for U:A. The minor groove is less variable; A:U and U:A base pairs are virtually identical stereochemically as are G:C and C:G base pairs (Seeman et al. 1976; Saenger 1984; Steiz 1990).
A comparison of the base pairs that comprise the stems of the K10 and Orb TLS elements reveal a remarkable conservation of the minor groove. Indeed, apart from an additional G:U base pair (bracketed in Fig. 3) near the bottom of the K10 TLS stem, the minor grooves of two stems are identical, i.e., all base pair changes are simple reversals, e.g., A:U for U:A. The major grooves of the two stems are less well conserved, varying at six positions (marked by asterisks in Fig. 3).
The variability of the major groove together with the fact that the major groove of RNA helices (which invariable adopt the A form) are very deep, narrow, and generally not accessible to amino acid side chains (Seemen et al. 1976; Saenger 1984; Steitz 1990; Frugier and Schimmel 1997) suggests that the major groove of the TLS stem does not form many or possibly any critical H-bonds with the localization machinery. To test this idea more rigorously, we made a series of seven TLS elements that collectively alter the stereochemistry of the entire major groove, while preserving that of the minor groove. This was accomplished by reversing the orientation of all of the base pairs that comprise the TLS stem, either individually, or in small to medium sized groups (Fig. 5). All of these mutations are predicted by the mFOLD program (Zuker 2003) to form the identical stem–loop structure as the wild-type K10 element and to be equally stable (see predicted ΔG values in Table 1). These TLS elements (rev mutants) were individually inserted into the 3′UTR of the Δ construct and analyzed in transgenic flies as before.
FIGURE 5.
Alterations of the major groove of the TLS stem do not significantly interfere with TLS activity. The wild-type K10 TLS is shown at the far Left for reference. The altered regions of the seven reversal (rev) mutants are shown to the Right of the wild-type TLS. Five of the seven reversal mutants exhibit wild-type transport and anterior localization, including rev8 −17 (see figure for representative in situ hybridization experiment) in which each of the last 10 base pairs are reversed. The other two reversal mutants, rev5 (top Right) and rev3 −7 (Table 1) exhibit moderate activity, distinguishable from wild-type only by their inability to mediate complete localization to the anterior cortex.
Five of the seven rev mutants, including one in which the entire bottom two-thirds of the helix is mutated, mediate wild-type transport and anterior localization (Table 1; Fig. 5). The other two rev mutants (rev5 and rev3–7), possess moderate activity, distinguishable from wild-type only by an inability to maintain persistent localization to the anterior cortex (Table 1; Fig. 5). Since mutation of the 5th base pair alone (the rev5 mutant) causes the same reduction in TLS activity as does simultaneous mutation of the 3rd through 7th base pairs (the rev3–7 mutant), all interactions between the localization machinery and the major groove of the TLS helix would appear to be directed at the 5th base pair. Given the slight negative affect that mutation of this base pair has on TLS activity, it seems unlikely that it makes a critical H-bond with the localization machinery. Instead, it might be involved in a hydrophobic or other weak interaction. Regardless of the nature of the interaction at the 5th base pair, it is clear that the major groove of the TLS stem does not, as a whole, play a major role in TLS activity.
TLS activity is influenced by base changes that alter the stereochemistry of the minor groove of the stem
As described above, the minor groove of the TLS stem is highly conserved between K10 and Orb, suggesting that it might provide critical sites of contact for the localization machinery. Consistent with this idea, the minor groove of RNA helices are wide and shallow and readily accessible to amino acid side chains (Saenger 1984). To determine if the minor groove of the TLS stem is critical for TLS activity, we individually changed the A:U base pairs at the 1st, 3rd, 5th, 8th, 13th, and 15th positions of the TLS stem to G:C. We also changed the G:C base pair at position 10 to A:U. We deliberately did not mutate large blocks of base pairs in these experiments so as to preserve the A:U versus G:C content of the helix, which is heavily biased (82%) toward A:U.
None of the seven minor groove mutations described above are predicted by mFOLD to alter the secondary structure of the TLS, although all are predicted to alter its stability (Table 1). In the case of the six A:U to G:C mutations the stability of the folded structure is predicted to be increased; the folding of the wild-type element has a predicted ΔG of −7.1 kcal/mol, while the folding of the A:U to G:C mutants have predicted ΔG values of −9.0 kcal/mol to −11.3 kcal/mol. Significantly, all of these ΔG values lie within the range defined by the native K10 and Orb TLS elements (Table 1). Thus, while an increase in stability could adversely affect TLS activity, e.g., by preventing an unfolding reaction that favorably maximizes contact between the TLS and the localization machinery, none of the single A:U to G:C mutations described here would appear to be candidates for such interference.
In contrast to the A:U to G:C mutations, the G:C to A:U mutation at position 10 is predicted to decrease the stability of the folded structure. Because the predicted decrease is large (28% drop in ΔG) and outside the range of any TLS variant known to possess wild-type or near wild-type activity (Table 1), we analyzed this mutation alone (i.e., in the context of otherwise wild-type stem) and in combination with an A:U to G:C mutation at position 8. The predicted ΔG of folding (−7.1 kcal/mol) of the double mutant is identical to that of the wild-type element. Importantly, the A:U to G:C mutation at position 8 on its own has no adverse affect on TLS activity (see below; Table 1).
We find that minor groove mutations at the 3rd, 5th, 10th, and 8th plus 10th base pairs strongly interfere with TLS activity (Table 1; Fig. 6). Transcripts containing TLS elements with mutations at these positions show no enrichment in the oocyte until about stage 6 and localization to the anterior cortex is very transient and/or diffuse. Consistent with the idea that these mutations directly interfere with the RNA localization activity of the TLS element, Northern blot analyses reveal no defects in RNA stability (Fig. 4C; data not shown).
FIGURE 6.

Alterations of the minor groove of the TLS can greatly reduce or eliminate TLS activity. The wild-type K10 TLS is shown at the far Left , with the arrows and numbers indicating those base pairs that were mutated individually or in pairs to alter the minor groove. For example, 3UA → CG denotes mutation of the third base pair of the stem from UA to CG. The middle panels show representative in situs to reporter transcripts containing TLS elements with minor groove mutations at the indicated base pairs. The far Right panels show representative in situs of the 3&5 TLS in which the 3rd and 5th base pairs were simultaneously mutated and of the 8&10 TLS in which the 8th and 10th base pairs were simultaneously mutated. The 3&5 TLS possesses no activity, while the 8&10 TLS possess weak activity, indistinguishable from that of the 10th base pair single mutation.
Each of the other four single-base pair mutations exhibit wild-type or near wild-type transport and anterior localization, with the exception of the A:U to G:C mutation at base pair position 13, which has a slight defect in anterior localization (Table 1; Fig. 6). We interpret the above data to mean that the localization machinery makes critical contacts with the minor groove of the TLS at some, but not all, base pairs. These contacts include, but are not necessarily limited to, the 3rd, 5th, and 10th base pairs. Based on their strong mutant phenotypes, we think it is likely that at least some of these contacts involve the formation of H-bonds.
Consistent with the idea that the 3rd, 5th, and 10th base pairs define three unique points of contact with the localization machinery, rather than one unique contact point that is dependent on all three base pairs, simultaneous mutation of the 3rd and 5th base pairs from A:U to G:C completely abolishes TLS activity (Table 1; Fig. 6).
TLS transport and anterior localization activity correlates well with K10 function
To determine the degree of TLS activity needed to support K10 function, we used an egg hatching assay (Serano and Cohen 1995a,b). This assay is carried out with eggs collected from flies that are homozygous for the K10LM00 null allele such that the only source of K10 protein is that produced from the transgene. In the absence of any transgene, or in the presence of K10 transgenes that lack the TLS, <1% of the collected eggs hatch (Serano and Cohen 1995a). Conversely, >99% of the eggs hatch when collected from K10LM00 homozygous females that contain a K10 transgene with an intact TLS element (Serano and Cohen 1995a). Because the K10-lacZ reporter construct used in the current study encodes nonfunctional K10 protein, we generated a new series of transgenes by recloning select TLS elements into a reporter construct that encodes full-length K10 protein.
We found that all of the TLS elements that exhibit wild-type to moderate RNA localization activity also exhibit wild-type (>99%) or nearly wild-type egg hatching activity (Table 1). Conversely, all TLS elements that exhibit weak or no RNA localization activity exhibit little or no (<1%–8%) egg hatching activity (Table 1). That TLS elements with “only” moderate RNA localization activity, i.e., that are unable to power strong localization to the anterior cortex, exhibit wild-type K10 gene function is not surprising given our previous finding that the distribution of K10 mRNA within the oocyte has no bearing on the localization of K10 protein to the oocyte nucleus and/or K10 function (Serano and Cohen 1995b). Rather, it is only important that K10 transcripts are localized (transported) to the oocyte.
The K10 and Orb TLS elements are highly conserved in other Drosophila species
In an attempt to define further the key features of the TLS, we took advantage of the recent postings of the complete genome sequences of Drosophila yakuba, D. ananassae, D. pseudoobscura, D. virilis, and D. mojavensis (available at http://www.fruitfly.org). Sequence alignments of the D. melanogaster K10 and Orb genes with their orthologs from these other species reveal high conservation of the TLS element (Fig. 7A). Such conservation is particularly striking at the level of the predicted secondary structure (Fig. 7A), which shows almost no variation through D. virilis and D. mojavenesis, which diverged from D. melanogaster 60–65 million years ago (MYA) (Powell 1997). Indeed, only the position and nature of the loop-proximal bulge is variable amongst the species. In the K10 elements of D. melanogaster, D. yakuba, and D. ananassae, this bulge falls after the 7th base pair, while in the other three examined species it falls after the 8th (Fig. 7A). The bulge is completely absent from Orb TLS elements, although a 2-nucleotide bubble (i.e., a mismatched base pair) is found at position 8 in D. mojavenis (Fig. 7B).
The primary sequence of the stem also shows conservation, consistent with our findings that the identity of at least some of the base pairs that comprise the stem is critical for TLS activity. In all, 7 base pairs are completely invariant, and five others vary in ways that only change the stereochemistry of the major groove (see above). Seven base pair changes (including four GU for AU substitutions) alter the stereochemistry of the minor groove, but none occur at positions (i.e., 3, 5, and 10) that our mutagenesis experiments showed are critical for TLS activity.
The TLS is not found in other Drosophila genes
Several hundred mRNAs in addition to K10 and Orb are known or thought to be transported into, and localized to the anterior cortex of, the oocyte (Dubowry and Macdonald 1998). As a first step in determining whether the TLS is responsible for the transport/anterior localization of any of these other mRNAs, we searched all predicted D. melanogaster transcripts for TLS-like sequences using the RNA-BOB program (see Materials and Methods; Fig. 7). Unlike standard (e.g., BLAST) searches, the RNABOB program allows one to define a sequence (or part of a sequence) in terms of its relationship to another sequence. This feature greatly increases one’s ability to identify helices and other structural motifs that are specified by two or more sequence elements, each with a large number of primary sequence possibilities. In our initial runs, we searched for all sequences that preserve the basic TLS secondary structure, without violating rules (essential features) established by our mutagenesis and phylogenic studies. For example, we allowed the loop to be as small as 5 base pairs and mispairing at position 8 of the helix, but disallowed base pairs that would alter the stereochemistry (relative to the wild-type K10 and Orb TLS elements) of the minor groove at positions 3, 5, and/or 10 of the stem. These searches identified the K10 and the Orb elements, but no others. Relaxation of our search criteria to include loops as small as 3 nucleotides and stems as short as 15 base pairs identified three candidate TLS elements (Fig. 7C).
None of the candidate elements identified in the above searches reside in genes whose transcripts are known to be transported into, or localized within, the oocyte, so we used a phylogenic approach to test their veracity. The expectation here was that bona fide TLS elements will confer important functional properties onto their genes and thus be conserved through evolution. Conversely, sequences that by mere chance resemble the TLS should not be conserved. As seen in Figure 7C, none of the candidate elements are conserved to nearly the same extent as are the K10 and Orb elements. In two cases (Fig. 7C), the sequences that define the elements are barely identifiable in D. ananassae and D. yakuba, which separated from D. melanogaster only 5–15 MYA (Powell 1997). In the third case, nonconservation of the element is apparent when the analysis is extended out to D. pseudoobscura, which separated from D. melanogaster ~40 MYA (Powell 1997). We conclude from these analyses that all three of the candidate elements are most likely specious in nature.
We have carried out additional searches with RNABOB using even more relaxed conditions, e.g., acceptance of GC base pairs at positions 3 or 5, or AU base pairs at position 10, and have identified >100 candidate elements. Similar phylogenic analyses of 50 randomly chosen candidates (data not shown) indicate they are also specious in nature and we conclude that the TLS element is, in all likelihood, unique to the K10 and Orb transcripts.
DISCUSSION
We showed previously that the TLS RNA localization element is required and sufficient for the transport into, and the anterior localization within, the oocyte of K10 transcripts. We further showed that conservation of the predicted stem–loop secondary structure is essential for TLS activity (Serano and Cohen 1995a). Here we have extended our studies of the TLS by examining its positional requirements within the mRNA and by probing to higher resolution its sequence and structural requirements.
Positional requirements of the TLS
Our data indicate that the TLS can mediate RNA localization within a variety of different sequence contexts and regardless of its position within the transcript. We show, for example, that the TLS retains full activity when placed in the 5′UTR or protein coding portion (i.e., in a non-spliced intron) of a K10-lacZ reporter transcript.
While the activity of the TLS is not affected by its position within the transcript, TLS activity can influence the activity of other RNA control elements in a position-dependent fashion. We have shown, for example, that the TLS strongly interferes with splicing when placed close to the splice donor site of the K10-lacZ reporter transcript (Fig. 2B). We suspect that such interference reflects poor binding of splicing factors to the RNA due to steric hindrance by the neighboring TLS element. Whether the TLS secondary structure is alone sufficient to hinder the binding of splicing factors to the RNA, or whether such hindrance requires the binding of proteins (the localization machinery) to the TLS is not clear from our data. That components of the RNA localization are present in the nucleus and available for binding is suggested by recent analyses of the proteins that mediate Vg1 mRNA localization in Xenopus oocytes (Kress et al. 2004) and by our previous studies (Karlin-McGinness et al. 1996), which showed that K10 transcripts form distinct particles (in a TLS-dependent manner) when overexpressed in nurse cells. Regardless of the nature by which the TLS interferes with splicing, our data indicate that the position of the TLS, and presumably other localization elements, within the transcript can affect the action of neighboring cis-elements that control other aspects of mRNA metabolism.
Sequence and structural requirements of the TLS
The data presented here add strong support to our previous finding that the ability of the TLS to mediate RNA localization is dependent on its ability to form a stem–loop secondary structure. The data further indicate that the overall size and shape of the stem–loop secondary structure is important. We show, for example, that a mutation that increases the size of the TLS loop by just 2 nucleotides greatly reduces TLS activity. Since the sequence of the loop can be completely changed or reduced in length by 1 or 3 nucleotides without adversely affecting TLS activity, it is unlikely that the loop makes critical contacts with the localization machinery or is otherwise itself important for TLS activity. Rather, it would seem that the loop is only needed to allow/stabilize the stem structure. In this scenario, the ideal loop is one that is small and unobtrusive. By extension, mutations (e.g., loop10) that increase the size of the loop could interfere with TLS activity by getting in the way of important interactions between the localization machinery and stem portion of the TLS structure. An alternative possibility, that the loop10 mutation interferes with TLS activity by promoting a competing fold, is not supported by secondary structure predictions with mFOLD (Zuker 2003).
TLS activity is also reduced upon removal of the two single nucleotide bulges. The observed reduction probably reflects a requirement of the loop distal bulge as only it is conserved between K10 and Orb. The base identity of this bulge is not conserved, indicating that it is not a source of base-specific contact with the localization machinery. Rather, the bulge is likely important for imparting a bend or some other structural change in the TLS that maximizes its contact with the localization machinery.
TLS activity is also strongly reduced by mutations that shorten the length of the stem by 3 or 6 base pairs. These mutations could interfere with TLS activity by reducing the stability of the stem–loop secondary structure—mFOLD predicts up to a 28% drop in the stability of the fold for these mutations—or by eliminating base pairs that themselves make important contacts with the localization machinery. Regardless of their mode of interference, these mutations set a lower limit for the length of the stem that is useful in searching for TLS-like elements in other mRNAs.
Previous studies on the recognition of other known RNA control elements by their cognate regulatory proteins invariable identify H-bond formation as essential (for review, see Hall 2002). Consistent with an essential role for H-bond formation in the recognition of the TLS by the localization machinery, several mutations that change the base pair composition of the stem, without changing the predicted overall structure or stability of the element, greatly reduce TLS activity. Significantly, only those base pair changes (e.g., AU for GC) that alter the stereochemistry of the minor groove, which is the wider and more accessible groove of RNA helixes, result in a decrease in TLS activity.
In addition to forming H-bonds with the localization machinery, functional groups in the minor groove could form intramolecular H-bonds with other parts of the TLS. Such interactions could generate higher-order folds, which may be important for stable binding by the localization machinery.
The TLS element is rare
While many hundreds of mRNAs are known or thought to exhibit K10- and Orb-like localization, extensive searches of the Drosophila genome indicate that the TLS element is unique to the K10 and Orb genes. How, then, is the K10-and Orb-like transport/localization of other mRNAs achieved? Sheer numbers alone provide a compelling argument that some localization machineries must localize multiple different mRNAs. Perhaps K10 and Orb are the exceptions to this rule, using a highly specialized, dedicated machinery while most other mRNAs use a much more generic machinery. While formally possible, this idea is not readily reconciled with our previous finding that the spatial and temporal dynamics of K10 RNA transport into the oocyte are indistinguishable from those of other transported mRNAs, when all are expressed under the control of the same promoter (Karlin-McGinness et al. 1996). A second possibility is that K10, Orb, and other transported mRNAs all bind to same localization machinery, but to different parts of it. In this scenario, only those mRNAs (e.g., K10 and Orb) that bind to the same part (surface) of the localization machinery would be expected to contain the same RNA localization element. This idea is consistent with a number of studies which show that RNA localization machineries are very large, containing a dozen or more protein subunits (e.g., see Wilhelm et al. 2000; Arn et al. 2003; Kress et al. 2004), Implicit in this idea is that RNA localization elements are a recent adaptation that have allowed mRNAs to be localized by parasitizing pre-existing motors and other proteins involved in intracellular trafficking.
MATERIALS AND METHODS
Fly genetics and transformations
All constructs were cloned into the pCaSpeR4 vector (Pirrotta 1988) for introduction into the Drosophila germline. P-element mediated transformation of w1118 flies was carried out as previously described (Serano and Cohen 1995a; Saunders and Cohen 1999). At least two lines were generated and analyzed for each construct. Most of the transgene lines were maintained as homozygous stocks. Transgenes that were homozygous lethal were maintained over CyO or TM3, Sb balancer chromosomes. The strong K10 loss-of-function allele, K10LM00, was maintained over the X chromosome balancer, FM7. Homozygous K10LM00 females survive to adulthood but are sterile, laying dorsalized eggs that do not hatch. Hemizygous males (K10LM00/Y) are viable and fertile. A complete description of all alleles and balancer chromosomes is found at http://flybase.bio.indiana.edu.
Whole-mount in situ hybridization and immunostaining
Enzyme-linked in situ hybridization to whole-mount ovaries was carried out according to Tautz and Pfeifle (1989) with the modifications described in Cheung et al. (1992). Digoxigenin-labeled DNA probes were made by the random priming method (Feinberg and Vogelstein 1983). The K10 and lacZ probes were as previously described (Serano and Cohen 1995a). The template for the intron probe was a 784 base pair BsaI SacI restriction fragment derived from the intron portion of a K10 genomic clone. The β-gal protein was immunolocalized with a rabbit anti-β-gal primary antibody (Organon Teknika Corp.) followed by incubation with the Vector Elite ABC reagent (Vector Labs) and staining with fastDAB (Sigma) according to our published procedures (Serano et al. 1995). Photographs were taken with a Zeiss Axiophot and digitized by scanning with a Nikon LS-3510 film recorder.
Northern blot analysis
RNA was prepared from transgenic ovaries and size fractionated on formaldehyde-containing agarose gels as previously described (Cohen and Meselson 1985). Hybridizations were performed at 42°C in premade hybridization buffer (Amicon Corp.) containing 50% formamid. Blots were washed for 30 min in several changes of 2× SSC, 0.1% SDS and then for 2 h in several changes of 0.1× SSC, 0.5% SDS. Hybridization probes were made by the random priming method (Feinberg and Vogelstein 1983) using α-32P-labeled-dGTP. DNA templates for these reactions consisted of gel-purified fragments corresponding to the complete K10 cDNA (K10 probe), or to the lacZ segment (lacZ probe) of the K10-lacZ reporter constructs (see below).
K10-lacZ constructs
The Δ and TLS constructs
The Δ construct was derived from a previous construct called KZK (Cheung et al. 1992), which has a tripartite architecture of 5′K10-lacZ-K10. The leading K10 segment is 2086 base pairs and includes 433 base pairs of 5′ flanking DNA and the first 1653 base pairs of the K10 cDNA (Cheung et al. 1992), where the K10 protein coding region begins at nucleotide position 192 of the cDNA. This segment is linked in-frame to the entire protein coding region, including the translation stop codon, of the Escherichia coli lacZ gene. The trailing K10 sequence is 2208 base pairs and includes all but the first 50 base pairs of the K10 3′UTR and the first 800 base pairs of 3′K10 flanking DNA. The Δ construct was derived from KZK by replacing the 308 base pair TLS-containing StuI–HpaI fragment in the middle of the K10 3′UTR with a BglII–XbaI linker sequence. The intron containing version of the Δ construct differs from Δ only in that the leading K10 sequence was derived completely from genomic DNA and thus contains the 867 base pair K10 intron. The TLS construct was made by inserting a synthetic copy of the wild-type TLS element into the BglII–Xba sites in the 3′UTR of the Δ construct. The top and bottom strands of the TLS elements were designed in such a way as to give BglII and XbaI sticky ends upon annealing to each other. The sequence of the TLS in this and all other constructs (Table 1) used in this study were verified by DNA sequencing.
The 5′ TLS and intronTLS constructs
The 5′TLS construct was made by inserting a synthetic copy of the TLS element into a naturally occurring XhoI site, 49 nucleotides upstream of the ATG translation start codon in the 5′UTR of the Δ construct. The intronTLS construct was made by replacing the 252 base pair BsaI–Pst fragment from the intron of the intron construct (see above) with a BglII–XbaI linker and then inserting a synthetic copy of the TLS element into those site. This insertion places the first nucleotide of the TLS stem 15 nucleotides away from the first nucleotide of the splice donor site. Several stop codons are located downstream of the TLS in the intron, precluding production of β-gal fusion protein from nonspliced transcripts.
Other TLS constructs
All other TLS constructs were made by inserting synthetic TLS elements carrying the desired alterations (see Table 1 and Results) into the BglII–XbaI sites in the 3′UTR of the Δ construct.
K10 constructs and egg hatching (rescue) assay
The starting construct for these experiments was a fully functional K10 minigene construct, called Kmini (Cheung et al. 1992). This construct includes the K10 nurse cell enhancer/promoter, the entire K10 protein coding region, the K10 3′UTR, and ~800 base pairs of 3′ flanking DNA. TLS elements of interest (see Table 1) were cloned into engineered BglII and XbaI sites of Kmini using a strategy very similar to that described above for the K10-lacZ constructs. The K10 transgenes were introduced into a homozygous K10LM00 mutant background using standard genetic crosses. For the rescue assay, five females of genotype K10LM00/K10LM00; transgene/transgene were mated to three to five wild-type males. Approximately 200 eggs were collected on yeasted apple plates and the percent that hatched was recorded.
Bioinformatics: pattern searches with RNABOB
All pattern searches were carried out with RNABOB version 2.1 as downloaded from http://selab.wustl.edu/cgi-bin/selab.pl?mode=software. Details of the search parameters are given in Figure 7 and/or are available upon request.
Acknowledgments
We thank Cherry Song, Eric Struckhoff, and Matthew Kimbrell for their help with the fly transformations. We also wish to extend thanks to Sean Eddy for making the RNABOB program available to us, and to Jiangwen Fang for help in downloading and recompiling the program to run on our Apple computer. Finally, we thank Vicki COrbin, Robert Ward, and members of the Cohen laboratory for helpful discussions and comments on the manuscript. R.S.C. was supported by grants from NIH, NSF and NSF-EPSCoR.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7218905.
REFERENCES
- Arn, E.A., Cha, B.J., Theurkauf, W.E., and Macdonald, P.M. 2003. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev. Cell 4: 41–51. [DOI] [PubMed] [Google Scholar]
- Bashirullah, A., Cooperstock, R.L., and Lipshitz, H.D. 1998. RNA localization in development. Annu. Rev. Biochem. 67: 335–394. [DOI] [PubMed] [Google Scholar]
- Bullock, S.L. and Ish-Horowicz, D. 2001. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414: 611–616. [DOI] [PubMed] [Google Scholar]
- Cheung, H.-K., Serano, T.L., and Cohen, R.S. 1992. Evidence for a highly selective RNA transport system and its role in establishing the dorsoventral axis of the Drosophila egg. Development 114: 653–661. [DOI] [PubMed] [Google Scholar]
- Cohen, R.S. and Meselson, M. 1985. Separate regulatory elements for the heat-inducible and ovarian expression of the Drosophila hsp26 gene. Cell 42: 737–746. [DOI] [PubMed] [Google Scholar]
- Dubowry, J. and Macdonald, P.M. 1998. Localization of mRNA to the Drosophila oocyte is common in Drosophila ovaries. Mech. Dev. 70: 193–195. [DOI] [PubMed] [Google Scholar]
- Duncan, J.E. and Warrior, R. 2002. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 12: 1927–1938. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P. and Vogelstein, B. 1983. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132: 6–13. [DOI] [PubMed] [Google Scholar]
- Frugier, M. and Schimmel, P. 1997. Subtle atomic group distribution in the RNA minor groove. Proc. Natl. Acad. Sci. 94: 11291–11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, K.B. 2002. RNA–protein interactions. Curr. Opin. Struct. Biol. 12: 283–288. [DOI] [PubMed] [Google Scholar]
- Huynh, J.R. and St. Johnston, D. 2004. The origin of asymmetry: Early polarisation of the Drosophila germline cyst and oocyte. Curr. Biol. 14: R438–R449. [DOI] [PubMed] [Google Scholar]
- Januschke, J., Gervais, L., Dass, S., Kaltschmidt, J.A., Lopez-Schier, H., St. Johnston, D., Brand, A.H., Roth, S., and Guichet, A. 2002. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 12: 1971–1981. [DOI] [PubMed] [Google Scholar]
- Karlin-McGinness, M., Serano, T.L., and Cohen, R.S. 1996. Comparative analysis of the kinetics and dynamics of K10, bicoid, and oskar mRNA localization in the Drosophila oocyte. Dev. Genet. 19: 238–248. [DOI] [PubMed] [Google Scholar]
- Kloc, M, Zearfoss, N.R., and Etkin, L.D. 2002. Mechanisms of sub-cellular mRNA localization. Cell 108: 533–544. [DOI] [PubMed] [Google Scholar]
- Kress, T.L., Yoon, Y.J., and Mowry, K.L. 2004. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J. Cell Biol. 165: 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz, V. and Schedl, P. 1994. Multiple cis-acting target sequences are required for Orb mRNA localization during Drosophila oogenesis. Mol. Cell. Biol. 14: 2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz, V., Ambrosio, L., and Schedl, P. 1992. The Drosophila Orb gene is predicted to encode sex-specific germline RNA-binding proteins and has localized transcripts in ovaries and early embryos. Development 115: 75–88. [DOI] [PubMed] [Google Scholar]
- Macdonald, P.M. 1990. bicoid mRNA localization: Phylogenetic conservation of function and RNA secondary structure. Development 110: 161–171. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V. 1988. Vectors for P-mediated transformation in Drosophila. In Vectors: A survey of molecular cloning vectors and their uses (eds. R.L. Rodriguez and D.T. Denhardt), pp. 437–456. Butterworths, Boston.
- Powell, J.R. 1997. Progress and prospects in evolutionary biology: The Drosophila model. Oxford University Press, New York.
- Saenger, W. 1984. Principles of nucleic acids, advanced texts in chemistry. Springer, New York.
- Saunders, C. and Cohen, R.S. 1999. The role of oocyte transcription, the 5′UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol. Cell 3: 43–54. [DOI] [PubMed] [Google Scholar]
- Seeman, N.C., Rosenberg, J.M., and Rich, A. 1976. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. 73: 804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serano, T.L. and Cohen, R.S. 1995a. A single small stem–loop structure mediates K10 mRNA localization. Development 121: 3809–3818. [DOI] [PubMed] [Google Scholar]
- ———. 1995b. Gratuitous mRNA localization in the Drosophila oocyte. Development 121: 3013–3021. [DOI] [PubMed] [Google Scholar]
- Serano, T.L., Cheung, H.-K., Frank, L.H., and Cohen, R.S. 1994. P element transformation vectors for studying Drosophila melanogaster oogenesis and early embryogenesis. Gene 138: 181–186. [DOI] [PubMed] [Google Scholar]
- Serano, T.L., Karlin-McGinness, M., and Cohen, R.S. 1995. The role of fs(1)K10 in the localization of the mRNA of the TGFα homolog gurken within the Drosophila oocyte. Mech. Dev. 51: 183–192. [DOI] [PubMed] [Google Scholar]
- Spradling, A.C. 1993. Developmental genetics of oogenesis. In The development of Drosophila melanogaster (eds. M. Bate and A. Martinez Arias ), pp. 1–70. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- Steiz, T.A. 1990. Structural studies of protein–nucleic acid interaction: The sources of sequence-specific binding. Q. Rev. Biophys. 23: 205–280. [DOI] [PubMed] [Google Scholar]
- Tautz, D. and Pfeifle, C. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98: 81–85. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J.E., Mansfield, J., Hom-Booher, N., Wang, S., Turck, C.W., Hazelrigg, T., and Vale, R.D. 2000. Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J. Cell Biol. 148: 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]