Abstract
One of the most frequently encountered problems in transposon-mediated transgenesis is low transformation frequency, often resulting from difficulty in expressing from injected plasmid DNA constructs adequate levels of transposase in embryos. Capped RNA corresponding to the spliced transcript of the Minos transposable element has been synthesized in vitro and shown to be an effective source of transposase protein for Minos transposon mobilization. Transposase produced by this mRNA is shown to catalyze excision of a Minos transposon from plasmid DNA in Medfly embryos. When injected into Drosophila or Medfly embryos, transposase mRNA leads to a several-fold increase in transformation efficiencies compared with injected plasmids expressing transposase. Also, frequent mobilization of a Minos transposon from the X chromosome into autosomes was demonstrated after injections of Minos transposase mRNA into pre-blastoderm Drosophila embryos. The high rates of transposition achieved with transposase mRNA suggest that this is a powerful system for genetic applications in Drosophila and other insects.
INTRODUCTION
Transgenic technology based on mobile elements is currently applied to diverse insect species of agricultural or medical importance. Over the past few years several Type II transposable elements have been used for germ line transformation in species other than their natural hosts, such as Minos (see below), mariner in Drosophila melanogaster, Drosophila virilis and Aedes aegypti (1–3); Hermes in D.melanogaster, Ae.Aegypti and Ceratitis capitata (4–6); hobo in D.virilis (7); piggyBac in D.melanogaster, C.capitata and Bombyx mori (8–10). Moreover, purified transposases from the Tc1, Mos1 and Himar1 elements, all members of the Tc1/mariner superfamily, can catalyze transposition of the respective transposons in vitro indicating that transposition of these elements may not require other host encoded factors (11–13).
Minos, a member of the Tc1 family of elements, was isolated from Drosophila hydei (14) and is currently used for routine germ line transformation of D.melanogaster and C.capitata (15–17) and more recently for germ line transformation of Anopheles stephensi (18). Using transient mobilization assays, Minos has also been shown to be active in embryos of D.melanogaster, A.stephensi and B.mori and in cell lines of D.melanogaster, Ae.aegypti, A.gambiae and Spodoptera frugiperda (19–21).
The standard methodology (22) for transposable element mediated transformation is based on co-injecting into pre-blastoderm embryos a mixture of two plasmids: one carrying a transposon, consisting of the gene of interest flanked by the inverted terminal repeats of the element, and a helper, expressing the cognate transposase. Transformed progeny of injected animals are detected by the expression of dominant marker genes incorporated into the transposon. For species in which microinjection of DNA is easy, low transformation frequency is not a major drawback. Nevertheless, for some species the experimental procedure is laborious either due to small numbers of available eggs or to other factors. In such cases, high transformation frequencies may be critical for success.
It is possible that one of the most important parameters in controlling the transformation frequency is the amount of active transposase supplied. In all cases of non-Drosophila insect transformation, the transposase gene was placed under the control of a Drosophila promoter (usually the one of the Hsp70 gene). Constitutive levels of expression from these promoters were sufficient to produce transformants in the insects tested (15,16,18). However, it is possible that the Drosophila promoters may not be active in all species, and therefore a ‘ready to use’ transposase source such as in vitro synthesized mRNA (23) or transposase protein (24) could be more versatile.
We report here that in vitro synthesized Minos mRNA is an improved Minos transposase source, which effectively substitutes for classical helper plasmids. The transposase encoded by Minos mRNA can mobilize Minos transposons from plasmids that are co-injected into embryos and leads to high frequencies of germ line transformation in both Drosophila and Medfly. Moreover, injection of mRNA into the pre-blastoderm embryos of a transgenic Drosophila line carrying an X-linked Minos insertion leads to very frequent re-mobilizations of the Minos transposon, which suggests an alternative approach for Minos-based enhancer trap methodologies in insects.
MATERIALS AND METHODS
Plasmids, mRNA synthesis and protein production
The transposon plasmids pMiw1, genetically marked with a Drosophila white minigene, and pMihsCcw, marked with a Medfly white minigene, were used in the past for the germ line transformation of Drosophila (15) and C.capitata (16), respectively, whereas the transposon plasmid pMiLRTetR(L) was used in excision assays (19). Plasmid pNB40ILTMi (see Fig. 1A), used as template in the in vitro transcription, was constructed as follows: a PstI–SalI fragment from helper plasmid pHSS6hsILM20 (19), containing the transposase ORF fused to Hsp70 5′-UTR sequences (nucleotides 1596–1716 of the deposited sequence; AC J01104), was transferred concomitantly with a SalI–NotI fragment of ∼700 nt derived from a Drosophila PS2 cDNA clone in a PstI–NotI digested PNB40 vector (25). The PS2 cDNA fragment includes the 3′-UTR sequences of the position-specific antigen 2 (PS2) from nucleotide position 5026 of the deposited sequence (DDBJ/EMBL/GenBank accession no. M19059).
Figure 1.
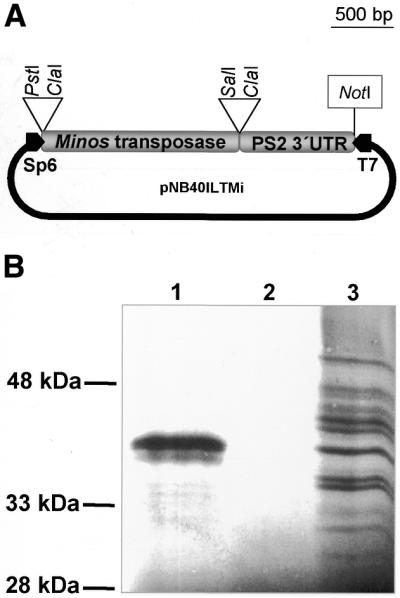
(A) pNB40ILTMi. Template for in vitro transcription. Sp6 promoter, represented as a black box arrow on the left, is used for Minos transposase mRNA synthesis. (B) SDS–PAGE and autoradiographic detection of in vitro translated peptides. Lane 1, in vitro transcribed Minos transposase mRNA is added as template in the translation reaction. The upper band corresponds to a peptide of the expected size for Minos transposase. The lower band presumably results from proteolytic cleavage of the main product. Lane 2, no RNA template is added; lane 3, control RNA (Luciferase) is added as a template. Multiple products presumably are due to degradation of the RNA template.
Minos mRNA was synthesized using the SP6 Cap-Scribe kit (Boehringer Mannheim) with 0.5 µg of pNB40ILTMi, linearized with NotI, as a DNA template. The in vitro RNA synthesis reaction was modified to achieve higher yields of transcript: additional polymerase (20 U) was added at the end of the 2 h incubation period, followed by another 2 h incubation. After completion of the synthesis, the DNA template was digested with 10 U RNAse-free DNAse I (Boehringer Mannheim) and the transcripts were precipitated by adding 5 M ammonium acetate and 2.5 vol of ethanol.
In vitro translation was performed with the Flexi Rabbit Reticulocyte System (Promega) after optimizing the reaction for the concentrations of template RNA (40 µg/ml of Minos mRNA), Mg+ (2 mM), K+ (80 mM) and DTT (2 mM). The addition of 0.4 mCi of 35S-methionine (Amersham International) per milliliter of translation reaction permits detection of the synthesized peptides by autoradiography.
Fly strains and germ line transformation
Pre-blastoderm embryos of the D.melanogaster y w67c23 recipient strain were injected (22) with 100 µg/ml of Minos transposase mRNA and 300 µg/ml of pMiw1 (15) in DEPC-treated water. For the transformation of C.capitata a previously described white strain was employed (16) and the mRNA and pMihsCcw concentrations were 100 and 300 µg/ml, respectively. In order to induce the expression of the w minigene from the Hsp70 promoter, pupae of G1 and further generations were exposed daily to a 39°C heat shock for 1 h.
The previously described line C58 (y w67c23, Mi{w+mC}17D) of D.melanogaster (15) was employed for the in vivo mobilization of a chromosomal Minos insertion.
Southern blot analysis
Genomic DNA from adult flies was digested to completion with EcoRI, subjected to agarose gel electrophoresis (∼10 µg per lane), blotted onto nitrocellulose membrane filters and hybridized with a 1 kb HhaI Minos probe as described (16).
Excision from extrachromosomal sites
Ceratitis capitata pre-blastoderm embryos were injected with a mixture of Minos transposase mRNA (100 µg/ml) and pMiLRTetR(L) (400 µg/ml) (19). DNA was prepared using a standard protocol (26) from pools of 50–60 embryos 18 h after injections and was subjected to PCR analysis (30–40 cycles, 94°C for 1 min, 55°C for 1 min, 72°C for 30 s). The oligonucleotide primers that were used anneal to sequences flanking the transposon [MiRhydei 5′-TGCTCCATTCT CTATGCT-3′ anneals to a D.hydei genomic sequence and MiLLorist 5′-CCAGCTGGCTTATCGAAA-3′ to a plasmid (Lorist) sequence].
RESULTS
In vitro transcribed RNA is translated in vivo to a functional transposase
The mRNA produced from the pNB40ILTMi vector (Fig. 1A) was tested by in vitro translation in a rabbit reticulocyte lysate. The mRNA was translated to a polypeptide of a molecular mass expected for Minos transposase (Fig. 1B). To test whether the mRNA is translated to a functional transposase in vivo, a rapid PCR-based transposon excision assay (19) was applied in Medfly embryos. The assay is based on the observation that following Minos excision the chromatid (or the donor plasmid) is frequently repaired by re-ligation inside the host cell (19,27).
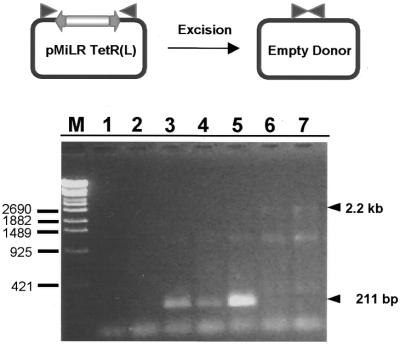
Medfly pre-blastoderm embryos were injected with either a mixture of mRNA and pMiLRTetR(L) plasmid or the plasmid alone and the DNA extracted from injected embryos was subjected to PCR analysis. Using appropriate DNA primers flanking the Minos transposon in the pMiLRTetR(L) plasmid (Fig. 2, top), a band of the expected 211 bp in size was amplified only from those embryos injected with the mRNA/pMiLRTetR(L) mixture. No such band was amplified from embryos injected only with the transposon plasmid (Fig. 2, bottom). These results clearly indicate that the injected mRNA is translated in Medfly embryos into functional transposase fully able of carrying out the excision of the Minos transposon from the co-injected plasmids.
Figure 2.
Minos transposon excision from a donor plasmid mediated by Minos transposase mRNA. Excision scheme and PCR results from Medfly embryos. Lane 1, no DNA (negative control of PCR). Lane 2, non-injected embryos. Lane 3, injected with Minos-mRNA/donor mix embryos. Lane 4, injected with Minos-mRNA/donor mix embryos. Lane 5, excision in S2 cells (positive control of the PCR). Lane 6, injected with donor plasmid. Lane 7, injected with donor plasmid. Excision results in a PCR product of 211 bp whereas the 2.2 kb band is produced by the amplification of an intact transposon sequence. The band at ∼1.2 kb is the result from ectopic priming from the donor plasmid sequences.
Increased frequencies of C.capitata germ line transformation using transposase mRNA
In previously reported experiments on Medfly germ line transformation (16,17), Minos transposase was supplied by the pHSS6hsMi2 helper plasmid, co-injected at concentrations ranging from 100 (16) to 300 µg/ml (17), together with Minos-based transposons. In the first case, a transformation frequency (expressed as the frequency of insertion events that could be distinguished by distinct eye colors) of 1.2% was obtained (five events/390 G0 adults) (16), whereas in the second case the transformation frequencies ranged from 0.7 to 3.5% (17).
To test the efficacy of in vitro transcribed mRNA as a source for transposase for transgenesis, approximately 2400 Medfly pre-blastoderm embryos of a white Medfly strain were injected with a mixture of RNA and the pMihsCcW transposon plasmid. Single-pair crosses of the Medfly are frequently unsuccessful. For this reason, the 382 adult Medflies that survived (G0 flies) were back-crossed to white Medflies in groups of 10 G0 males to 20 w virgin females (20 cages) and 20 G0 females to 10 w males (nine cages). The progeny (G1 flies) were scored for non-white eye phenotype. Table 1 shows a summary of the results and Table 2 shows the results from individual crosses. Among the 67 966 G1 flies a total of 665 phenotypic revertants of the white-eye mutation were recovered from 22 of the cages. The 284 of the non-white G1 flies derived from the same cage (C39) and were falling into two distinct eye color phenotypes. Apart from C39 where the frequency of G1 transformants was relatively high (7.4%), the rest of the G0 cages varied between 2.4 and 0.055%, with only three of the cages giving just one transformed G1 fly. Furthermore, the phenotypic variations observed in the non-white flies were extensive (from pale yellow to almost wild-type with color variegation occasionally), whereas the number of distinct phenotypes among the progeny of the same cage varied from one to eight.
Table 1. An overview of the C.capitata germ line transformation experiment.
| Injected embryos | Hatched larvae | Sex | G0 flies (crosses) | G1 progeny | Non-white G1 flies | Different non-white eye phenotypes |
|---|---|---|---|---|---|---|
| ∼2400 | 592 | Males | 202 (20) | 53 666 | 576 | 38 |
| Females | 180 (9) | 14 300 | 89 | 23 |
Table 2. Presentation of the 22 G0 crosses of C.capitata that gave at least one transformant among their progeny.
| G0 males cage number | Total G1s | Non-white G1s | Percentage of non-white G1s | Different non-white eye phenotypes | G0 females cage number | Total G1s | Non-white G1s | Percentage of non-white G1s | Different non-white eye phenotypes |
|---|---|---|---|---|---|---|---|---|---|
| C39 | 3819 | 284 | 7.4 | 2 | C52 | 2017 | 50 | 2.5 | 8 |
| C53 | 1963 | 48 | 2.4 | 5 | C44 | 2019 | 25 | 1.2 | 5 |
| C45 | 261 | 6 | 2.3 | 2 | C55 | 2434 | 6 | 0.3 | 2 |
| C47 | 2112 | 41 | 1.9 | 5 | C60 | 1068 | 2 | 0.2 | 2 |
| C46 | 3248 | 45 | 1.4 | 3 | C42 | 1802 | 3 | 0.2 | 3 |
| C51 | 2244 | 29 | 1.3 | 6 | C62 | 957 | 1 | 0.1 | 1 |
| C48 | 3623 | 43 | 1.2 | 4 | C66 | 1190 | 1 | 0.1 | 1 |
| C38 | 4459 | 30 | 0.7 | 1 | C50 | 1822 | 1 | 0.05 | 1 |
| C58 | 3493 | 15 | 0.4 | 2 | |||||
| C43 | 3711 | 13 | 0.3 | 2 | |||||
| C64 | 2716 | 9 | 0.3 | 2 | |||||
| C40 | 2718 | 8 | 0.3 | 1 | |||||
| C59 | 2275 | 3 | 0.1 | 1 | |||||
| C41 | 2432 | 2 | 0.1 | 2 |
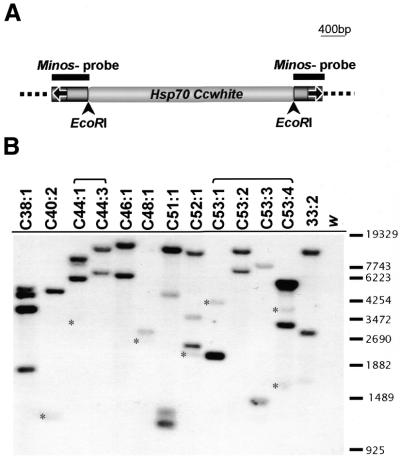
The expression of the white gene in transformants is sensitive to position effects (28) and, therefore, the phenotypic variation observed among siblings is indicative of different integration events, or multiple insertions. This was supported by molecular analysis. Twelve representative G1 males were backcrossed to w females and the DNA of their progeny was used for Southern analysis. Six of the male G1s were derived from different G0 cages whereas six (two and four each) were derived from two cages, but showed distinct phenotypes. Blotted DNA was hybridized with a probe encompassing the Minos sequences included in the MihsCcW transposon (Fig. 3A). Each integration event is expected to give two chimeric fragments, each consisting of one of the transposon ends fused to flanking genomic sequences. As shown in Figure 3B, all flies, even those deriving from the same G0 cage (C44:1, C44:3 from cage C44 and C53:1, C53:2, C53:3, C53:4 from cage C53), contain distinct insertions. Seven of the examined G2s carried a single insertion of the MihsCcw transposon, whereas the rest (C38:1, C44:1, C51:1, C52:1 and C53:4) contained additional bands indicating the presence of a second insertion. In most cases it is evident that the two chimeric fragments do not hybridize with the probe to the same extent. We assume that this is partly due to the fact that the Minos sequences included in the two chimeric fragments are unequal in size.
Figure 3.
(A) The MihsCcw transposon. Medfly white cDNA and Hsp70 (promoter and terminator) sequences are shown in white. Black arrows indicate the EcoRI restriction sites. Black bars above the map indicate the Minos sequences that were used as probe for the analysis of transformants. (B) Southern blot analysis of C.capitata transformants. DNA from G2-transformed flies and the w recipient strain was digested with EcoRI and hybridized with the Minos probe (see Materials and Methods). Brackets indicate phenotypically distinct G1 flies derived from the same G0 cage. Small asterisks mark faint bands. The lane marked as 33:2 serves as a positive control and contains DNA from the 33:2 transformed line of C.capitata (15).
These results clearly indicate that G1 flies from the same cage with different eye phenotypes represent independent integration events. Therefore, an estimation of transformation frequencies can be based on phenotypic criteria. Based on this criterion, the transformation frequency is ∼16% (61 distinct phenotypes per 382 G0 adults), but given the fact that ∼40% of the transformants that were analyzed (five out of 12) carry two insertions, the overall integration frequency in Medfly using transposase mRNA and a plasmid-based transposon may approach 22%.
Transformation frequency in D.melanogaster using Minos transposase mRNA
In previous transformation experiments of D.melanogaster, Minos transposase was supplied from the same Hsp70–Minos transposase construct either inserted in the genome of the recipient strain or in the pHSS6hsMi2 helper plasmid. In these experiments, the mean transformation frequency expressed as a percentage of fertile G0s giving transformed progeny was 2.1% (15).
In a test of mRNA as a source of transposase for Drosophila transformation, approximately 800 embryos of the y w67c23 strain were injected with a mixture of in vitro synthesized Minos RNA and the pMiw1 transposon (15). The 154 adults that survived (G0 generation) were backcrossed to w flies. Male G0 flies were crossed individually with four w females, whereas female G0s were crossed in groups of two with three w males, except for six females, which were crossed individually. Tables 3 and 4 show the results of this experiment. At least six of these flies (two males and four females) were sterile; the 38 452 progeny of the fertile G0 flies were screened for the presence of non-white phenotypes. A total of 691 G1 flies with colored eyes were recovered from 35 out of 114 different crosses. Thus, the overall transformation frequency expressed as a fraction of G0s with transformed progeny is ∼23.6% (35/148 adults). This is 10 times higher than the previously reported frequency with a DNA Minos helper (15).
Table 3. An overview of the D.melanogaster germ line transformation experiment.
| Injected embryos | Hatched larvae | Sex | G0 flies (crosses) | Fertile G0s (crosses) | G1 progeny | Non-white G1 flies | Different non-white eye phenotypes |
|---|---|---|---|---|---|---|---|
| ∼800 | 299 | Males | 78 (78) | 76 (76) | 26 981 | 483 | 30 |
| Females | 76 (41) | ? (38) | 11 471 | 208 | 30 |
Table 4. Presentation of the 35 G0 crosses of D.melanogaster that had at least one transformant among their progeny.
| G0 males vial number | Total G1s | Non-white G1s | Percentage of non-white G1s | Different non-white eye phenotypes | G0 females vial number | Total G1s | Non-white G1s | Percentage of non-white G1s | Different non-white eye phenotypes |
|---|---|---|---|---|---|---|---|---|---|
| D3M:4 | 208 | 90 | 43.3 | 3 | D1F:12 | 150 | 23 | 15.3 | 2 |
| D3M:45 | 199 | 54 | 27.1 | 2 | D3F:14 | 204 | 26 | 12.7 | 3 |
| D1M:2 | 387 | 79 | 20.4 | 1 | D3F:3 | 406 | 36 | 8.9 | 3 |
| D3M:20 | 382 | 61 | 16.0 | 3 | D3F:6 | 249 | 22 | 8.8 | 3 |
| D1M:33 | 276 | 43 | 15.6 | 2 | D1F:4 | 152 | 13 | 8.6 | 1 |
| D1M:5 | 296 | 36 | 12.2 | 3 | D1F:16 | 200 | 13 | 6.5 | 2 |
| D1M:21 | 382 | 30 | 7.9 | 1 | D3F:11 | 479 | 30 | 6.3 | 3 |
| D1M:25 | 475 | 32 | 6.7 | 1 | D1F:15 | 144 | 7 | 4.9 | 2 |
| D3M:36 | 312 | 13 | 4.2 | 2 | D1F:20 | 447 | 19 | 4.3 | 2 |
| D3M:35 | 368 | 15 | 4.1 | 2 | D1F:13 | 176 | 7 | 4.0 | 2 |
| D3M:30 | 349 | 13 | 3.7 | 2 | D3F:4 | 270 | 6 | 2.2 | 2 |
| D3M:7 | 245 | 7 | 2.9 | 1 | D1F:1 | 108 | 1 | 0.9 | 1 |
| D3M:6 | 207 | 2 | 1.0 | 1 | D3F:16a | 196 | 1 | 0.5 | 1 |
| D3M:3 | 217 | 2 | 0.9 | 1 | D3F:9 | 503 | 2 | 0.4 | 1 |
| D1M:24 | 421 | 2 | 0.5 | 1 | D1F:14 | 279 | 1 | 0.4 | 1 |
| D1M:3 | 292 | 1 | 0.3 | 1 | D3F:8 | 353 | 1 | 0.3 | 1 |
| D1M:1 | 307 | 1 | 0.3 | 1 | |||||
| D3M:1 | 379 | 1 | 0.3 | 1 | |||||
| D3M:41 | 611 | 1 | 0.2 | 1 |
aD3F:16 has only one female G0.
As expected, w+ G1 flies from the same cross often differed in eye color phenotype. Southern analysis verified that, as in Medfly, distinct phenotypes correspond to different integration events (data not shown). Based on these differences, G1 transformants were ranked into 60 distinct groups. Therefore, the overall integration rate, expressed as number of phenotypically distinct events divided by the number of fertile G0 flies, can be as high as 40.5% (60 distinct phenotypes per 148 fertile flies). Similar insertion efficiencies are calculated if only progeny of G0 males, which were crossed individually, are analyzed: nineteen out of the 76 fertile G0 males gave w+ progeny, i.e. a transformation frequency of 25%. Eleven of those G0 males gave progeny with indistinguishable phenotypes, five gave progeny falling in two distinct phenotypes and three in three phenotypes. Therefore, G0 males showed an integration rate of up to 39.5% (30 events per 76 fertile males).
Mobilization of a Minos transposon using Minos transposase mRNA
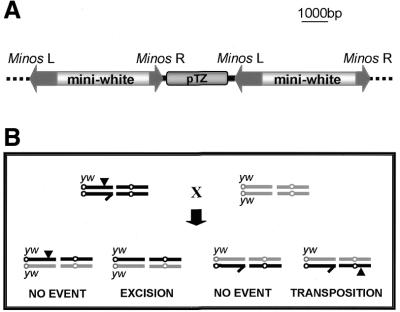
Transposase produced by a Hsp70–Minos helper construct inserted in two different chromosomal sites of D.melanogaster was previously shown to mobilize Minos transposon insertions in somatic cells and in the germ line of Drosophila (27). We have used one of these insertions, designated C58, to test the ability of Minos transposase mRNA injected into pre-blastoderm embryos to mobilize the transposon in the germ line. The C58 insertion is at cytological position 17D and comprises two Miw1 transposons arranged in tandem and separated by a full copy of the plasmid vector (15), a structure that has probably resulted from homologous recombination of two plasmids after injection, followed by transposition of the compound double transposon (Fig. 4A).
Figure 4.
(A) Structure of the C58 insertion. (B) Mating schemes for the detection of excision and transposition of the stable Minos insertion (black arrowhead) in the C58 strain. For more information see Materials and Methods.
Two different concentrations of mRNA, 50 and 100 µg/ml, were tested for their efficacy in inducing (i) excision of the compound transposon and (ii) transposition of either (or both) of the transposon copies to an autosome. Microinjected male flies carrying the C58 insertion were backcrossed to w flies and G1 progeny were screened for exceptional eye phenotypes. Loss of the compound transposon in the germ line of the injected (G0) males was detected as female progeny having reverted to the w phenotype. Transpositions to an autosome were detected as exceptional sons with non-white eye phenotypes (Fig. 4B). The results of this analysis are shown in Table 5. Partial transposon loss (e.g. excision of one of the two transposons from the compound C58 insertion) was also detectable as female progeny with eye coloration fainter that the heterozygous C58 phenotype (data not shown); these events were not analyzed further. No excisions or transpositions were detected in control crosses with non-injected flies. Approximately 16% of males injected with 100 µg/ml mRNA had at least one total excision event (w phenotype) among their female progeny and ∼32% (14 out of 44) gave at least one exceptional w+ son among their progeny. Non-white eye phenotypes, deriving from different and in two cases from the same cross, varied considerably, presumably due to strong position effects. In the previous reported re-mobilization experiments the transposition frequencies from the X chromosome to autosomes varied between 1 and 12% (27). The substitution of the helper chromosome by microinjected mRNA results in a pronounced increase in transposition efficiency (31.8%).
Table 5. Mobilization of the Minos chromosomal insertion.
| RNA (µg/ml) | Fertile G0 males | Percentage of G0 males with white-eyed daughters | Total female G1s screened | Percentage of G1 females with white eyes | Percentage of G0 males with non-white eyed sons | Total male G1s screened | Percentage of G1 males with non-white eyes |
|---|---|---|---|---|---|---|---|
| 100 | 44 | 15.9 | 8301 | 0.4 | 31.8 | 7808 | 0.9 |
| 50 | 50 | 6.0 | 9627 | 0.1 | 6.0 | 8053 | 0.04 |
When the concentration of the injected mRNA was decreased to 50 µg/ml both excision and transposition frequencies dropped to 6%, revealing an effect of mRNA concentration on the efficiency of transposition (Table 5). This result suggests that the system may not be saturated even at 100 µg/ml mRNA. Taking into account that no effect was observed on the viability and fecundity of the embryos injected with 100 µg/ml mRNA, it is possible that higher RNA concentrations may lead to even higher re-mobilization efficiencies.
DISCUSSION
For mobile elements of the Tc1/mariner family, the presence of a transposon and the corresponding transposase during early embryogenesis is considered to be necessary and sufficient for mobilization of the transposon. For several mobile elements of the Tc1/mariner superfamily, increasing amounts of transposase enhance transpositional activity but after a given concentration which depends on the respective element, the transposition is either saturated or inhibited (12,15,29).
Transformation efficiency, expressed as the percentage of injected individuals that give transformed progeny, is a crucial parameter in designing strategies for transgenesis, especially for species that are difficult to breed or have eggs that are difficult to inject. Depending on the species and the transformation system, the outcome may vary widely among insects. For example, transformation rates of 1–3.5 and 3–5% have been reported for the Medfly with Minos and with piggyBac, respectively (9,16,17), 5.5% for the malaria mosquito with Minos (18), 4 and 8% for the mosquito Ae.aegypti with mariner and Hermes, respectively (3,5), and 2% for the silkworm B.mori with piggyBac (10). Apart from the transformation procedure itself, the efficiency of transformation depends in all cases on the levels of transposase in germ line nuclei and, possibly, the size of the transposon. Our results demonstrate that for Minos, substitution of chromosomal or plasmid helpers with mRNA encoding transposase proves more efficient in promoting the mobilization of tested transposons.
The introduction of in vitro synthesized Minos transposase mRNA as a helper in D.melanogaster and C.capitata boosts transformation rates in both species. The overall transformation frequency in D.melanogaster (22.7%) makes Minos-based transformation comparable in efficiency to P element mediated transformation (30). Transformation rates in C.capitata represent minimum estimates because G0 Medflies are routinely tested in small groups under our laboratory conditions, on account of low fecundity of single pair matings. In this study, at least 14 of a total of 202 G0 male flies (20 cages) yielded transformed progeny resulting in a conservative estimate of the transformation rate of ∼6.9%. The corresponding rates from cages containing G0 females are ∼4.4% (eight of nine cages, containing a total of 180 G0 females; see Table 5).
Integration rates, expressed as the frequency of appearance of different phenotypically detectable events, are an additional criterion of efficiency of transformation and actually more informative than transformation rates, because they indicate the number of different independent lines that can be obtained from a given transformation experiment. Using Minos transposase mRNA, overall integration rates were 38.9 and 15.9% in Drosophila and the Medfly, respectively. Southern analysis showed that the different eye phenotypes present among the G1 progeny from the same cage represent independent integration events in the germ line either from different G0s carrying single insertions or from a single G0 with multiple insertions. Double insertions of the transposon in the progeny of five of the 12 G1s examined suggest that multiple integration events in the germ line of individual G0s are not rare (9,16). It is notable that some of these additional insertions behave as ‘silent’ events, i.e. there is no w+ phenotype associated with them. This is consistent with the observation that the white gene is subject to strong position effects (28), which can result in complete suppression of the phenotype (31). Our observation that out of 17 insertions only 12 were detectable through the eye phenotype, suggests that in Medfly transformation with the MihsCcw transposon, up to 30% of the insertions may have remained undetected. Consequently, transformation frequencies using the white marker are somewhat underestimated, and higher success rates can be expected with markers that may be less susceptible to position effects, such as the Green Fluorescent Protein (8,32).
The stability of genomic Minos insertions in the absence of transposase suggests that there is no interaction between Minos and other related mobile elements of the Tc1/mariner family that are present in the D.melanogaster and the Medfly genomes (15; I.Livadaras and C.Savakis, unpublished results). Conveniently, insertions can be mobilized upon expression of Minos transposase from a chromosomal position (15,27). Active Minos transposase encoded by in vitro synthesized mRNA induces mobilization of either one or both transposons present in the compound insertion in C58 transgenic flies. The apparent efficiencies of marker loss due to excision and transposition to autosomes are ∼16 and ∼32%, respectively. Because excision is a prerequisite for transposition in Type II elements, transposition rates would be expected to be at most equal to excision rates. The apparent discrepancy can be explained by a combination of the following: first, our assay only scores excision of both elements that are present in C58 as ‘excisions’, whereas excisions of only a single element of the compound transposon can lead to re-integration of the excised element. Secondly, a number of excisions remain undetectable, because the X chromosome is occasionally lost upon transposon mobilization, presumably as a result of unsuccessful repair. This effect can be detected in the progeny of transposing females, which contain unexpectedly high frequencies of XO males (27).
Mobilization of Minos transposons from chromosomal positions using a genomic source of Minos transposase showed that the frequencies of excision and transposition is partly dependent on the helper chromosome (27). The use of an RNA helper instead of a chromosomal source of transposase not only increases transposition frequencies (>2-fold compared with the most active chromosomal source) but, more importantly, also simplifies the procedure of transposon re-mobilization since there is no need for elaborate mating schemes with strains expressing transposase, marked chromosomes and other genetic tools (15,27,33). Mutagenesis screens, gene tagging and enhancer trapping are some of the techniques where high rates of mobilization are necessary. Even in Drosophila where the P element is routinely used for genetic analysis (34–37), it is unlikely that a complete sampling of the genome will be achieved using only this element; P element genomic insertion sites display a wide variation in their receptivity to insertion, and many loci in the Drosophila genome may not be accessible to P at all (38,39). Moreover, in non-Drosophila species of economic or of public health interest, RNA-induced transposon mobilization will allow genetic manipulations without the need of transgenic strains expressing transposase from marked chromosomes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Stefan Oehler for critically reading the manuscript. This work was supported by a European Union INCO grant (ERBIC18CT960100) to C.S. M.G.K. was partially supported by a Graduate Fellowship (EPEAEK) of the Greek Ministry of Education.
REFERENCES
- 1.Lidholm D.A., Lohe,A.R. and Hartl,D.L. (1993) The transposable element mariner mediates germline transformation in Drosophila melanogaster. Genetics, 134, 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lohe A.R. and Hartl,D.L. (1996) Germline transformation of Drosophila virilis with the transposable element mariner. Genetics, 143, 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coates C.J., Jasinskiene,N., Miyashiro,L. and James,A.A. (1998) Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc. Natl Acad. Sci. USA, 95, 3748–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brochta D.A., Warren,W.D., Saville,K.J. and Atkinson,P.W. (1996) Hermes, a functional non-Drosophilid insect gene vector from Musca domestica. Genetics, 142, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jasinskiene N., Coates,C.J., Benedict,M.Q., Cornel,A.J., Rafferty,C.S., James,A.A. and Collins,F.H. (1998) Stable transformation of the yellow fever mosquito, Aedes aegypti, with the Hermes element from the housefly. Proc. Natl Acad. Sci. USA, 95, 3743–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michel K., Stamenova,A., Pinkerton,A.C., Franz,G., Robinson,A.S., Gariou-Papalexiou,A., Zacharopoulou,A., O’Brochta,D.A. and Atkinson,P.W. (2001) Hermes-mediated germ-line transformation of the Mediterranean fruit fly Ceratitis capitata. Insect Mol. Biol., 10, 155–162. [DOI] [PubMed] [Google Scholar]
- 7.Lozovskaya E.R., Nurminsky,D.I., Hartl,D.L. and Sullivan,D.T. (1996) Germline transformation of Drosophila virilis mediated by the transposable element hobo. Genetics, 142, 173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handler A.M. and Harrell Ii,R.A. (1999) Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol. Biol., 8, 449–457. [DOI] [PubMed] [Google Scholar]
- 9.Handler A.M., McCombs,S.D., Fraser,M.J. and Saul,S.H. (1998) The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proc. Natl Acad. Sci. USA, 95, 7520–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura T., Thibert,C., Royer,C., Kanda,T., Abraham,E., Kamba,M., Komoto,N., Thomas,J.L., Mauchamp,B., Chavancy,G., Shirk,P., Fraser,M., Prudhomme,J.C. and Couble,P. (2000) Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat. Biotechnol., 18, 81–84. [DOI] [PubMed] [Google Scholar]
- 11.Vos J.C., De Baere,I. and Plasterk,R.H. (1996) Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev., 10, 755–761. [DOI] [PubMed] [Google Scholar]
- 12.Tosi L.R. and Beverley,S.M. (2000) cis and trans factors affecting Mos1 mariner evolution and transposition in vitro, and its potential for functional genomics. Nucleic Acids Res., 28, 784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lampe D.J., Churchill,M.E. and Robertson,H.M. (1996) A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J., 15, 5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 14.Franz G. and Savakis,C. (1991) Minos, a new transposable element from Drosophila hydei, is a member of the Tc1-like family of transposons. Nucleic Acids Res., 19, 6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loukeris T.G., Arca,B., Livadaras,I., Dialektaki,G. and Savakis,C. (1995) Introduction of the transposable element Minos into the germ line of Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 92, 9485–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loukeris T.G., Livadaras,I., Arca,B., Zabalou,S. and Savakis,C. (1995) Gene transfer into the medfly, Ceratitis capitata, with a Drosophila hydei transposable element. Science, 270, 2002–2005. [DOI] [PubMed] [Google Scholar]
- 17.Christophides G.K., Livadaras,I., Savakis,C. and Komitopoulou,K. (2000) Two medfly promoters that have originated by recent gene duplication drive distinct sex, tissue and temporal expression patterns. Genetics, 156, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catteruccia F., Nolan,T., Loukeris,T.G., Blass,C., Savakis,C., Kafatos,F.C. and Crisanti,A. (2000) Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature, 405, 959–962. [DOI] [PubMed] [Google Scholar]
- 19.Klinakis A.G., Loukeris,T.G., Pavlopoulos,A. and Savakis,C. (2000) Mobility assays confirm the broad host-range activity of the Minos transposable element and validate new transformation tools. Insect Mol. Biol., 9, 269–275. [DOI] [PubMed] [Google Scholar]
- 20.Catteruccia F., Nolan,T., Blass,C., Muller,H.M., Crisanti,A., Kafatos,F.C. and Loukeris,T.G. (2000) Toward Anopheles transformation: Minos element activity in anopheline cells and embryos. Proc. Natl Acad. Sci. USA, 97, 2157–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu K., Kamba,M., Sonobe,H., Kanda,T., Klinakis,A.G., Savakis,C. and Tamura,T. (2000) Extrachromosomal transposition of the transposable element Minos occurs in embryos of the silkworm Bombyx mori. Insect Mol. Biol., 9, 277–281. [DOI] [PubMed] [Google Scholar]
- 22.Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- 23.Raz E., van Luenen,H.G., Schaerringer,B., Plasterk,R.H. and Driever,W. (1998) Transposition of the nematode Caenorhabditis elegans Tc3 element in the zebrafish Danio rerio. Curr. Biol., 8, 82–88. [DOI] [PubMed] [Google Scholar]
- 24.Coates C.J., Jasinskiene,N., Morgan,D., Tosi,L.R., Beverley,S.M. and James,A.A. (2000) Purified mariner (Mos1) transposase catalyzes the integration of marked elements into the germ-line of the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol., 30, 1003–1008. [DOI] [PubMed] [Google Scholar]
- 25.Brown N.H. and Kafatos,F.C. (1988) Functional cDNA libraries from Drosophila embryos. J. Mol. Biol., 203, 425–437. [DOI] [PubMed] [Google Scholar]
- 26.Holmes D.S. and Bonner,J. (1973) Preparation, molecular weight, base composition and secondary structure of giant nuclear ribonucleic acid. Biochemistry, 12, 2330–2338. [DOI] [PubMed] [Google Scholar]
- 27.Arca B., Zabalou,S., Loukeris,T.G. and Savakis,C. (1997) Mobilization of a Minos transposon in Drosophila melanogaster chromosomes and chromatid repair by heteroduplex formation. Genetics, 145, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazelrigg T., Levis,R. and Rubin,G.M. (1984) Transformation of white locus DNA in Drosophila: dosage compensation, zeste interaction, and position effects. Cell, 36, 469–481. [DOI] [PubMed] [Google Scholar]
- 29.Lampe D.J., Grant,T.E. and Robertson,H.M. (1998) Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics, 149, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Brochta D.A. and Handler,A.M. (1988) Mobility of P elements in Drosophilids and Nondrosophilids. Proc. Natl Acad. Sci. USA, 85, 6052–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirrotta V., Steller,H. and Bozzetti,M.P. (1985) Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J., 4, 3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berghammer A.J., Klingler,M. and Wimmer,E.A. (1999) A universal marker for transgenic insects. Nature, 402, 370–371. [DOI] [PubMed] [Google Scholar]
- 33.Robertson H.M., Preston,C.R., Phillis,R.W., Johnson-Schlitz,D.M., Benz,W.K. and Engels,W.R. (1988) A stable genomic source of P element transposase in Drosophila melanogaster. Genetics, 118, 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooley L., Kelley,R. and Spradling,A. (1988) Insertional mutagenesis of the Drosophila genome with single P elements. Science, 239, 1121–1128. [DOI] [PubMed] [Google Scholar]
- 35.Wilson C., Pearson,R.K., Bellen,H.J., O’Kane,C.J., Grossniklaus,U. and Gehring,W.J. (1989) P-element-mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev., 3, 1301–1313. [DOI] [PubMed] [Google Scholar]
- 36.Bellen H.J., O’Kane,C.J., Wilson,C., Grossniklaus,U., Pearson,R.K. and Gehring,W.J. (1989) P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev., 3, 1288–1300. [DOI] [PubMed] [Google Scholar]
- 37.O’Kane C.J. and Gehring,W.J. (1987) Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl Acad. Sci. USA, 84, 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spradling A.C., Stern,D., Beaton,A., Rhem,E.J., Laverty,T., Mozden,N., Misra,S. and Rubin,G.M. (1999) The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics, 153, 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith D., Wohlgemuth,J., Calvi,B.R., Franklin,I. and Gelbart,W.M. (1993) hobo enhancer trapping mutagenesis in Drosophila reveals an insertion specificity different from P elements. Genetics, 135, 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]