Abstract
Proper assembly of large protein–RNA complexes requires sequential binding of the proteins to the RNA. The signal recognition particle (SRP) is a multiprotein–RNA complex responsible for the cotranslational targeting of proteins to biological membranes. Here we describe the crystal structure at 2.6-Å resolution of the S-domain of SRP RNA from the archeon Methanococcus jannaschii. Comparison of this structure with the SRP19-bound form reveals the nature of the SRP19-induced conformational changes, which promote subsequent SRP54 attachment. These structural changes are initiated at the SRP19 binding site and transmitted through helix 6 to looped-out adenosines, which form tertiary RNA interaction with helix 8. Displacement of these adenosines enforces a conformational change of the asymmetric loop structure in helix 8. In free RNA, the three unpaired bases A195, C196, and C197 are directed toward the helical axis, whereas upon SRP19 binding the loop backbone inverts and the bases are splayed out in a conformation that resembles the SRP54-bound form. Nucleotides adjacent to the bulged nucleotides seem to be particularly important in the regulation of this loop transition. Binding of SRP19 to 7S RNA reveals an elegant mechanism of how protein-induced changes are directed through an RNA molecule and may relate to those regulating the assembly of other RNPs.
Keywords: RNP assembly, protein–RNA interactions, signal recognition particle, SRP RNA, X-ray structure
INTRODUCTION
The signal recognition particle (SRP) is a ribonucleoprotein complex (RNP) that coordinates targeting of nascent secretory and membrane proteins to the translocation apparatus in the endoplasmic reticulum or the plasma membrane of bacteria (for recent reviews, see Keenan et al. 2001; Nagai et al. 2003; Sauer-Eriksson and Hainzl 2003; Doudna and Batey 2004; Wild et al. 2004). SRP binds to the ribosome and the hydrophobic signal sequences of the nascent polypeptides as they emerge from the ribosome and targets the ribosome-nascent chain–SRP complex to the membrane by an interaction between SRP and its receptor (SR). In the presence of the translocon, the signal peptide is released into the translocon channel and SRP dissociates from SR primed for another cycle of protein targeting. SRP is found in all organisms, but its subunit composition varies considerably between the three domains of life. Mammalian SRP consists of 7S RNA (or SRP RNA) and six proteins, of which four, SRP19, SRP54, and the heterodimer SRP68/72, bind to the S domain of 7S RNA (7S.S RNA). The RNA component of archaeal SRP has a similar size and a highly conserved secondary structure with its mammalian counterpart, but in archaea only SRP19 and SRP54 homologs have been identified so far (Eichler and Moll 2001; Zwieb and Eichler 2002; Rosenblad et al. 2003). A minimal set of SRP subunits exists in eubacteria comprising a 4.5S RNA and a SRP54 homolog, also called Ffh (Poritz et al. 1990; Ribes et al. 1990).
SRP54, the only SRP protein component present in all organisms, plays the main role in the targeting event. The NG domains of SRP54 harbor GTPase activity and directly interact with the SRP receptor at the membrane, while the C-terminal M domain (SRP54M) binds to signal sequences and mediates high-affinity interaction with 7S.S RNA (Lütcke et al. 1992; Zopf et al. 1993).
The secondary structure of 7S.S RNA is approximately Y-shaped with helices 6 and 8 forming the two apical stems capped by tetraloops. Helix 8 consists of three short regular helical segments connected by a symmetric and an asymmetric internal loop. Sequence and secondary structures of the two loops show strong similarities in all organisms, and they comprise the binding site for the SRP54 M domain. The structures of 4.5 S RNA and 4.5 S RNA bound to the M domain from Escherichia coli show the symmetric loop to be rigid in structure, whereas the asymmetric loop displays considerable flexibility and undergoes a dramatic structural rearrangement upon SRP54 binding (Schmitz et al. 1999; Batey et al. 2000; Jovine et al. 2000). In the SRP54M–RNA complex structure, the asymmetric loop is held in an unusual conformation, in which the bases of the long strand are fully exposed on the outside of the RNA helix. Three of the loop bases are stacked and a universally conserved adenosine, corresponding to A195 in Methanococcus jannaschii, together with the symmetric loop forms a virtually contiguous recognition surface for the SRP54 M domain (Batey et al. 2000).
SRP biogenesis in higher eukaryotes involves sequential binding of SRP19 and SRP54 proteins to 7S RNA. In vitro reconstitution experiments of mammalian SRP showed that SRP19 must bind to 7S RNA before SRP54 can do so (Walter and Blobel 1983). Recent in vivo studies support a model in which mammalian SRP19 assembles with 7S RNA in the nucleus or nucleolus, followed by SRP54 binding to the nascent SRP in the cytoplasm (Politz et al. 2000). Archaeal SRP54, in contrast, has an intrinsic affinity for 7S RNA, although the presence of SRP19 significantly enhances SRP54 attachment (Bhuiyan et al. 2000; Diener and Wilson 2000). Chemical protection studies of human and archaeal SRP suggested that binding of SRP19 to 7S.S RNA leads to a restructuring of helices 6 and 8, causing localized changes in the asymmetric loop (Diener and Wilson 2000; Rose and Weeks 2001). Crystal structures of the S-domain RNA bound to SRP19 revealed that SRP19 binds to the tetraloops of helices 6 and 8, clamps them together, and induces extensive interactions between them (Hainzl et al. 2002; Oubridge et al. 2002). In the archeon M. jannaschii, looped-out adenosines in helix 6 seem to stabilize the asymmetric loop in an SRP54-binding competent state. In humans, helix 6 acts as the acceptor helix in an A-minor motif that forms only after SRP54M binding. The expulsion of adenosines in the short strand of the asymmetric loop and the collapse of the loop form an RNA platform in the long strand similar to that observed in E. coli (Kuglstatter et al. 2002).
To better understand the underlying mechanism by which SRP19 promotes SRP54 binding to 7S RNA, knowledge of the free 7S RNA structure is indispensable, but which to date has been unavailable. Here we present the crystal structure of free 7S.S RNA from M. jannaschii. Comparison of this structure with the SRP19-bound variant (Hainzl et al. 2002) leads us to propose a novel mechanism of long-range structural rearrangements mediated by an inducible RNA structure.
RESULTS
SRP19 facilitates SRP54 binding to 7S.S RNA in M. jannaschii
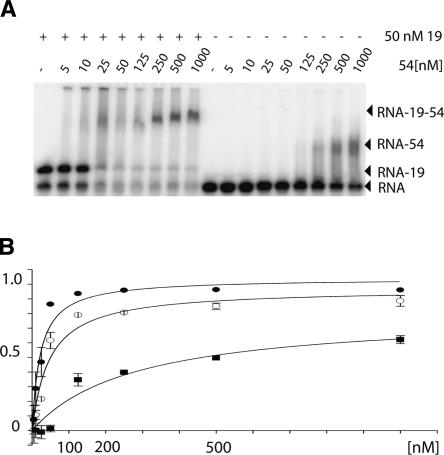
Native gel mobility shift assays were used to determine the binding constants of M. jannaschii SRP54 to 7S.S RNA. Twenty-five hundredths of a nanometer of radiolabeled 7S.S RNA was preincubated with or without 50 nM SRP19 followed by incubation with increasing amounts of SRP54. Protein–RNA complexes were separated from unbound RNA by native gel electrophoresis and binding constants were deduced from the SRP54 concentration at which 50% of RNA was bound. As measured by this technique, binding constants of the M. jannaschii SRP54 to 7S.S RNA were estimated at 25 nM and 500 nM, respectively, in the presence and absence of SRP19 (Fig. 1).
FIGURE 1.
Quantitative analysis of SRP54 binding to 7S RNA. (A) Example of the result from a typical gel mobility shift experiment. (B) Plot of the fractional saturation [7S.S RNA SRP54-bound] / [7S.S RNA total] versus [SRP54]. Mean binding values from independent measurements are shown for the 7S.S RNA–SRP54–SRP19 complex (filled circles), the 7S.S RNA–SRP54–SRP19 double mutant complex (open circles), and the 7S.S RNA–SRP54 complex (squares). The dissociation constant Kd was estimated from the SRP54 concentration at a fractional saturation of 0.5.
In a model of the M. jannaschii SRP19–SRP54M–7S.S RNA complex, side chains of His57 and Trp58 situated in loop L3 of SRP19 come into close proximity with a conserved arginine residue, Arg401, of the SRP54M domain. This suggests that in M. jannaschii direct protein–protein interactions occur between SRP19 and SRP54. To determine to what extent SRP19-induced enhancement of SRP54 attachment depends on structural changes in the RNA only, we analyzed the binding constants of SRP54 in the presence of an H57A/ W58A double mutant in the same set of experiments done for the SRP19 wild-type protein and estimated it to be 50 nM. Taken together, the binding data showed that M. jannaschii SRP54 has an intrinsic affinity for its cognate 7S.S RNA in vitro, as has been previously shown for other archaeal species (Bhuiyan et al. 2000; Diener and Wilson 2000). Protein–protein interactions contribute to the stability of the M. jannaschii SRP19–SRP54–7S.S RNA complex; however, the cooperative assembly of SRP appears to be predominantly driven by the formation of a new RNA structure, which defines the SRP54 recognition surface.
Overview of the 7S.S RNA structure
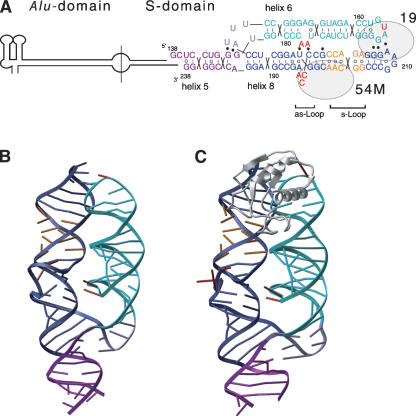
The 2.6 Å X-ray structure of a 101-nucleotide 7S.S RNA fragment of M. jannaschii comprising helices 6 and 8 and part of helix 5 was determined by molecular replacement (Table 1; Fig. 2A,B; crystal packing interactions are depicted in Fig. 3). In the RNA structure, helices 6 and 8 lie side by side. Helix 8 stacks coaxially onto helix 5 forming a continuous helical subdomain, which connects to helix 6 via a three-way junction. Helices 6 and 8 are held in close proximity by tertiary RNA interactions at two distinct sites. At the tips of helices 6 and 8, conserved bases of their respective tetraloops and closing base pairs clamp the helices together by forming a network of base and backbone interactions. At the second site, two consecutive adenosines, A176 and A177, loop out of helix 6 and form A-minor motif interactions (Doherty et al. 2001; Nissen et al. 2001) with the base pairs flanking the bulged nucleotides in the asymmetric loop in helix 8. Bases situated in the short strand of the asymmetric loop base pair continuously, while in the long strand, continuous base stacking is interrupted and the bases of the three unpaired nucleotides, A195, C196, and C197, are pointing toward the interior of the helix. The overall arrangement of the helices and many of the tertiary RNA interactions present in free M. jannaschii 7S.S RNA are similar to the ones present in the SRP19 bound complex (Fig. 2C) (Hainzl et al. 2002). A more detailed analysis, however, revealed significant structural differences, which allowed us to propose a mechanism by which SRP19 creates a new RNA structure at the SRP54 binding site.
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| Wavelength (Å) | 1.081 |
| Resolution range (Å)a | 20.0–2.6 (2.7–2.6) |
| Total number of observations | 78,570 |
| Unique reflectionsa | 15,645 (1632) |
| Completeness (%)a | 98.9 (98.6) |
| <I/σ(I)>a | 8.85 (1.78) |
| Rmerge (%)a | 10.7 (96.0) |
| Refinement | |
| aValues in parantheses are for the highest-resolution shell. | |
| Resolution range (Å) | 20.0–2.6 |
| Reflections in Rwork | 14,857 |
| Reflections in Rfree | 783 |
| Rwork (%) | 25.3 |
| Rfree (%) | 29.5 |
| Mean temperature factor (Å2) | 39.2 |
| R.m.s. deviations | |
| bond lengths (Å ) | 0.020 |
| bond angles (3) | 3.270 |
| No. of atoms | |
| RNA | 2169 |
| Water oxygens | 142 |
FIGURE 2.
Structure of 7S.S RNA. (A) Secondary structure of 7S RNA with the approximate positions of SRP19 and SRP54M indicated. The nucleotide sequence and 2D-topology diagram of the 7S.S RNA fragment used for crystallization are given. The numbering corresponds to full-length M. jannaschii 7S RNA. Lines between bases indicate WC base pairs, open circles noncanonical pairs, and filled circles tertiary RNA–RNA interactions. Nucleotides of particular interest are highlighted in red. (B) Ribbon representation of free M. jannaschii 7S.S RNA colored as in A. (C) 7S.S RNA in complex with SRP19 (PDB code 1LNG; Hainzl et al. 2002).
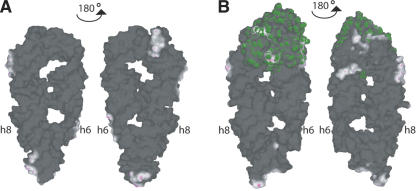
FIGURE 3.
Crystal packing interactions. The distal part of helix 6 and the asymmetric loop of helix 8 are not influenced by crystal packing interactions. Solvent-contactable surfaces of free M. jannaschii 7S.S RNA (A) and in complex with SRP19 (B) are shown. The SRP19 surface is colored in green. Colored in dark are areas of the molecules in the crystal, which are too far away from any atoms outside the surface to be bonded. White or light areas are close enough for hydrophobic van der Waals interactions. Magenta areas are close enough for hydrogen bonds. Crystals of 7S.S RNA and 7S.S RNA–SRP19 were grown in fairly similar conditions. 7S.S RNA crystallized with end-to-end stacking that was dictated by complementary interactions between the 5′-overhanging nucleotides of successive molecules. Three more regions of crystal packing contacts include nucleotides in the proximal helix 6 (U183, U184, G148, and G149), in the symmetric loop of helix 8 (A203, G204, G205, and A220), and helix 8 tetraloop (G210, A212, and G213). In the 7S.S RNA–SRP19 complex, adjacent RNA molecules are stacked head-to-end comprising noncanonical base interactions of the 5′-overhanging nucleotides with G210 and A211 of helix 8 tetraloop. The remaining lattice contacts include nucleotides in the symmetric loop of helix 8 (A203, G204, G205, and C206) and in the distal helix 6 (U159, G168, G169, and A170) as well as SRP19–SRP19 interactions.
Induced fit of helix 6
At one end of free M. jannaschii 7S RNA, the tetraloops of helices 8 (209-GGAA-212, GNRA-type, where N is any nucleotide, and R is A or G), and 6 (163-GUAG-166, GNAR-type) are in close proximity and interact via an intricate network of base-specific and phosphodiester-backbone contacts. The GGAA loop in helix 8 has the previously described compact fold (Jucker et al. 1996). The GUAG sequence of helix 6 adopts what we refer to as the “closed-loop” conformation (Fig. 4A). The second base, U164, has a 3′-endo pucker and its base points toward the minor groove roughly coplanar with the A165–G166 stacking pair of the tetraloop 3′-side. The first tetraloop base, G163, is turned into the major groove so that its 2-amino group contacts the phosphate group of G166, but no direct bond to the base of G166 is formed. The two stacked bases, A165 and G166, are wedged into the minor groove of helix 8 and make tertiary contacts with bases and backbone atoms of the helix 8 tetraloop and its closing base pair. The base of A165 forms a symmetric A–A base pair with the invariant A212 at the fourth position of the GGAA tetraloop and directly beneath, G166 forms a symmetric G–G pair with the guanine of the tetraloop-closing G213–C208 pair.
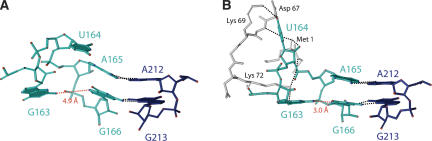
FIGURE 4.
Induced fit of the helix 6 tetraloop. The helix 6 tetraloop in (A) free 7S.S RNA and (B) the 7S.S RNA–SRP19 complex (pdb code 1LNG; Hainzl et al. 2002). The overlays are based on the nucleotides A212 and G213 in helix 8. Amino acids of SRP19 interacting with G163 and U164 are shown in light grey and hydrogen bonds between the protein and RNA are indicated. Also shown are hydrogen bonds within the A165–A212 and G166–G213 pairs. The distance between the N2 atom of G163 and the N7 atom of G166 in the two structures is shown in red.
When bound to SRP19, the tetraloops are held together not only via tertiary RNA interactions, but also by SRP19 that bridges both helices (Hainzl et al. 2002). The SRP19-binding site on helix 8 is situated in the distal minor groove. Contacts made from residues in loop L3 of SRP19 lead to no significant structural changes in the binding site. In contrast, significant structural changes are induced in the helix 6 tetraloop when SRP19 residues interact with the widened major groove at the tip of helix 6. As previously shown in the 7S.S RNA–SRP19 complex, the helix 6 tetraloop has an “open-loop” conformation, i.e., U164 is looped-out with its sugar pucker converted to the 2′-endo form (Fig. 4B; Hainzl et al. 2002). This surface-exposed position of U164 is extensively stabilized by pseudo Watson-Crick interactions with residues Met1, Asp67, and Lys69 of SRP19. Within the loop, a rotation of G163 toward the minor groove causes changes in the hydrogen bonding. The N1 hydrogen of G163 is now bonded to the O1P atom of G166, and while the N2 atom still forms a bond to the O1P atom of G166, it forms also a direct bond with the base atom N7 of G166. The base of G163 and both bases of the loop closing U162–G167 pair are stabilized by a large hydrogen-bonding network comprising the conserved lysine residues Lys19 and Lys72 of SRP19.
The induced fit of the helix 6 tetraloop upon SRP19 binding causes a marked difference in the tetraloop–tetraloop interface including a newly formed hydrogen bond between the phosphate oxygen of G167 and the 2′OH hydrogen of G215. The tertiary base contacts involving A165 and G166 are preserved in both the free- and the SRP19-bound RNA forms, but the two bases show large variations in their orientations. In the binary complex, these bases are approximately coplanar with their base partners in helix 8, while in the free RNA they approach helix 8 at a steep angle deviating from coplanarity by 30–35° in both the symmetric A–A and G–G interactions (Fig. 4A). The SRP19-induced displacement of atoms in helix 6 extends over the entire distal region and causes a substantial repositioning relative to helix 8 (Fig. 5). When not bound to SRP19, elevated temperature factors indicate a high degree of intrinsic flexibility in this part of helix 6, which becomes highly ordered upon SRP19 interaction (Fig. 6).
FIGURE 5.
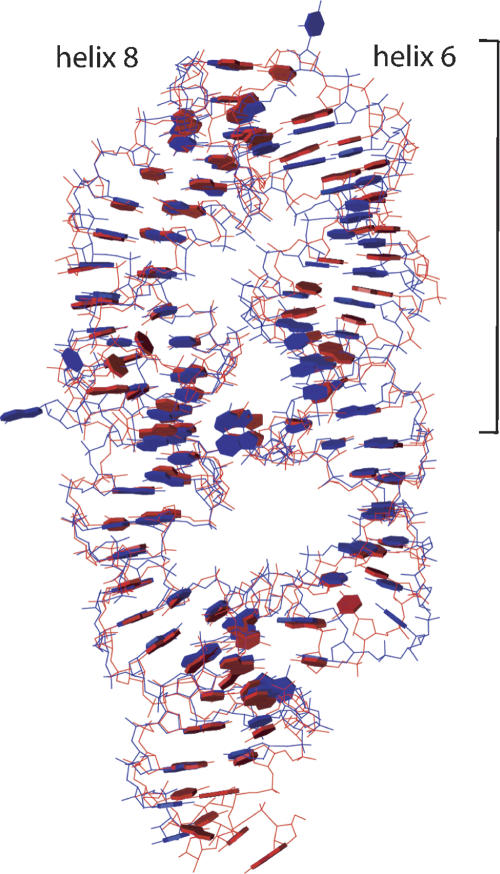
An overlay of 7S.S RNA in free form (red) and SRP19-bound form (blue) shows the displacement of the distal helix 6. The overlay is based on helix 8.
FIGURE 6.
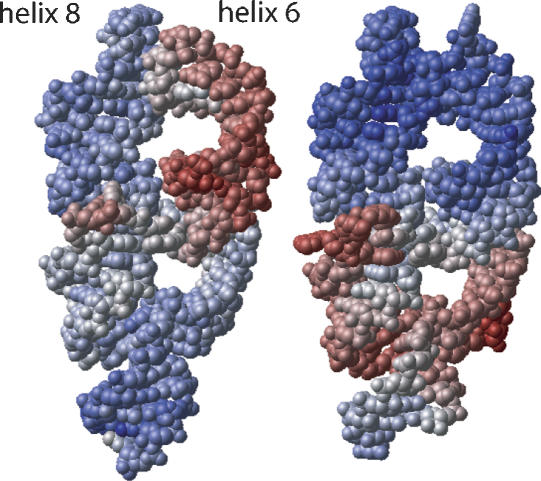
Space-filling representation of 7S.S RNA in free form (left) and SRP19-bound form (right), in the same view as in Figure 5. The atoms are colored in a continuum according to their individual temperature factors (B-factors): the smallest values are blue and the highest values are red. The minimum B-factor values are ~10 Å 2 in free RNA and ~5 Å 2 in the SRP19-bound complex. Maximum values are ~100 Å 2 in both structures. Mean B-factor values are 39.2 Å 2 and 38.4 Å 2, respectively. Elevated temperature factors of nucleotides around the three-way junction of the SRP19-bound RNA indicate flexibility, which is likely required for subsequent SRP54NG recognition.
In free and SRP19-bound 7S.S RNA, adenines A176 and A177 dock into the minor groove of helix 8 forming A-minor type interactions (Doherty et al. 2001; Nissen et al. 2001) with the base pairs flanking the 195-ACC-197 bulge. The N3 atom of A176 binds to the 2′OH group of U224 while the N1 atom of A177 binds to the 2′OH group of C198. The repositioning of the distal helix 6 upon SRP19 binding results in subtle but significantly altered positions of these two adenines relative to helix 8. The backbone of the two bulged adenosines adopts a more pronounced S-shape, tilting the furanose rings to near coplanarity with the helical bases. Consequently, the attached bases displace their N1 respective N3 imino groups by as much as 1.8 Å and enforce this displacement on the interacting riboses in helix 8 accordingly.
Backbone inversion of the asymmetric loop
SRP54 binds to the symmetric- and asymmetric-internal loops of helix 8. The distal part of helix 8, comprising the symmetric loop, is a rigid structure during SRP assembly. In contrast, the asymmetric loop is flexible and undergoes a dramatic structural rearrangement upon SRP54 binding. When bound to SRP54M, this loop is held in an unusual conformation with its backbone inverted and the bases stably stacked on the outside of the helix (Batey et al. 2000; Kuglstatter et al. 2002). Likewise, in free- and SRP19-bound 7S.S RNA of M. jannaschii the distal helix 8 with the symmetric loop maintains its conformation with overall low-temperature factors (Fig. 6). However, free-and SRP19-bound 7S.S RNA clearly deviate in the geometry of their asymmetric loop (Fig. 7A,B).
FIGURE 7.
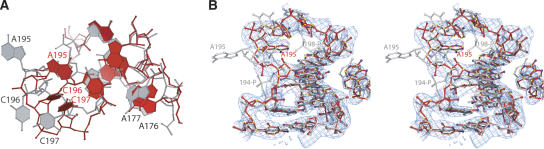
Asymmetric loop conformations in the absence and presence of SRP19. (A) The asymmetric loop structures in free RNA (red) and SRP19-bound RNA (light grey) are superimposed based on nucleotides G190–C192 and G226–C228, situated proximal to the asymmetric loop. (B) The 2|FO|–|FC| electron density map, contoured at 1.3-σ level over the finally refined coordinates, shows the quality of the electron density at the asymmetric loop in free RNA.
In free 7S.S RNA of M. jannaschii, the bulged bases 195-ACC-197 in the long strand of the asymmetric loop are well defined in the electron density; however, elevated temperature factors indicate some degree of mobility. The three unpaired bases are pointing toward the interior of the helix. A195 is coplanar with bases in the helix, whereas C196 and C197 form a stacking pair with their base planes almost perpendicular to A195. The cytosines tilt in the major groove against the helical axis so that the N4 atom of C197 is positioned 3.1 Å from the phosphate oxygen of C218 above the loop. A cross-strand A-stack motif is situated immediately below the bulge, comprising the WC G192–G226 pair followed by a sheared G193–A225 pair and a reversed Hoogsteen A194–U224 pair. G193 has a 2′-endo sugar pucker, while all other sugars have 3′-endo conformations. The 2′-endo pucker causes an extended backbone structure of G193 with a C1–C1 distance of 8.4 Å in the G193:A194 base step, and a pronounced stagger in the reversed Hoogsteen A194–U224 pair. The phosphodiester backbone in the 5′-strand aborts the A-forms trajectory at G193 and reverses the chain direction at A195 to form the overall smoothly curved A195-C197 bulge.
In SRP19-bound RNA, the displacement of C198, enforced by A177, opens up the major groove above the bulge by ~1 Å . This displacement is made possible by a small shift of nucleotides C198–C201. As a result, the distance between the N4 atom of C197 and the 2′OH atom of C218 is too far to maintain a stable hydrogen bond. Release of this bulge–helix association probably adds necessary flexibility to the bulge. The furanoses in the bulge are rotated around their C3′–C4′ bonds moving them out of the major groove. This causes a marked overwinding of the backbone, and leads to the formation of tight S-turns of the bulged backbone with the attached bases splayed out on the surface. With this backbone conformation, the rise of the A194:C198 stacking pair flanking the bulged nucleotides is too small and must be compensated for by the pucker conversion of G193 to the 3′-endo form. The backbone in the 5′-strand contracts accordingly; the C1–C1 distance in the G193:A194 base step is now 5.5 Å and has a structure more typical of A-form RNA. The cross-strand A-motif becomes internally stabilized in the SRP19–7S.S RNA complex by intensified stacking and hydrogen-bonding interactions.
DISCUSSION
Previous structural studies of the 7S.S RNA-bound complexes of SRP19 and SRP19-SRP54M suggested a mechanism by which SRP19 promotes the binding of SRP54 to 7S RNA in humans (Kuglstatter et al. 2002; Oubridge et al. 2002). However, the lack of high-resolution structures of free 7S RNA and the apparent difference in SRP19 dependency in the mammalian and archaeal systems indicated the need for further structural analysis. With the structure of free 7S.S RNA of M. jannaschii and the previously solved structure of M. jannaschii RNA in complex with SRP19, we have now obtained detailed insight into the molecular function of SRP19 in archaea.
In mammals, the lack of protection for the RNA in the absence of SRP19 suggests that in the free 7S RNA, helix 6 swings away from helix 8 (Siegel and Walter 1988; Rose and Weeks 2001). However, RNase susceptibility of 7S RNA in the archeon Archaeoglobus fulgidus indicates that the RNA has a compact fold prior to SRP19 binding (Yin et al. 2004). Our structure of free 7S.S RNA from M. jannaschii shows that helices 6 and 8 are in close contact, stabilized by tertiary RNA interactions. These interactions involve conserved bases in the tetraloop–tetraloop interface and two unpaired adenosines of helix 6 that protrude into the minor groove of the asymmetric loop of helix 8. It is probable that the unpaired bases found in many other archaeal helix 6 sequences form tertiary interactions similar to the ones found in M. jannaschii and thereby stabilize a close association of helices 6 and 8 independently of SRP19.
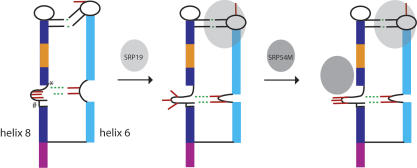
A comparison of the free- and SRP19-bound 7S.S RNA structures of M. jannaschii reveals the mechanism by which SRP19 directs restructuring of the RNA in a region roughly 40 Å away from its binding site (Fig. 8). When SRP19 binds to the tips of helix 6 and 8, only the binding site on helix 6 becomes substantially restructured. SRP19 interaction with 7S RNA consists therefore of a rigid docking of helix 8 and an induced fit of helix 6. Most prominently, the second base U164 in the helix 6 tetraloop is looped-out and bases three and four, which form tertiary interactions with helix 8, become reorientated. A recent comparison of tetraloop structures showed that different loop sequences display a large variation in the orientation of the last three bases (Correll and Swinger 2003). These loop bases are frequently used in the assembly of RNA tertiary structure and their structural variation is thought to allow for a larger repertoire of RNA receptors (Doherty et al. 2001; Nissen et al. 2001). SRP19 interaction with 7S RNA demonstrates that a reorientation of these bases plays an important role in guiding the relative position of donor to receptor RNA.
FIGURE 8.
Model of SRP assembly in M. jannaschii. Binding of SRP19 to 7S RNA causes an induced fit of the helix 6 tetraloop and a repositioning of the distal helix 6 relative to helix 8. The altered positions of A176 and A177 stabilize an asymmetric loop structure with the bases A195–C197 surface exposed. These bases become subsequently stacked upon interaction with SRP54M. Thus, the stabilization of a bulge structure, which resembles the one in the SRP54-bound form, accounts for the enhanced affinity of SRP54 for 7S RNA in the presence of SRP19. The positions of C198 (*) and the G193:A194 base step (#) situated in the long strand of the asymmetric loop are indicated. Important tertiary RNA contacts are indicated in green. The RNA helical segments and nucleotides are colored as in Figure 2.
Molecular dynamics simulations of a hairpin molecule comprising a GNRA-type tetraloop showed that the stem maintains a stable structure independent of the conformational change that take place at the loop (Sorin et al. 2002). In helix 6, however, such structural independence of the tetraloop and stem is most likely overcome by SRP19 bridging the helix 6 tetraloop and adjacent stem through direct contacts with both elements (Hainzl et al. 2002). Moreover, the distal helix 6 appears to provide enough intrinsic flexibility to transmit the structural changes initiated at the SRP19-binding site. The altered position of the adenosines A176 and A177 triggers a profound conformational change in the long strand of the asymmetric loop in helix 8. In free RNA, the three unpaired bases A195, C196, and C197 are directed toward the helical axis, whereas upon SRP19 binding the loop backbone inverts and the bases are on the outside of the RNA helix (Hainzl et al. 2002), similar to the SRP54-bound form (Batey et al. 2000). Elevated temperature factors of the bulged nucleotides A195, C196, and C197 indicate higher mobility and suggest that a small fraction of the RNA molecules in the crystal have these bases surface-exposed like in the SRP19-bound form. Presumably, these SRP19-independent loop transitions in free 7S RNA may account for the intrinsic affinity for SRP54 seen in M. jannaschii and other archaea. However, when SRP19 is bound, the fraction of surface-exposed bases in the asymmetric loop is increased and consequently SRP54 binding is enhanced, in good agreement with the biochemical data (Diener and Wilson 2000; Rose and Weeks 2001). Nucleotides surrounding the bulged bases seem to play a critical role chaperoning this loop transition. Most strikingly, G193 in the cross-strand A-stack converts its sugar pucker depending on SRP19 interaction. The backbone contraction and extension caused by the pucker switch effectively counterbalances the backbone displacement imposed by the shifted tertiary interactions and is well suited to accommodate the different bulge structures.
Based on chemical protection data of human free 7S RNA and crystal structures of human 7S.S RNAs bound to SRP19 and the SRP19-SRP54 M domain, it was suggested that the main function of SRP19 is to position helix 6 and 8 next to each other (Kuglstatter et al. 2002; Oubridge et al. 2002). Upon SRP54M binding to helix 8, the asymmetric loop collapses and two adenosines of the short strand loop out from helix 8 and form A-minor motif interactions with helix 6. However, already in free form, Methanococcus SRP RNA helices 6 and 8 are stabilized in a parallel fashion by tertiary RNA interactions. The main function of SRP19 in this organism therefore seems to be to alter the RNA interaction geometries and, consequently, cause an asymmetric loop conformation that resembles the one in the SRP54-bound form.
A general feature of protein–RNA recognition is costabilization of novel binding surfaces. To date, little is known about the detailed mechanism of how protein-induced changes are directed through the RNA molecule. The binding of SRP19 to 7S RNA reveals a remarkable mechanism, which might be similar to those regulating the assembly of other RNPs.
MATERIALS AND METHODS
Preparation of SRP RNA
The 101-nucleotide (nt) RNA fragment corresponding to nt-G138–G238 of M. jannaschii 7S RNA was transcribed in vitro and purified as described (Hainzl et al. 2002). Briefly, the 7S.S RNA was annealed in water by denaturation at 80°C followed by snap cooling on ice. The annealed RNA was purified on a Mono Q (Amersham Pharmacia Biotech) and dialyzed against a buffer containing 10 mM Tris-HCl, pH 7.5, and 5 mM MgCl2.
Gel mobility shift assays
Wild-type and mutant M. jannaschii SRP19 were prepared as described (Hainzl et al. 2002). M. jannaschii srp54 DNA comprising the NGM domains was cloned into the pNZ8048 vector and expressed in Lactococcus lactis cells. The SRP54 protein was purified by chromatography on Mono S, Heparin Sepharose, and Superdex 75 columns (Amersham Pharmacia Biotech) and stored at −80°C. For binding experiments 7S.S RNA was 5′-labeled with [α-32P]ATP using T4 polynucleotide kinase (BioLabs). Binding reactions were performed at a RNA concentration of 0.25 nM, 50 nM SRP19, and 0–2 μM SRP54 in a buffer containing 10 mM Tris-Cl, pH 7.5, 250 mM KCl, 5 mM MgCl2, 5% glycerol, 1% β-mercaptoethanol, and 1.0 mg/mL tRNA to prevent unspecific binding. Binding reactions were incubated after addition of SRP19 for 15 min at RT and for 1 h at 37°C after SRP54 addition before analysis on 4% native polyacrylamide gels (1× Tris-Acetate-EDTA, 5% glycerol) at room temperature (RT). Gels were dried and visualized using a Molecular Dynamics phosphorimager system and the supplied software (ImageQuant). The formation of SRP54 complexes was estimated by quantifying the bands of the free RNA and the SRP19–RNA complex. Binding curves show the best fit of the equation: Y = Bmax•X / (Kd + X). Bmax is the total concentration of 7S.S RNA, X is the concentration of free SRP54, and Kd is the equilibrium dissociation constant.
Structure determination and refinement
The M. jannaschii 7S.S RNA (3 mg/mL) was crystallized by hanging-drop vapor-diffusion against 32%–35% polyethylene glycol 400, 0.1 M Tris-HCl, pH 8.5, 0.2 M Na3 citrate. Crystals (0.4 × 0.4 × 0.5 mm3) were cryocooled and diffraction data were measured using synchrotron radiation at beam line I711 MAX lab (Lund, Sweden). Diffraction data from one crystal were processed using the program XDS (Kabsch 1993). The crystals belong to space group P43212 with cell parameters a = 62.11 Å , b = 62.11 Å , c = 247.52 Å , with one molecule in the asymmetric unit and with a solvent content of 67.5%.
The 7S.S RNA structure of the M. jannaschii SRP19-bound complex (PDB accession codes 1LNG; Hainzl et al. 2002) was used in molecular replacement searches with the program CNS (Brünger et al. 1998) and X-ray data from 10.0 to 4.0 Å resolution. The model was built in O (Jones et al. 1991) and refined by CNS and REFMAC (Collaborative Computational Project 1994), using the maximum likelihood residual, anisotropic scaling, bulk-solvent correction, and atomic displacement parameter refinement using the translation, libration, screw-rotation (TLS) method (Murshudov et al. 1997; Winn et al. 2001) against all data from spacings between 20 and 2.6 Å . The Rwork and Rfree for the final model are 25.3% and 29.5%, respectively. Data collection and refinement statistics are summarized in Table 1. Figure 2, B and C, Figure 4, A and B, and Figure 6 were prepared with ICM (Abagyan et al. 1994); Figures 5 and 7A with MOLMOL (Koradi et al. 1996); and Figure 7B with O and Molray (Harris and Jones 2001).
Coordinate accession codes
Coordinates and structure factors for the SRP RNA structure were deposited with the PDB entry number 1Z43.
Acknowledgments
We thank T. Bergfors for critical reading of the manuscript. This work was supported by the Swedish Research Council (K5104–1098) and European Union (CLK3-CT-2000–00082).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2080205.
REFERENCES
- Abagyan, R.A., Totrov, M.M., and Kuznetsov, D.N. 1994. ICM—A new method for protein modelling and design. Application to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 15: 488–506. [Google Scholar]
- Batey, R.T., Rambo, R.P., Lucast, L., Rha, B., and Doudna, J.A. 2000. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287: 1232–1239. [DOI] [PubMed] [Google Scholar]
- Bhuiyan, S.H., Gowda, K., Hotokezaka, H., and Zwieb, C. 2000. Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res. 28: 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project N. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D50: 760–763. [DOI] [PubMed] [Google Scholar]
- Correll, C.C. and Swinger, K. 2003. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 Å resolution. RNA 9: 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, J.L. and Wilson, C. 2000. Role of SRP19 in assembly of the Archaeoglobus fulgidus signal recognition particle. Biochemistry 39: 12862–12874. [DOI] [PubMed] [Google Scholar]
- Doherty, E.A., Batey, R.T., Masquida, B., and Doudna, J.A. 2001. A universal mode of helix packing in RNA. Nat. Struct. Biol. 8: 339–343. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A. and Batey, R.T. 2004. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 73: 539–557. [DOI] [PubMed] [Google Scholar]
- Eichler, J. and Moll, R. 2001. The signal recognition particle of Archaea. Trends Microbiol. 9: 130–136. [DOI] [PubMed] [Google Scholar]
- Hainzl, T., Huang, S., and Sauer-Eriksson, A.E. 2002. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature 417: 767–771. [DOI] [PubMed] [Google Scholar]
- Harris, M. and Jones, T.A. 2001. Molray—A web interface between O and the POV-Ray ray tracer. Acta Crystallogr. D Biol. Crystallogr. 57: 1201–1203. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Jovine, L., Hainzl, T., Oubridge, C., Scott, W.G., Li, J., Sixma, T.K., Wonacott, A., Skarzynski, T., and Nagai, K. 2000. Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA. Structure Fold. Des. 8: 527–540. [DOI] [PubMed] [Google Scholar]
- Jucker, F.M., Heus, H.A., Yip, P.F., Moors, E.H., and Pardi, A. 1996. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 264: 968–980. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26: 795–800. [Google Scholar]
- Keenan, R.J., Freymann, D.M., Stroud, R.M., and Walter, P. 2001. The signal recognition particle. Annu. Rev. Biochem. 70: 755–775. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- Kuglstatter, A., Oubridge, C., and Nagai, K. 2002. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat. Struct. Biol. 9: 740–744. [DOI] [PubMed] [Google Scholar]
- Lütcke, H., High, S., Romisch, K., Ashford, A.J., and Dobberstein, B. 1992. The methionine-rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J. 11: 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53: 240–255. [DOI] [PubMed] [Google Scholar]
- Nagai, K., Oubridge, C., Kuglstatter, A., Menichelli, E., Isel, C., and Jovine, L. 2003. Structure, function and evolution of the signal recognition particle. EMBO J. 22: 3479–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B., and Steitz, T.A. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 98: 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge, C., Kuglstatter, A., Jovine, L., and Nagai, K. 2002. Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol. Cell 9: 1251–1261. [DOI] [PubMed] [Google Scholar]
- Politz, J.C., Yarovoi, S., Kilroy, S.M., Gowda, K., Zwieb, C., and Pederson, T. 2000. Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. 97: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poritz, M.A., Bernstein, H.D., Strub, K., Zopf, D., Wilhelm, H., and Walter, P. 1990. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science 250: 1111–1117. [DOI] [PubMed] [Google Scholar]
- Ribes, V., Romisch, K., Giner, A., Dobberstein, B., and Tollervey, D. 1990. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell 63: 591–600. [DOI] [PubMed] [Google Scholar]
- Rose, M.A. and Weeks, K.M. 2001. Visualizing induced fit in early assembly of the human signal recognition particle. Nat. Struct. Biol. 8: 515–520. [DOI] [PubMed] [Google Scholar]
- Rosenblad, M.A., Gorodkin, J., Knudsen, B., Zwieb, C., and Samuelsson, T. 2003. SRPDB: Signal Recognition Particle Database. Nucleic Acids Res. 31: 363–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer-Eriksson, A.E. and Hainzl, T. 2003. S-domain assembly of the signal recognition particle. Curr. Opin. Struct. Biol. 13: 64–70. [DOI] [PubMed] [Google Scholar]
- Schmitz, U., Behrens, S., Freymann, D.M., Keenan, R.J., Lukavsky, P., Walter, P., and James, T.L. 1999. Structure of the phylogenetically most conserved domain of SRP RNA. RNA 5: 1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, V. and Walter, P. 1988. Binding sites of the 19-kDa and 68/72-kDa signal recognition particle (SRP) proteins on SRP RNA as determined in protein-RNA “footprinting.” Proc. Natl. Acad. Sci. 85: 1801–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin, E.J., Engelhardt, M.A., Herschlag, D., and Pande, V.S. 2002. RNA simulations: Probing hairpin unfolding and the dynamics of a GNRA tetraloop. J. Mol. Biol. 317: 493–506. [DOI] [PubMed] [Google Scholar]
- Walter, P. and Blobel, G. 1983. Disassembly and reconstitution of signal recognition particle. Cell 34: 525–533. [DOI] [PubMed] [Google Scholar]
- Wild, K., Halic, M., Sinning, I., and Beckmann, R. 2004. SRP meets the ribosome. Nat. Struct. Mol. Biol. 11: 1049–1053. [DOI] [PubMed] [Google Scholar]
- Winn, M.D., Isupov, M.N., and Murshudov, G.N. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57: 122–133. [DOI] [PubMed] [Google Scholar]
- Yin, J., Huang, Q., Pakhomova, O.N., Hinck, A.P., and Zwieb, C. 2004. The conserved adenosine in helix 6 of Archaeoglobus fulgidus signal recognition particle RNA initiates SRP assembly. Archaea 1: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf, D., Bernstein, H.D., and Walter, P. 1993. GTPase domain of the 54-kD subunit of the mammalian signal recognition particle is required for protein translocation but not for signal sequence binding. J. Cell Biol. 120: 1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb, C. and Eichler, J. 2002. Getting on target: The archaeal signal recognition particle. Archaea 1: 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]