Abstract
Metazoan replication-dependent histone mRNAs do not have a poly(A) tail but end instead in a conserved stem–loop structure. Efficient translation of these mRNAs is dependent on the stem–loop binding protein (SLBP). Here we explore the mechanism by which SLBP stimulates translation in vertebrate cells, using the tethered function assay and analyzing protein–protein interactions. We show for the first time that translational stimulation by SLBP increases during oocyte maturation and that SLBP stimulates translation at the level of initiation. We demonstrate that SLBP can interact directly with subunit h of eIF3 and with Paip1; however, neither of these interactions is sufficient to mediate its effects on translation. We find that Xenopus SLBP1 functions primarily at an early stage in the cap-dependent initiation pathway, targeting small ribosomal subunit recruitment. Analysis of IRES-driven translation in Xenopus oocytes suggests that SLBP activity requires eIF4E. We propose a model in which a novel factor contacts eIF4E bound to the 5′ cap and SLBP bound to the 3′ end simultaneously, mediating formation of an alternative end-to-end complex.
Keywords: histone mRNA, histone hairpin-binding protein (HBP), translation initiation, oocyte maturation, translational control, poly(A) tail
INTRODUCTION
The stem–loop binding protein (SLBP), or histone hairpin-binding protein (HBP), is an RNA-binding protein essential for the coordination of histone gene expression with DNA synthesis. A failure to ensure appropriate replication-dependent histone gene expression in somatic cells results in the inhibition of cell-cycle progression, defects in cell division, and changes in gene expression. SLBP binds to the conserved stem–loop structure located in the 3′ untranslated region (3′ UTR) of histone mRNA (Wang et al. 1996; Martin et al. 1997). SLBP is an essential protein (Sullivan et al. 2001; Kodama et al. 2002; Pettitt et al. 2002; Zhao et al. 2004) that is part of a multifactor complex that binds to the histone mRNA 3′ UTR and contains the U7 snRNP and the zinc-finger protein ZFP100 (Müller and Schümperli 1997; Marzluff and Duronio 2002). This complex mediates histone-specific mRNA processing that results in mature mRNAs that do not contain a poly(A) tail but end in the stem–loop structure. The stem–loop and SLBP are also involved in translation of histone mRNAs (Gallie et al. 1996; Ling et al. 2002; Sanchez and Marzluff 2002). SLBP is associated with polyribosomes, dependent on the presence of intact histone mRNA (Whitfield et al. 2004; S. Andrews and B. Müller, unpubl. results), and can activate the translation of reporter mRNAs containing the stem–loop structure (Ling et al. 2002; Sanchez and Marzluff 2002).
Interestingly, the production of SLBP and histones is also regulated during development but, in contrast to regulation in somatic tissues, it is not coupled to DNA replication (Wang et al. 1999; Allard et al. 2002). In Xenopus laevis, replication-dependent histone mRNAs are accumulated during the early stages of oogenesis and are maintained in an inactive state. During oocyte maturation, translation of these stored mRNAs ensures that sufficient histones are present prior to the midblastula transition (Woodland 1980), when zygotic transcription begins. Xenopus oocytes express two SLBP species (Wang et al. 1999). xSLBP1 is the homolog of mammalian SLBP, can stimulate translation (Sanchez and Marzluff 2002), and is present throughout oogenesis, increasing approximately twofold at oocyte maturation (Wang et al. 1999). xSLBP2 is similar to xSLBP1 only in the RNA-binding domain, is degraded at oocyte maturation, and is not present in somatic cells. Histone mRNAs are mainly bound to xSLBP2 during oogenesis and since it cannot stimulate translation (Sanchez and Marzluff 2002), it is thought to maintain them in a translationally silent state. Upon maturation, xSLBP2 is exchanged for xSLBP1, which is released from the nucleus and activates translation of histone mRNAs (Wang et al. 1999; Sanchez and Marzluff 2002).
It is well established that the translation of polyadenylated mRNAs is stimulated by the interaction of poly(A)-binding proteins such as PABP1 and ePABP with the poly(A) tail (Gorgoni and Gray 2004; Wilkie et al. 2005). It is proposed that PABP contacts factors at the 5′ end of the mRNA, thereby increasing the recruitment of ribosomal subunits. One important interaction is between PABP and the scaffolding factor eIF4G, which in turn interacts with the cap-binding protein eIF4E. This interaction is thought to simultaneously increase the affinity of eIF4E for the cap and of PABP for the poly(A) tail. eIF4G then recruits the small ribosomal subunit by binding eIF3 (for reviews, see Mangus et al. 2003; Gorgoni and Gray 2004). PABP also interacts with termination factors, suggesting a role for PABP in ribosome recycling (Mangus et al. 2003; Gorgoni and Gray 2004). SLBP may fulfill a role similar to PABP in the translation of histone mRNAs. However, the mechanism by which SLBP stimulates translation is not yet clear. In a heterologous yeast system, human SLBP stimulates translation of reporter genes dependent on the presence of initiation factors eIF4G and eIF3 (Ling et al. 2002). An interaction between eIF4G and SLBP was also detected in mammalian cell extracts. Additionally, sequences in the N terminus and in the C terminus have been suggested to be required for SLBP activity (Ling et al. 2002; Sanchez and Marzluff 2002).
Here we investigate the mechanism by which SLBP stimulates translation in Xenopus oocytes. We show that SLBP affects translation initiation and that its ability to stimulate translation increases dramatically during oocyte maturation. Using a directed yeast two-hybrid approach we found that it interacts with translation factors eIF3 and Paip1. We confirmed these interactions in mammalian cell extracts and mapped the interaction with eIF3h to the SLBP RNA-binding domain and with Paip1 to the C-terminal domain. However, the N-terminal region is sufficient for stimulation of translation. This, together with an IRES-based approach, indicates that other factors are required for SLBP-mediated histone mRNA translation. We present experimental evidence that these factors act early in translation initiation and propose that this may be mediated by a novel specific factor that functions through eIF4E.
RESULTS
SLBP translational activity is increased by oocyte maturation
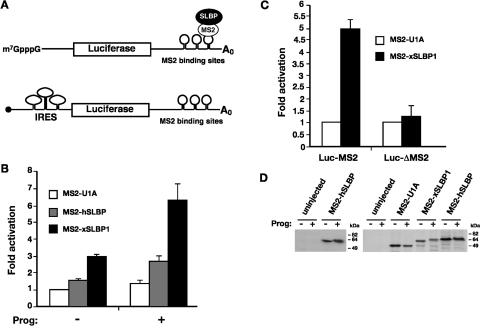
It has been previously reported that Xenopus SLBP1 (xSLBP1) and human SLBP (hSLBP) can stimulate translation in Xenopus stage VI oocytes and in yeast, respectively (Ling et al. 2002; Sanchez and Marzluff 2002). Since histone mRNA translation is normally activated during oocyte maturation (Woodland 1980), we compared the translational activity of both human and Xenopus SLBP1 in stage VI versus mature Xenopus oocytes, using the tethered function assay (Gray et al. 2000; Fig. 1A). This eliminates interference from the endogenous xSLBP proteins present in oocytes (xSLBP1 and xSLBP2). A fusion of MS2 to the RNA-binding protein U1A, which does not activate translation, was used as a negative control. Figure 1B shows that tethered xSLBP1 stimulates expression of an m7GpppG (m7G)-capped luciferase reporter approximately threefold compared to MS2-U1A in stage VI oocytes. The injected reporter mRNAs remain stable throughout the time course of this assay (Gray et al. 2000), indicating that the effects of xSLBP1 occur at the level of translation, consistent with previous results (Ling et al. 2002; Sanchez and Marzluff 2002). While weaker, the effect of tethered hSLBP was reproducible, stimulating translation between 1.4 and 1.7 times compared to MS2-U1A. Interestingly, the translational activity of both hSLBP and xSLBP1 increased significantly when oocytes were matured by treatment with progesterone: up to threefold for the human and sevenfold for the Xenopus protein (Fig. 1B). This increase is specific, since no stimulation of a luciferase reporter lacking the MS2 RNA-binding sites was observed in mature oocytes, as shown for MS2-xSLBP1 (Fig. 1C). Importantly, translation of the polyadenylated β-galactosidase control reporter did not notably change during maturation (average of β-galactosidase activity of progesterone-treated samples is 0.93 times that of untreated samples; data not shown), and the increase in stimulation was not due to increased expression of the fusion proteins (Fig. 1D). Interestingly, the mobility of SLBPs was reduced in progesterone-treated samples, presumably due to protein phosphorylation (Müller et al. 2000). Taken together these results suggest that SLBP translational activity is upregulated during oocyte maturation.
FIGURE 1.
SLBP activity increases during oocyte maturation. (A) Cartoon illustrating the tethered function assay. Luciferase reporter mRNAs contain binding sites for the MS2 coat protein within its 3′ UTR (Luc-MS2); SLBP is expressed as a fusion to MS2 coat protein. Binding of the coat protein tethers SLBP to the mRNA. The luciferase reporters have an m7GpppG (m7G) cap, an ApppG (ApG, black circle) cap, or contain a viral internal ribosome entry site (IRES) in their 5′UTRs, as indicated. An m7G-capped polyadenylated β-galactosidase mRNA (β-Gal) that contains no MS2 sites is utilized as an internal control. (B) In vitro transcribed m7G-capped Luc-MS2 and β-Gal mRNAs were co-injected into the cytoplasm of Xenopus stage VI oocytes expressing MS2-U1A, MS2-hSLBP, or MS2-xSLBP1. After micro-injection, half of the oocytes were incubated overnight in media containing 10 μg/mL of progesterone (prog). Luciferase activity was normalized to β-galactosidase activity and the fold activation relative to MS2-U1A (– progesterone) was plotted, with MS2-U1A values set to one. The average of at least four independent experiments is shown. (C) Stage VI oocytes expressing MS2-U1A or MS2-xSLBP1 were co-injected with m7G-capped Luc-MS2 or a luciferase reporter lacking MS2-binding sites (Luc-ΔMS2) and β-Gal mRNAs. Subsequently, oocytes were incubated overnight in media containing 10 μg/mL of progesterone. The average of three independent experiments is shown. (D) In vitro transcribed MS2-U1A, MS2-xSLBP1, or MS2-hSLBP were micro-injected into stage VI oocytes that were incubated overnight in the absence (−) or presence (+) of 10 μg/mL of progesterone. Oocytes were analyzed by Western blotting using anti-hSLBP antibody (left panel) or were labeled overnight with 50 μCi/mL 35S-methionine and immunoprecipitated with anti-MS2 antibody (right panel). Autoradiography of SDS-PAGE after fluorography is shown.
SLBP stimulates translation initiation
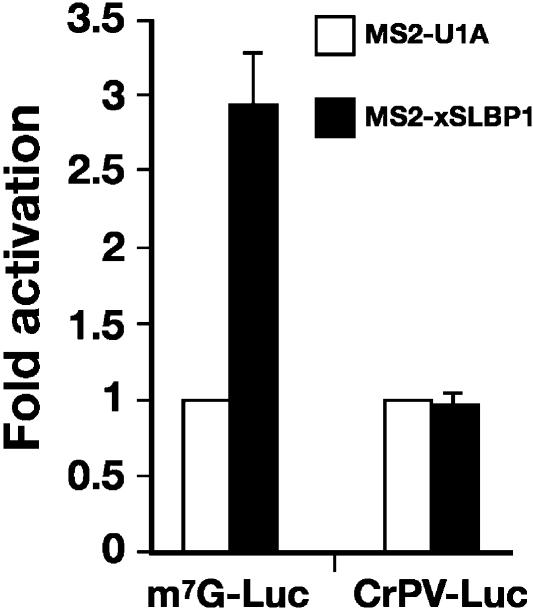
Although it is established that SLBP can stimulate translation, the mechanism by which this is achieved is less clear. In order to determine whether SLBP affects translation initiation or a downstream event, we compared translation stimulation by xSLBP1 of an m7G-capped luciferase reporter with a reporter containing the Cricket Paralysis Virus internal ribosome entry site (CrPV IRES). This IRES is known to bind the ribosome directly, without a requirement for any of the canonical translation initiation factors, by interacting with the P-site of the ribosome (Wilson et al. 2000). Although the CrPV IRES mRNA is translated less efficiently than m7G-capped mRNA in oocytes, luciferase activity of both reporters in oocytes expressing MS2-U1A was normalized to 1, allowing for direct comparison of SLBP-mediated translational activation. As shown in Figure 2, tethered xSLBP1 stimulates translation of m7G-capped luciferase but, in contrast, does not stimulate translation of the CrPV IRES reporter. This suggests that xSLBP1 functions at the level of translation initiation and that it requires the activity of translation initiation factors. MS2-xSLBP1 was also unable to stimulate translation of the CrPV IRES in mature oocytes, suggesting a common mechanism in both developmental stages (data not shown).
FIGURE 2.
SLBP stimulates initiation of translation. m7G-capped Luc-MS2 reporter (m7G-Luc) or an ApG capped reporter containing CrPV IRES within its 5′ UTR (CrPV-Luc) was co-injected with β-Gal mRNA into stage VI oocytes expressing MS2-U1A or MS2-xSLBP1. Luciferase activity was normalized to β-galactosidase activity and the fold activation relative to MS2-U1A was plotted, with MS2-U1A values set to one. The average of two independent experiments is shown.
SLBP interacts with eIF3 and Paip1
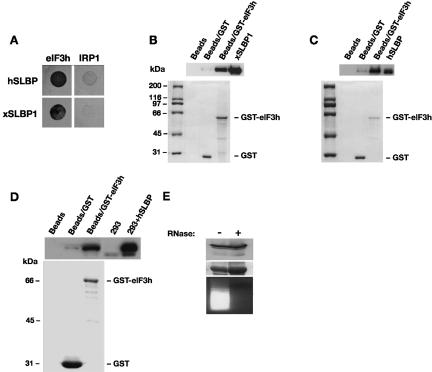
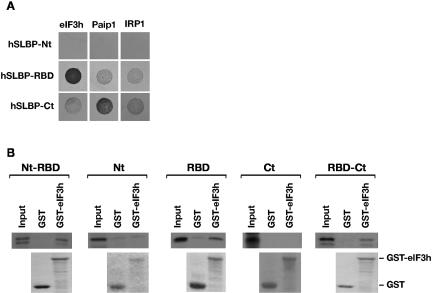
Our results suggest that one or more canonical translation initiation factors is required for SLBP-mediated translation stimulation. To identify the factors that may be involved, we performed a directed yeast two-hybrid screen using hSLBP and xSLBP1 as bait and a panel of vertebrate translation factors, including eIF4G and eIF3 subunits, and also eIF1, eIF1A, eIF2, eIF4A, eIF4B, eIF4E, eIF4H, eIF5, eIF5A, and eIF5B (data not shown). Since SLBP showed a degree of self-activation of the lacZ reporter, we selected for interaction on media lacking histidine and containing 25 mM 3-aminotriazole (3-AT). The iron regulatory protein 1 (IRP1), an RNA-binding protein with a defined role in mRNA specific translation repression, was used as negative control. The growth pattern on 3-AT-containing media indicated a clear interaction between both human and Xenopus SLBP and subunit h (or p40) of eIF3 (Fig. 3A). Importantly, this protein was also isolated in a yeast two-hybrid screen of a human cDNA library using hSLBP as bait (data not shown), highlighting the potential importance of this interaction. We performed GST-pulldown assays with purified GST-eIF3h and SLBP proteins to confirm the interaction between the SLBPs and eIF3h. Figure 3B and 3C show that both human and Xenopus SLBPs were specifically enriched when GST-eIF3h was coupled to glutathione beads, but not when GST-eIF3h was omitted or replaced by GST alone. Importantly, this shows that the interaction between eIF3h and SLBP is direct and does not require the presence of other proteins. To determine whether eIF3 and SLBP interact in mammalian cell extracts, a GST-based approach was taken. HA-tagged human SLBP was expressed in HEK293 cells and whole-cell extracts were incubated in the presence of GST-eIF3h. Figure 3D shows that hSLBP is specifically co-isolated with GST-eIF3h. The interaction between eIF3h and hSLBP in cell extracts is not mediated by RNA, as treatment of the extract with RNase A did not abrogate the interaction (Fig. 3E).
FIGURE 3.
SLBP interacts with eIF3. (A) Yeast two-hybrid assay in MaV99 strain using hSLBP or xSLBP1 fused to GAL4 DNA-binding domain (DB) and eIF3 subunit h or IRP1 fused to GAL4 activation domain (AD). IRP1 is used as a negative control. Transformed colonies were diluted and plated on selective media lacking histidine and containing 25 mM 3-AT. (B) GST-pulldown assay with GST-eIF3h fusion protein and recombinant xSLBP1. GST-eIF3h or GST bound to glutathione sepharose beads or beads alone were incubated with His-tagged xSLBP1. Samples were analyzed by Western blotting using anti-His antibody (top panel) or visualized by staining with Coomassie brilliant blue (bottom panel). xSLBP1 indicates recombinant protein input control (1/30th reaction volume). Note that GST is in excess over GST-eIF3h, indicating that the enrichment of xSLBP1 observed is due to eIF3h. (C) GST-pulldown assay with GST-eIF3h and recombinant hSLBP. The assay was performed as in B except that recombinant hSLBP was used and detected with an anti-hSLBP antibody. hSLBP indicates recombinant protein input control (1/30th reaction volume). (D) Assays were performed and analyzed as in C except that extracts from HEK293 cells expressing HA-tagged hSLBP were used. 293 and 293+ hSLBP are input controls (1/30th reaction volume). Proteins were visualized by Western analysis using an anti-hSLBP antibody (top panel) or by staining with Coomassie brilliant blue (bottom panel). (E) The interaction between eIF3h and hSLBP is not dependent on RNA. Pulldown assays were performed in parallel with D, except that one sample was pretreated with RNase A (+). hSLBP was visualized by Western blotting (top panel) and GST-eIF3h by staining with Coomassie brilliant blue (middle panel). The efficiency of the RNaseA treatment was confirmed by staining extracts with ethidium bromide after agarose gel electrophoresis (bottom panel).
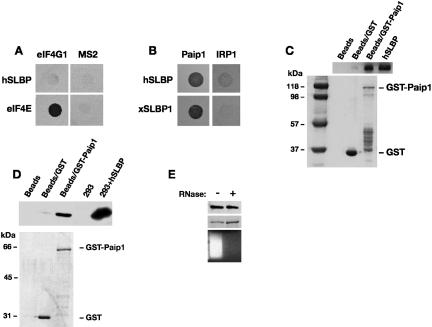
Interestingly, yeast eIF4G was shown to be genetically required for hSLBP to stimulate the translation of reporter mRNAs in yeast and human eIF4G was found in complexes with SLBP in mammalian extracts (Ling et al. 2002). However, these studies did not address whether the co-isolation of these proteins was mediated by other factors. Surprisingly, no interaction between SLBP and eIF4G was identified in the directed yeast two-hybrid analysis, despite experimental controls indicating that the proteins were active in the yeast two-hybrid system (Fig. 4A). This suggests that eIF4G and SLBP do not interact directly, although they may be present in the same complex. However, vertebrate cells contain an additional protein, Paip1, which shares considerable homology with the C-terminal two thirds of eIF4G and has been proposed to mediate the translation of polyadenylated mRNAs, similar to the role of eIF4G (Craig et al. 1998). We therefore examined whether Paip1, which is not present in yeast but is expressed in Xenopus oocytes (Gray et al. 2000), could interact with SLBP. Figure 4B shows that, in contrast to eIF4G, Paip1 interacts with both human and Xenopus SLBP1. This interaction is direct as it can be detected in pulldown assays using purified proteins (Fig. 4C). Furthermore, the hSLBP-Paip1 interaction can be detected in mammalian whole-cell extracts. hSLBP was specifically enriched with GST-Paip1 (Fig. 4D) and this interaction was not mediated by RNA (Fig. 4E).
FIGURE 4.
SLBP interacts with Paip1. (A) Yeast two-hybrid assay in L40ura− strain using eIF4G1 or MS2 fused to LexA-DB and hSLBP or eIF4E fused to GAL4-AD. eIF4E and MS2 are used as positive and negative controls, respectively. Transformed colonies were diluted and plated on selective media lacking histidine and containing 10 mM 3-AT. (B) Yeast two-hybrid assay in MaV99 strain using hSLBP or xSLBP1 fused to GAL4-DB and Paip1 or IRP1 fused to GAL4-AD. Transformed colonies were diluted and plated on selective media lacking histidine and containing 25 mM 3-AT. (C) GST-pulldown assays with GST or a GST-Paip1 fusion protein and recombinant hSLBP were performed and analyzed as described in Figure 3B and 3C. hSLBP indicates input control. Proteins were visualized by Western blotting with an anti-hSLBP antibody (top panel) or by staining with Coomassie brilliant blue (bottom panel). (D) GST-pulldown assay with HA-hSLBP expressed in HEK293 cells. Assays were performed and analyzed as in Figure 3D, except that GST-Paip1 was used. (E) The interaction between Paip1 and hSLBP is not dependent on RNA. Assays were performed and analyzed as in Figure 3E.
eIF3 and Paip1 bind to different regions of SLBP
The interaction of hSLBP with eIF3h and Paip1 was mapped to determine whether it corresponds to the same domains that were previously implicated in the activation of translation (Ling et al. 2002; Sanchez and Marzluff 2002). The human SLBP protein was divided in three portions: N terminus (Nt, amino acids 1–120), RNA-binding domain (RBD, amino acids 121–203), and C terminus (Ct, amino acids 204–270). Directed yeast two-hybrid analysis with eIF3h revealed that this subunit specifically binds the RBD of hSLBP (Fig. 5A). In contrast, Paip1 was found to interact with the C-terminal region of hSLBP (Fig. 5A). The interaction between the RBD of hSLBP and eIF3h was confirmed in GST-pulldown assays, using 35S-methionine-labeled hSLBP domains and GST-eIF3h. Figure 5B shows that all domains containing the RBD were enriched in pulldown assays with GST-eIF3h, whereas the N-terminal or C-terminal domain alone showed no enrichment, mirroring the results of the yeast two-hybrid analysis. Thus the interaction of eIF3 and Paip1 with SLBP does not correlate with regions of SLBP previously ascribed to stimulate translation in Xenopus.
FIGURE 5.
eIF3 interacts with RBD and Paip1 interacts with C terminus of hSLBP. (A) Yeast two-hybrid assay in MaV99 strain using hSLBP domains fused to GAL4-DB and eIF3h, Paip1, or IRP1 fused to GAL4-AD. (Nt) amino acids 1–120; (RBD) amino acids 121–203; (Ct) amino acids 204–270. Transformed colonies were diluted and plated on selective media lacking histidine and containing 25 mM 3-AT. (B) GST-pulldown assays with GST or GST-eIF3h fusion and 35S-methionine-labeled hSLBP domains. (Top panel) Autoradiography of SDS-PAGE after fluorography. (Bottom panel) Coomassie blue stained SDS-PAGE. Input is 1/80th reaction volume.
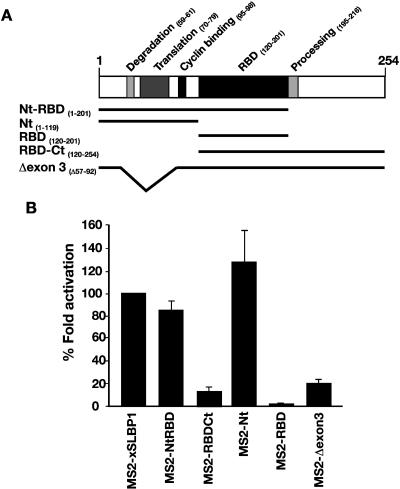
The N terminus of xSLBP1 is required to activate translation
A sequence of 10 residues in the N-terminal domain of xSLBP1 (residues 70–79) was described as necessary to activate translation of reporter RNAs in vitro and in Xenopus stage VI oocytes (Sanchez and Marzluff 2002), although a requirement for the C terminus of hSLBP in yeast has also been suggested (Ling et al. 2002). The finding that interactions with neither eIF3 nor Paip1 apparently explain the function of the N-terminal domain led us to reexamine the regions required for translation. Since increased activity of SLBP was observed in mature oocytes compared to stage VI, the ability of different domains to stimulate translation was examined in mature oocytes, where histone mRNAs are normally translated. The tethered function assay was utilized, as it allows separation of the role of protein domains in translation from their RNA-binding activity. The Xenopus SLBP1 protein was divided into the three main domains, Nt, RBD, and Ct, and MS2 fusions were constructed (Fig. 6A). When analyzed in mature oocytes, MS2-NtRBD maintained an activity similar to full-length xSLBP1, while RBD-Ct was unable to appreciably stimulate translation of the luciferase reporter (Fig. 6B). A further analysis of Nt and RBD revealed that full translational activity was retained by Nt, while RBD showed no activity. These data are in agreement with data reported previously in stage VI oocytes (Sanchez and Marzluff 2002). To further investigate whether the N-terminal region identified by Marzluff and colleagues also represents the only active region in mature oocytes, a construct was created taking advantage of a natural splice isoform of SLBP, which is present in human and mouse (Modrek et al. 2001; A. Schaller, D. Schümperli, and B. Müller, unpubl. observation). This splice form deletes exon 3, which contains the 10 amino acid activation domain, while maintaining the open reading frame. An MS2-xSLBP1 fusion with an exon 3 deletion, encompassing amino acids 57–92 (Fig. 6A), was constructed and tested in the tethered function assay. As shown in Figure 6B, this construct was unable to stimulate translation of the luciferase reporter. This supports the idea that exon 3 in the N terminus is required for translation stimulation by xSLBP1 and confirms that interactions with eIF3 and Paip1 are not sufficient for translational activity in either mature or immature oocytes. Furthermore, as deletion of exon 3 does not interfere with the binding to histone mRNA (data not shown), it is possible that this SLBP form may act as an inhibitor of histone gene expression.
FIGURE 6.
Tethered N-terminal domain of xSLBP1 stimulates translation in Xenopus mature oocytes. (A) Representation of xSLBP1 with its functional domains (Wang et al. 1996; Dominski et al. 1999; Sanchez and Marzluff 2002; Zheng et al. 2003). Beneath, the xSLBP1 sequences (indicated by black lines) included in MS2-fusion constructs are depicted. (B) m7G capped Luc-MS2 and β-Gal mRNAs were micro-injected in stage VI oocytes expressing MS2-U1A, MS2-xSLBP1, or the fusion constructs indicated in A. Oocytes were incubated in media containing 10 μg/mL of progesterone. Luciferase activity was normalized to β-galactosidase activity and the fold activation of MS2-xSLBP1 was set to 100%. Fold activation of the deletion constructs is plotted as a percentage of MS2-xSLBP1. The average of at least three independent experiments is shown.
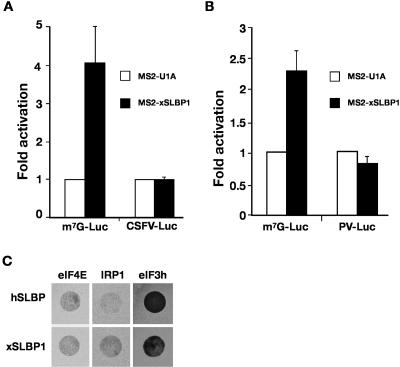
xSLBP1 stimulates an early step in translation initiation
The above results do not completely exclude a role for Paip1 and eIF3 in SLBP-mediated translation; however, they reveal that different factor(s) that interact with the N-terminal domain must have a more dominant function. Our results with the CrPV IRES implicate an important role of an initiation factor in this process. To determine the step in translation initiation at which SLBP acts and to gain insights into which initiation factors are involved, we performed tethered function assays in stage VI oocytes expressing MS2-xSLBP1 with luciferase reporters containing viral IRESs that show different factor requirements (Ostareck et al. 2001). The experiments were also performed in mature oocytes and equivalent results were obtained (data not shown). Initially, we analyzed the classical swine fever virus (CSFV) IRES, whose activation does not require eIF4E, eIF4G, eIF4A, eIF4B, eIF1, and eIF1A (Pestova et al. 1998). We found that MS2-xSLBP1 was unable to stimulate translation directed by this IRES (Fig. 7A), suggesting that one or more of these factors are required and that 40S recruitment or scanning is promoted by SLBP. To further delineate the requirements for SLBP-mediated stimulation, two additional reporter mRNAs were tested, which contained either the poliovirus (PV) IRES or a nonfunctional ApppG (ApG) cap. The analysis of PV IRES showed that xSLBP1-mediated activation of this reporter was completely abrogated (Fig. 7B). Since this IRES utilizes all the canonical initiation factors except eIF4E (Belsham and Jackson 2000), this implies that xSLBP1 affects 40S joining through eIF4E. This is consistent with a marked reduction in the level of stimulation of ApG-capped reporter mRNA by xSLBP1 (data not shown). However, we were unable to detect a direct interaction between SLBP and eIF4E in the yeast two-hybrid approach, despite these proteins being expressed and active in the experimental system (Fig. 7C).
FIGURE 7.
xSLBP1 does not stimulate CSFV or PV IRES reporters. (A) m7G-capped Luc-MS2 (m7G-Luc) or ApG capped CSFV IRES-containing Luc-MS2 (CSFV-Luc) mRNAs were co-injected with β-Gal mRNA into stage VI oocytes expressing MS2-U1A or MS2-xSLBP1. Luciferase activity was normalized to β-galactosidase activity and the fold activation over MS2-U1A was plotted. The average of at least three independent experiments is shown. (B) Tethered function analysis was performed as in A except that ApG-capped PV IRES-containing Luc-MS2 (PV-Luc) was used. The average of seven independent experiments is shown. (C) Yeast two-hybrid assay in MaV99 strain using hSLBP or xSLBP1 fused to GAL4-DB and eIF4E, IRP1, or eIF3h fused to GAL4-AD. Transformed colonies were diluted and plated on selective media lacking histidine and containing 25 mM 3-AT.
DISCUSSION
Replication-dependent histone mRNAs differ from other metazoan mRNAs in that they do not end in a poly(A) tail but in a conserved stem–loop structure. This structure binds the protein SLBP that stimulates translation of histone mRNAs (Ling et al. 2002; Sanchez and Marzluff 2002). An attractive hypothesis is that SLBP could be the functional homolog of PABP, directing circularization of histone mRNAs through the interaction with factors at the 5′ end. This idea is supported by the genetic requirement of eIF4G and eIF3 for its activity in yeast (Ling et al. 2002). In this article we have shown that SLBP targets translation initiation and investigated the physical interaction of SLBP with translation initiation factors. We found that it interacts with eIF3 and with the eIF4G-homolog Paip1. However, interactions with neither Paip1 nor eIF3 seem to underlie the effects of SLBP in translation. Importantly, translation regulation by xSLBP1 appears to target the initial cap-binding step, which is critical for recruitment of the small ribosomal subunit.
Interaction of SLBP with eIF3 and Paip1
Our investigation of the interaction between SLBP and human eIF3 extends previous observations (Ling et al. 2002). We show that this interaction is direct and identify the subunit of eIF3 involved as subunit h (previously called p40 in human) (Fig. 3). This subunit is not part of the core eIF3 complex conserved in yeast, suggesting that it may have a regulatory function (Hershey and Merrick 2000). It is tempting to speculate that one of its possible roles is in the regulation of specific mRNAs. However, our mapping of the region of interaction between eIF3h and hSLBP shows that it binds to the RBD (Fig. 5), a domain that is not sufficient for translation activation (Fig. 6; Ling et al. 2002; Sanchez and Marzluff 2002).
Previous work had suggested a genetic requirement for eIF4G and shown eIF4G to be present in complexes with hSLBP (Ling et al. 2002). However, we could not detect a direct interaction between eIF4GI and hSLBP or xSLBP1. Although a negative result in the yeast two-hybrid system cannot formally rule out a direct interaction with eIF4GI, it is noteworthy that both SLBP and eIF4GI fusion proteins used in this analysis were capable of mediating protein–protein interactions with other factors (Fig. 4). A second eIF4G, eIF4GII, has been described (Gradi et al. 1998) and we cannot exclude an interaction with this protein. However, the functional homology between eIF4GI and eIF4GII and the failure of SLBP to stimulate a PV IRES makes this unlikely. Conversely, a direct interaction with Paip1 was detected by yeast two-hybrid and confirmed in whole-cell extracts (Fig. 4). Paip1 is not conserved in yeast and shares considerable homology with the C-terminal two thirds of eIF4G. Given the hypothesis of SLBP being a functional homolog of PABP, an interaction with Paip1 appears interesting, as this protein has been suggested to play a role in poly(A)-mediated translation and mRNA stability by contributing to the circularization of mRNA (Craig et al. 1998). The SLBP Paip1-binding domain is in the C-terminal region (Fig. 5), which had been suggested to contribute to the translational activity of human SLBP (Ling et al. 2002). However, this domain cannot stimulate translation in Xenopus oocytes (Fig. 6; Sanchez and Marzluff 2002).
Characterization of SLBP activity during oogenesis
Previous analysis in yeast had indicated that both N- and C-terminal sequences contributed to the translational activity of hSLBP (Ling et al. 2002), whereas analysis in stage VI oocytes suggested that sequences in the N terminus were sufficient (Sanchez and Marzluff 2002). In our analysis, the use of the tethered function assay allowed the dissection of the domain functions, separating those required for RNA binding from those required for translational activation per se. We were therefore able to examine the role of each domain singularly.
Histone mRNAs are normally stored inactive in the late stages of oogenesis and their translation is activated at meiotic maturation (Woodland 1980). Interestingly, when the activity of full-length SLBP in stage VI and mature oocytes was compared, a significant increase in SLBP activity was found (Fig. 1). Although overall translation rate rises following oocyte maturation (Richter et al. 1982), the tethered function assay and relative controls ensure that the effects we observe are specific for SLBP. Xenopus oocytes express a second SLBP protein, xSLBP2. During oogenesis xSLBP2 is the main protein bound to histone mRNAs in the cytoplasm and it maintains them in a repressed state (Wang et al. 1999; Sanchez and Marzluff 2002). At maturation, xSLBP2 is degraded, xSLBP1 is released from the nucleus, and it substitutes xSLBP2 on histone mRNAs, activating their translation (Wang et al. 1999; Sanchez and Marzluff 2002). Our results suggest that in addition to the presence or localization of a specific SLBP protein, translation of histone mRNAs may be regulated also by an increased activity of xSLBP1. Since this increase cannot be due to changes in RNA binding of SLBP, it may be explained by modification of the translational activity of the protein or of its partner(s) or by de novo expression of a new partner at maturation. Our analysis of xSLBP1 domains in mature oocytes (Fig. 6), combined with that of Marzluff and colleagues in stage VI oocytes (Sanchez and Marzluff 2002), indicates that the N-terminal domain mediates translation by xSLBP1 in both stages. This suggests that the same translation factor(s) are likely to be utilized in both developmental time points and that a variation of their expression levels or activity may occur. Meiotic maturation involves the activation of various signal transduction pathways that activate kinases (Schmitt and Nebreda 2002). Phosphorylation of SLBP has been described during the cell cycle, where it targets the protein for degradation in mammalian cells (Zheng et al. 2003) and for nuclear relocation in Drosophila embryos (Lanzotti et al. 2004). Mouse SLBP appears to be phosphorylated during oocyte maturation (Allard et al. 2002) and xSLBP1 can be phosphorylated in Xenopus egg extracts by addition of cyclinB/cdc2, of which it is a substrate in vitro (Müller et al. 2000). This modification does not appear to alter binding to the stem–loop or RNA processing but it is currently unknown whether it can alter its translational activity. The small mobility shift of both MS2-hSLBP and MS2-xSLBP1 following progesterone treatment indicates that the MS2 fusion proteins retain the ability to be phosphorylated (Fig. 1D). This raises the possibility that the increase in their translational activity may be due to this modification. Additionally, it is interesting to note that phosphorylation of translation factors, including eIF4E, has been described during oocyte maturation, and phosphorylation of eIF4E is associated with an increase in its activity (Morley and Pain 1995). It is therefore reasonable to imagine that modification of eIF4E and/or xSLBP1 during oocyte maturation contributes to the increase in histone mRNA translation.
A model for SLBP-mediated stimulation
SLBP has been suggested to be a molecular mimic of PABP (Ling et al. 2002; Sanchez and Marzluff 2002). Cytoplasmic polyadenylation during oocyte maturation is thought to enhance translation by providing an extended scaffold for the recruitment of additional molecules of PABP, thus increasing end-to-end complex formation. As a consequence, regulators that activate translation through PABP and whose target mRNAs are thought to contain multiple binding sites are responsive to the number of MS2 sites in tethered function assays (B. Collier, B. Gorgoni, C. Loveridge, H. Cooke, and N.K. Gray, in prep.). In contrast, stimulation by hSLBP and xSLBP1 is not enhanced by the presence of multiple MS2 binding sites, as a reporter mRNA containing only one MS2 binding site was not stimulated less efficiently than a reporter with three MS2 sites (data not shown). Consistent with this observation, PABP and SLBP appear to stimulate translation by different mechanisms.
While we show that SLBP can interact with eIF3 and Paip1, these interactions are not sufficient to stimulate translation and may even play no role. First, the tethering of deletion constructs containing the interaction domains (RBD, Δexon 3, RBD-Ct) fails to stimulate translation (Fig. 6). We cannot formally exclude defects in structural folding of these deletion mutants, although the mimic of the natural isoform missing exon 3 is likely to be correctly folded. Additionally, SLBP fails to stimulate translation of IRESs that utilize eIF3 but do not utilize eIF4E, specifically PV and CSFV IRESs (Fig. 7; Pestova et al. 1998; Belsham and Jackson 2000). Nevertheless, these results do not rule out a contribution of eIF3, for instance by stabilizing interactions formed between SLBP and other factors at the 5′ end (see below and Fig. 8). Likewise, Paip1 functions by interacting with eIF4A (Craig et al. 1998), which is required for PV IRES translation (Pause et al. 1994), suggesting that it does not play a predominant role in SLBP-mediated translation. Moreover, unlike eIF3, the role of Paip1 in translation is less clear and recent results have suggested an important function of this protein in mediating mRNA stability (Grosset et al. 2000). Thus, Paip1 might play a role in stabilizing histone mRNAs.
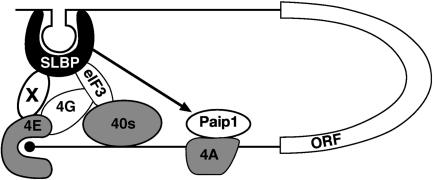
FIGURE 8.
Model for SLBP-mediated translation. SLBP binds the stem–loop structure in the 3′ UTR of histone mRNAs. SLBP interacts with a novel factor X that in turn binds to eIF4E at the m7GpppG cap (indicated as a black circle), leading to circularization of histone mRNA. Additionally, SLBP interacts with subunit h of eIF3 and with Paip1. Paip1 binds eIF4A and is predicted to bind eIF3. These interactions may play a minor role by either stabilizing end-to-end complexes containing SLBP-X-eIF4E or by aiding the recruitment of the small ribosomal subunit (40S). eIF4G may or may not be present in the complex (see Discussion). (ORF) Open reading frame. The figure is schematic and does not depict all the factors involved in translation initiation nor is it meant to indicate the full extent of RNA–protein and protein–protein interactions.
Analyses of IRES-containing reporters suggest that SLBP targets an early stage in the translation initiation pathway (Figs. 2, 7). SLBP was not able to stimulate translation through any of the IRESs tested, including PV, indicating that it utilizes factors involved in cap-dependent rather than cap-independent pathways. The finding that translation driven by an ApG cap is reduced compared to m7G-capped messages is consistent with this idea (data not shown). These pathways vary mainly in the way the small ribosomal subunit is recruited; thus SLBP influences the initial cap-binding step that facilitates the binding of the small ribosomal subunit. We propose that SLBP stimulates translation through eIF4E. SLBP however does not appear to contact eIF4E directly (Fig. 7C). We therefore favor a model in which a protein factor interacts simultaneously with both SLBP and eIF4E linking the two ends of the mRNA (Fig. 8). Several lines of evidence suggest that this factor is unlikely to be eIF4G, in contrast to PABP-mediated translation. First, we were unable to detect a direct interaction between SLBP and eIF4G (Fig. 4A). Second, PV IRES translation is dependent on the presence of eIF4G (Belsham and Jackson 2000; Svitkin et al. 2001) but SLBP is unable to stimulate PV IRES-mediated translation, indicating that this factor does not play a predominant role. This is in contrast to PABP1, which is known to function through eIF4G and can stimulate translation through the PV IRES (N.K. Gray, unpubl. results), supporting the idea that neither eIF4G I nor II is involved in SLBP activity. Although Paip1 is a homolog of eIF4G, it does not interact with eIF4E (Craig et al. 1998) and cannot therefore be the factor bridging SLBP and eIF4E. These indications lead us to conclude that the functional interaction between SLBP and eIF4E in translation initiation is mediated by a yet unidentified protein factor.
Several proteins that interact with eIF4E have been described including eIF4G, 4E-BPs, and mRNA-specific regulators such as maskin and Cup (von der Haar et al. 2004). These proteins share a common 4E-binding motif and bind to the same region of eIF4E. One possibility is that the proposed factor contains this motif, although this may preclude interactions with eIF4G that are important in recruiting the small ribosomal subunit via eIF3. However, SLBP can bind directly to eIF3 and Paip1, and these factors or the novel factor may allow small ribosomal subunit recruitment in the absence of eIF4G. Alternatively, the proposed factor may interact with a different region of eIF4E, allowing its normal interaction with eIF4G, and the SLBP-eIF3 interaction may serve to stabilize complexes formed at the 5′ end, as depicted in Figure 8. The identification of this factor is a priority of future investigations, enabling a more complete understanding of the mechanism by which SLBP stimulates translation.
MATERIALS AND METHODS
Plasmids
MS2-fusion proteins
pMS2-U1A (Gray et al. 2000) and pMSPN (Wilkie et al. 2005) have been previously described. For production of MS2-SLBP fusion proteins, the Xenopus SLBP1 open reading frame was amplified by PCR with primers upstream of initiation and downstream of termination codon: TGAAAGCTAGCGAGGTCACCTGC and CTCGAGCTTCTGGTAAAGGACTGTGC. After insertion into pGEM-T easy, the xSLBP1 fragment was released using NheI and XhoI and inserted into pMSPN. The hSLBP open reading frame was inserted into a modified version of pMSPN, obtained by replacing the XbaI-NcoI fragment with TCTAGAAATTTTGGTTAACTT TAAGGAGATATAACATGG, resulting in the destruction of the NcoI site but preserving the initiation codon. hSLBP was ligated into the unique XhoI site, resulting in a MS2-hSLBP fusion containing an additional alanine prior to the hSLBP initiation codon. The Xenopus SLBP1 N-terminus/RBD fragment was amplified with CGAATTCGAGGTCACCTGCTCAGGGAT and CGGATCCTTA AGGAGGGTCCCACTGATGTA, the RBD/C-terminus fragment with CGAATTCATGGAAACTGACCCAGCTGTA and CGGATC CGACTGTGCGTTAACTAAGCAA, the N-terminal domain with CGAATTCGAGGTCACCTGCTCAGGGAT and CGGATCCCTC TCCATGTGTAGATGAATC, and the RBD with CGAATTCATG GAAACTGACCCAGCTGTA and CGGATCCTTAAGGAGGGTC CCACTGATGTA. The resulting fragments, shown in Figure 6A, were inserted into pGEM-T easy (Promega). For ligation into pMSPN, pGEM derivatives were linearized with BamHI, treated with Klenow, and then released using EcoRI. The fragments were inserted into pMSPN cleaved with SpeI and Klenow treated, followed by restriction with MfeI. The exon 3 sequences were deleted from xSLBP1 using Quick Change site-directed mutagenesis (Stratagene) with primers AAGAACCTCAAGCGAGGCCTGACA GATACAGGAGGAAACTTCTGATTAAT and ATTAATCAGAAG TT TCCTCCTGTATCTGTCAGGCCTCGCTTGAGGTTCTT.
Reporter plasmids
pJK350 (Evans et al. 1994), pLG-MS2, and pLGENB1 (Gray et al. 2000) have been previously described. pPV.IRES-luc (poliovirus IRES) (Bergamini et al. 2000) and pCSFV-CAT (classical swine fever virus IRES) (Ostareck et al. 2001) were a gift from Dr. Matthias Hentze (EMBL, Heidelberg), and pEJ4 (cricket paralysis virus IRES-luciferase) was a gift from Drs. Eric Jan and Peter Sarnow (Stanford University). To generate IRES-luciferase-MS2 reporter constructs, the PV IRES was amplified from pPV. IRES-luc with oligonucleotide primers ATCATAAAGCTTGGGAACAAAA GCTGGGTACCGGGG and TCTCTGGCATGCGAGAATCTGACG and the PCR product was digested with HindIII and SphI and ligated into pLG-MS2 cut HindIII and SphI. The CSFV-IRES was amplified from pCSFV-CAT with CAGTCAAAGCTTCGATCCGTCGACA AGGTTAGCTC and CAGTCAAAGCTTCCCGGTTCCTCCACTCC CACTGG and the PCR product was digested with HindIII and ligated into pLG-MS2 cut HindIII. To generate CrPV-luciferase-MS2, the MS2-binding sequence was amplified from pLG-MS2 with CAGTCAGTTGATCACTCGAGTCCGTTGAGAAGAAGATCA CTGG and TTCGGGGATCCTAGATGTTAAAATGAGC. The PCR product was digested with BclI and BamHI and ligated into the BamHI site of pEJ4.
Yeast plasmids
LexA-MS2, pACT-IRP1 (SenGupta et al. 1996), pACT-Paip1 (Gray et al. 2000), and pACT-hSLBP (Martin et al. 1997) have been previously described. pACT-eIF4E was a gift from Simon Morley (University of Sussex, Brighton) and pBTM-eIF4G1 was a gift from Joe Lewis (University of Edinburgh). To generate pGADT7-eIF3h, a GAL4-activation domain fusion of human eIF3h, the coding sequence was amplified using CATGCAGAATT CATGGCGTCCCGCAAGGAAGGTAC and CATGCAGGATCCT TAGTTGTTGTATTCTT GAAGAGCCTGGGCC. The PCR product was digested with restriction enzymes EcoRI and BamHI and ligated into pGADT7 (Clontech) cut with EcoRI and BamHI. To generate GAL4 DNA-binding fusions of SLBPs, pAS-hSLBP was constructed by inserting the 1184-bp hSLBP cDNA NcoI fragment into the NcoI site of pAS-1 (Clontech; Durfee et al. 1993). pGBKT7-xSLBP1 was made by releasing xSLBP1 open reading frame from pxSLBP1-TNT (a gift from Professor W. Marzluff, University of North Carolina, Chapel Hill) with EcoRI and ligating into pGBKT7 (Clontech) cut with EcoRI. To produce GAL4 DNA-binding domain fusions of hSLBP domains, the N terminus/RBD fragment was amplified using the oligonucleotide primers GGATCCTAATACGACTCACTATAGGGA GAATCCACCATGGCCTGCCGCCCGCGAAGC and CGGGATCC TTAAGACTCCTTTAATCAG, the RBD/C terminus fragment was amplified with CGGAATTCACCATGGCCTGCCGCCCGC and TA GGATCCTTAT CTTCCGCTGGAGGA, the N-terminal domain with GGATCCTAATACGACTCACTATAGGGAGAATCCACCATGGCC TGCCGCCCGCGAAGC and TAGGATCCTTAATAGACTCCTTT GAA, the RBD fragment with CGGAATTCACCATGGCCTGC CGCCCGC and CGGGATCCTTAAGACTCCTTTAATCAG, and the C-terminal domain with GGATCCTAATACGACTCACTATAG GGAGAATCCACCATGAGAAGGATGTGATTTGCAAGA and TAG GATCCTTATCTTCCGCTGGAGGA. PCR products were inserted into pGEM-T easy. Fragments were released from pGEM-T easy with BamHI and EcoRI and inserted into pGBKT7 cut with BamHI and EcoRI.
Protein expression plasmids
For in vitro translation, hSLBP fragments were released from pGEM-T easy constructs described above, using EcoRI, and were inserted into pTNT (Promega). pGEX-4T1 and pGEX-4T2 are from Amersham Biosciences, pcDNA3-Paip1 and pGEXHMK-Paip1 (Craig et al. 1998) were a gift from Professor N. Sonenberg (McGill University, Montreal) and xSLBP1-TNT was a gift from Professor W. Marzluff (University of North Carolina, Chapel Hill). pcDNA3-HA-SLBP was produced by inserting the 1184-bp hSLBP cDNA NcoI fragment into pcDNA3-HA modifed to express proteins with an N-terminal MAYPYDVPDYASLE tag. pGEX-eIF3h was produced by inserting the eIF3h open reading frame into pGEX-4T-2 using XhoI (Schaller 1999).
The sequence of all PCR products was verified by DNA sequencing.
Tethered function assays
Tethered function assays were performed as described (Gray et al. 2000). Briefly, in vitro transcribed m7G-capped RNAs encoding MS2-fusion proteins were micro-injected into Xenopus stage VI oocytes. After 6 h incubation, the m7G-capped luciferase-MS2 or luciferase-ΔMS2, or the ApG-capped luciferase-MS2, PV-lucifer-ase-MS2, CSFV-luciferase-MS2 or CrPV-luciferase-MS2 reporter mRNAs were co-injected with a polyadenylated β-galactosidase reporter that does not contain MS2-binding sites and acts as a control for variations in injection efficiency or in translational activity between oocytes. Oocytes were incubated overnight before collection. For maturation experiments, after the second micro-injection stage VI oocytes were treated overnight with 10 μg/mL progesterone (Sigma) and maturation was scored by the appearance of a white spot on the animal pole. Luciferase activity was normalized for β-galactosidase activity and values obtained with the different SLBP fusions were divided by values of MS2-U1A, thus obtaining the fold of translational activation. Error bars on graphs represent standard error in all cases.
Antibodies
Polyclonal anti-SLBP antibodies have been previously described (Zhao et al. 2004). Polyclonal anti-MS2 antibody was a gift from Professor Peter Stockley (University of Leeds). Mouse monoclonal anti-(His)6 antibody was from Promega. Anti-rabbit and anti-mouse IgGs coupled to horseradish peroxidase were from Sigma and Amersham Biosciences.
Immunoprecipitation
Xenopus oocytes were incubated overnight with 50 μCi/mL of 35S-methionine (ICN) in the absence or presence of 10 μg/mL progesterone. Oocytes were lysed in IP lysis buffer (10μL/oocyte; 150 mM NaCl, 50 mM Tris-HCl at pH 8, 1% NP40, protease inhibitors). Extracts were cleared by centrifugation and 20 μL of lysate were added to 200 μL of lysis buffer and incubated for 1 h at 4°C with an anti-MS2 antibody (1:100). Thirty microliters of Protein-G sepharose beads (Amersham Pharmacia Biotech) were added to the lysate and mixed for 1 h at 4°C. Beads were washed three times with IP lysis buffer and bound material eluted in 20 μL of SDS gel loading dye prior to SDS-PAGE analysis.
SDS-PAGE and Western blotting
Samples were separated by 8% (for MS2-fusion proteins) or 10% SDS-PAGE. Proteins were detected either by staining with Coomassie brilliant blue or by chemiluminescence (Amersham Pharmacia Biotech) after electro-transfer to Hybond-P membrane (Amersham Pharmacia Biotech) and probing with anti-hSLBP (1/5000) or anti-His6 (1/1000). Horseradish peroxidase-conjugated anti-rabbit and anti-mouse IgGs were used as secondary antibodies (1/5000). 35S-methionine-labeled proteins were detected by fluorography using 2′-5′-diphenyloxazole.
Yeast two-hybrid analysis
Yeast two-hybrid analyses were performed with strain Mav99 (Vidal et al. 1996) and L40ura− as described (Zhang et al. 1999). Transformed yeast cells were diluted and plated on media lacking leucine, tryptophan, and histidine and supplemented with 10 or 25 mM 3-AT.
Production of recombinant SLBP proteins
The production and purification of M(H)6-LEA-tagged recombinant hSLBP using Ni-NTA resin (Qiagen) was described earlier (Michel et al. 2000). For the expression of Histagged xSLBP1 the cDNA was amplified using CAGGGCTCGAGGTCATGTC and TAAAGGA CTCGAGGTTAACTAACG, and the amplification product was cleaved with XhoI and inserted into pFASTBAc modified to express proteins with M(H)6-LEA tag. Recombinant baculovirus was produced using the Bac-to-Bac system (Invitrogen), and protein production and purification was as for hSLBP. Protein concentrations were measured using Bradford assay (Biorad) with BSA as a standard. Human SLBP and Xenopus SLBP1 concentrations were 0.32 mg/mL and 1.38 mg/mL, respectively.
In vitro synthesis of 35S-methionine labeled SLBP fragments
hSLBP fragments in pTNT were produced using the wheat germ lysate coupled TNT system (Promega) supplemented with 35S-methionine. hSLBP N terminus/RBD and RBD/C terminus fragments were produced as described (Michel et al. 2000).
Cell line and protein expressions
HEK293 cells were grown in DMEM supplemented with 10% fetal calf serum (FCS) (Helena Biosciences) under standard conditions (37°C, 5% CO2). HEK293 cells were transfected in six-well plates with 5 μg/well pcDNA3-HA or pcDNA3-HAhSLBP using Lipofectamine 2000 (Invitrogen) and incubated for 24 h. Cells were harvested with 170 μL cold NP40 lysis buffer (150 mm NaCl, 1% NP-40, and 50 mM Tris-HCl at pH 8.0) and lysed by sonication at 4°C. Debris was removed by 15 min centrifugation at 16,000g at 4°C. Protein concentration in cleared lysates was determined by Bradford assay and lysates were analyzed by Western blotting or used in GST-pulldown assays.
GST-pulldown assays
BL21 pGEX-eIF3h were grown to an OD600 of 0.4–0.6 at 37°C and then expression was induced with 0.1 mM IPTG for 90 min. Two milliliters bacteria were harvested by centrifugation and stored at −80°C. Cell pellets were resuspended in buffer A (PBS with 1 mM phenyl methyl sulfonyl fluoride, 0.5 mM dithiothreitol, proteinase inhibitor cocktail [Roche]) and cells were lysed by sonication at 4°C and supplemented with 0.5% Triton X-100 (v/v). The insoluble protein was enriched by 10 min centrifugation at 10,000g at 4°C. Protein was resuspended in 300 μL 8 M urea/PBS and the urea concentration was then reduced stepwise by addition of 1/4 volumes of buffer A at room temperature until it was below 2.5 M. This was then added to glutathione sepharose beads. BL21 pGEXHMK-PAIP1 was grown and induced as described except that induction with 0.2 mM IPTG was for 3 h. GST was expressed in BL21 pGEX-4T-1 as described. Cells were lysed and soluble proteins separated from debris by centrifugation as above and proteins from 2-mL aliquots were used in pulldown assays. GST or GST-fusion proteins were mixed with 100 μL of 50% (v/v) glutathione 4B sepharose beads (Amersham Biosciences) in PBS for 1 h at room temperature. The beads were washed three times with 300 μL buffer A and then mixed with 300 μL buffer A containing either 1 μg recombinant hSLBP or xSLBP1, 300 μg HEK293 extract, or 30–40 μL 35S protein premixed with 300 μg BL21 lysate for 30 min at 4°C. Reactions were incubated for 1 h at room temperature. Subsequently the beads were washed three times with buffer A and resuspended in 25 μL SDS-PAGE loading buffer.
Acknowledgments
We are grateful to Nahum Sonenberg, William Marzluff, Matthias Hentze, Peter Sarnow, Simon Morley, and Joe Lewis for the generous gift of plasmids and to Peter Stockley for the kind gift of anti-MS2 antibody. We are grateful to Marc Vidal and Marvin Wickens for the MaV99 and L40ura−yeast strains, respectively. We thank William Richardson, Diane Lawson, Xiujie Zhao, and Susan McKillop-Smith for the preparation of constructs and Ross Anderson for technical assistance with the collection of oocytes, the IMAGE consortium for clones, and Sandy Bruce for preparation of the figures. Javier Cáceres, Joel Smith, and Ian Stansfield are acknowledged for critical reading of the manuscript. B.M. is funded by Tenovus Scotland and the Wellcome Trust; S.A. is funded by a BBSRC studentship; N.K.G. is supported by a MRC CDA award; and B.G. is supported by an AICR award to N.K.G.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7281305.
REFERENCES
- Allard, P., Champigny, M.J., Skoggard, S., Erkmann, J.A., Whitfield, M.L., Marzluff, W.F., and Clarke, H.J. 2002. Stem-loop binding protein accumulates during oocyte maturation and is not cell-cycle-regulated in the early mouse embryo. J. Cell Sci. 115: 4577–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham, G.J. and Jackson, R.J. 2000. Translation initiation on pico-rnavirus RNA. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 869–900. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bergamini, G., Preiss, T., and Hentze, M.W. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6: 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, A.W., Haghighat, A., Yu, A.T., and Sonenberg, N. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392: 520–523. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Zheng, L.X., Sanchez, R., and Marzluff, W.F. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19: 3561–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee, T., Becherer, K., Chen, P.L., Yeh, S.H., Yang, Y., Kilburn, A.E., Lee, W.H., and Elledge, S.J. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 7: 555–569. [DOI] [PubMed] [Google Scholar]
- Evans, T.C., Crittenden, S.L., Kodoyianni, V., and Kimble, J. 1994. Translational control of maternal gIp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell 77: 183–194. [DOI] [PubMed] [Google Scholar]
- Gallie, D.R., Lewis, N.J., and Marzluff, W.F. 1996. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 24: 1954–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni, B. and Gray, N.K. 2004. The role of cytoplasmic poly(A)-binding proteins in regulating gene expression: A developmental perspective. Brief Funct. Genomics & Proteomics 3: 125–141. [DOI] [PubMed] [Google Scholar]
- Gradi, A., Imataka, H., Svitkin, Y.V., Rom, E., Raught, B., Morino, S., and Sonenberg, N. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, N.K., Coller, J.M., Dickson, K.S., and Wickens, M. 2000. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19: 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosset, C., Chen, C.Y., Xu, N., Sonenberg, N., Jacquemin-Sablon, H., and Shyu, A.B. 2000. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103: 29–40. [DOI] [PubMed] [Google Scholar]
- Hershey, J.W.B. and Merrick, W.C. 2000. Pathway and mechanism of initiation of protein synthesis. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 33–88. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kodama, Y., Rothman, J.H., Sugimoto, A., and Yamamoto, M. 2002. The stem-loop binding protein CDL-1 is required for chromosome condensation, progression of cell death and morphogenesis in Caenorhabditis elegans. Development 129: 187–196. [DOI] [PubMed] [Google Scholar]
- Lanzotti, D.J., Kupsco, J.M., Yang, X.C., Dominski, Z., Marzluff, W.F., and Duronio, R.J. 2004. Drosophila stem-loop binding protein intra-cellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Mol. Biol. Cell 15: 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, J., Morley, S.J., Pain, V.M., Marzluff, W.F., and Gallie, D.R. 2002. The histone 3′-terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eukaryotic initiation factor 4G (eIF4G) and eIF3. Mol. Cell. Biol. 22: 7853–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus, D.A., Evans, M.C., and Jacobson, A. 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223.1–223.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, F., Schaller, A., Eglite, S., Schümperli, D., and Müller, B. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16: 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff, W.F. and Duronio, R.J. 2002. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell. Biol. 14: 692–699. [DOI] [PubMed] [Google Scholar]
- Michel, F., Schümperli, D., and Müller, B. 2000. Specificities of Caenorhabditis elegans and human hairpin binding proteins for the first nucleotide in the histone mRNA hairpin loop. RNA 6: 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29: 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, S.J. and Pain, V.M. 1995. Hormone-induced meiotic maturation in Xenopus oocytes occurs independently of p70s6k activation and is associated with enchanced initiation factor (eIF)-4F phosphorylation and complex formation. J. Cell Sci. 108: 1751–1760. [DOI] [PubMed] [Google Scholar]
- Müller, B. and Schümperli, D. 1997. The U7 snRNP and the hairpin binding protein: Key players in histone mRNA metabolism. Semin. Cell Dev. Biol. 8: 567–576. [DOI] [PubMed] [Google Scholar]
- Müller, B., Link, J., and Smythe, C. 2000. Assembly of U7 small nuclear ribonucleoprotein particle and histone RNA 3′ processing in Xenopus egg extracts. J. Biol. Chem. 275: 24284–24293. [DOI] [PubMed] [Google Scholar]
- Ostareck, D.H., Ostareck-Lederer, A., Shatsky, I.N., and Hentze, M.W. 2001. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell 104: 281–290. [DOI] [PubMed] [Google Scholar]
- Pause, A., Méthot, N., Svitkin, Y., Merrick, W.C., and Sonenberg, N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J., and Hellen, C.U. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus mRNAs. Genes & Dev. 12: 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt, J., Crombie, C., Schümperli, D., and Müller, B. 2002. The Caenorhabditis elegans histone hairpin-binding protein is required for core histone gene expression and is essential for embryonic and postembryonic cell division. J. Cell Sci. 115: 857–866. [DOI] [PubMed] [Google Scholar]
- Richter, J.D., Wasserman, W.J., and Smith, L.D. 1982. The mechanism for increased protein synthesis during Xenopus oocyte maturation. Dev. Biol. 89: 159–167. [DOI] [PubMed] [Google Scholar]
- Sanchez, R. and Marzluff, W.F. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell Biol. 22: 7093–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, A. “Post-transcriptional regulation of histone gene expression: Functional characterisation of the RNA-binding protein HBP and of the cleavage site sequence.” Ph.D. thesis, University of Bern, Switzerland.
- Schmitt, A. and Nebreda, A.R. 2002. Signalling pathways in oocyte meiotic maturation. J. Cell Sci. 115: 2457–2459. [DOI] [PubMed] [Google Scholar]
- SenGupta, D.J., Zhang, B., Kraemer, B., Pochart, P., Fields, S., and Wickens, M. 1996. A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Natl. Acad. Sci. 93: 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, E., Santiago, C., Parker, E.D., Dominski, Z., Yang, X., Lanzotti, D.J., Ingledue, T.C., Marzluff, W.F., and Duronio, R.J. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes & Dev. 15: 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y.V., Imataka, H., Khaleghpour, K., Kahvejian, A., Liebig, H.D., and Sonenberg, N. 2001. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA 7: 1743–1752. [PMC free article] [PubMed] [Google Scholar]
- Vidal, M., Brachmann, R.K., Fattaey, A., Harlow, E., and Boeke, J.D. 1996. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein–protein and DNA–protein interactions. Proc. Natl. Acad. Sci. 93: 10315–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar, T., Gross, J.D., Wagner, G., and McCarthy, J.E. 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 11: 503–511. [DOI] [PubMed] [Google Scholar]
- Wang, Z.F., Whitfield, M.L., Ingledue 3rd, T.C., Dominski, Z., and Marzluff, W.F. 1996. The protein that binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes & Dev. 10: 3028–3040. [DOI] [PubMed] [Google Scholar]
- Wang, Z.F., Ingledue, T.C., Dominski, Z., Sanchez, R., and Marzluff, W.F. 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: Implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol. 19: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, M.L., Kaygun, H., Erkmann, J.A., Townley-Tilson, W.H., Dominski, Z., and Marzluff, W.F. 2004. SLBP is associated with histone mRNA on polyribosomes as a component of the histone mRNP. Nucleic Acids Res. 32: 4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie, G.S., Gautier, P., Lawson, D., and Gray, N.K. 2005. Embryonic poly(A)-binding protein stimulates translation in germ cells. Mol. Cell. Biol. 25: 2060–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Pestova, T.V., Hellen, C.U., and Sarnow, P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520. [DOI] [PubMed] [Google Scholar]
- Woodland, H.R. 1980. Histone synthesis during the development of Xenopus. FEBS Lett. 121: 1–10. [DOI] [PubMed] [Google Scholar]
- Zhang, B., Kraemer, B., SenGupta, D., Fields, S., and Wickens, M. 1999. Yeast three-hybrid system to detect and analyze interactions between RNA and protein. Methods Enzymol. 306: 93–113. [DOI] [PubMed] [Google Scholar]
- Zhao, X., McKillop-Smith, S., and Müller, B. 2004. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 117: 6043–6051. [DOI] [PubMed] [Google Scholar]
- Zheng, L., Dominski, Z., Yang, X.C., Elms, P., Raska, C.S., Borchers, C.H., and Marzluff, W.F. 2003. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell Biol. 23: 1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]