Abstract
We identified the first archaeal tRNA ribose 2′-O-methylase, aTrm56, belonging to the Cluster of Orthologous Groups (COG) 1303 that contains archaeal genes only. The corresponding protein exhibits a SPOUT S-adenosylmethionine (AdoMet)-dependent methyltransferase domain found in bacterial and yeast G18 tRNA 2′-O-methylases (SpoU, Trm3). We cloned the Pyrococcus abyssi PAB1040 gene belonging to this COG, expressed and purified the corresponding protein, and showed that in vitro, it specifically catalyzes the AdoMet-dependent 2′-O-ribose methylation of C at position 56 in tRNA transcripts. This tRNA methylation is present only in archaea, and the gene for this enzyme is present in all the archaeal genomes sequenced up to now, except in the crenarchaeon Pyrobaculum aerophilum. In this archaea, the C56 2′-O-methylation is provided by a C/D sRNP. Our work is the first demonstration that, within the same kingdom, two different mechanisms are used to modify the same nucleoside in tRNAs.
Keywords: archaea, tRNA, ribose 2′-O-methylation, sRNP, Cm56
INTRODUCTION
All types of cellular RNAs contain modified nucleosides, and among them, tRNAs present the most extensive nucleoside modifications (Bjork 1995). To date, at least 99 different modified nucleosides have been identified in the many naturally occurring RNAs analyzed (see http://medstat.med.utah.edu/RNAmods/). The specific contributions that individual modifications make to tRNA function are generally thought to involve maintenance of the structural integrity of the tRNA when the modifications are located outside the anticodon region, while modifications within and around the anticodon are proposed to play a direct role in increasing translational efficiency and/or fidelity, and in specific cases, to serve as recognition determinants for aminoacyl-tRNA synthetases (Sampson and Uhlenbeck 1988; Agris 1996; Davis 1998; Grosjean and Benne 1998). The type of chemical alteration of a nucleoside, as well as the pattern of tRNA modification depend on the origin of the tRNA molecule (Sprinzl et al. 1998). Many of the tRNA modifications are highly conserved across the three phylogenetic domains (Grosjean et al. 1995; Auffinger and Westhof 1998; McCloskey and Crain 1998; Motorin and Grosjean 1998; Sprinzl et al. 1998). The archaeal tRNAs sequenced so far (assembled at http://www.staff.uni-bayreuth.de/~btc914/search/index.html) contain common modifications such as D, Ψ, Cm, Gm, m1G, m7G, m1A and m6A similar to the modifications of the eubacterial and eukaryotic tRNAs. In addition, they contain modifications that are specific to archaea, either in their position and/or nature, such as m1Ψ54, archaeosin15, Cm56 (Auffinger and Westhof 1998) and several hypermodified nucleosides. Some characteristic nucleosides are modified both in the base and by methylation of the 2′ hydroxyl group in ribose, like m2,2G, at position 10 or 26 (Armengaud et al. 2004) that is 2′-O-methylated only in the thermophile archaea (Noon et al. 2003). These extra methylations may help to stabilize the tridimensional structure of tRNAs at the high temperature (100°C) or high salt life conditions of these extremophiles (Agris et al. 1973; Kowalak et al. 1994; McCloskey et al. 2001).
Little is known about archaeal tRNA modification machineries, as no genes encoding specific archaeal tRNA 2′-O-methylases, or pseudo-uridylases had been identified until now. A variety of tRNA modification activities were demonstrated to be present in archaeal cell extracts (Grosjean et al. 1995; Constantinesco et al. 1999b), and only a few genes encoding mainly tRNA base methylases have been identified (Constantinesco et al. 1999a; Bai et al. 2000; Bjork et al. 2001; Pierrel et al. 2003; Armengaud et al. 2004; Roovers et al. 2004).
Ribose-methylation is one of the most frequent RNA modifications. In eukaryotes and bacteria, tRNA ribose methylation is catalyzed exclusively by site-specific methyltransferase (Mtase) proteins that recognize both sequence and structure within the unmodified precursor tRNA substrate (Droogmans et al. 1986; Hori et al. 1998; Cavaille et al. 1999; Pintard et al. 2002). These site-specific RNA 2′-O-ribose methyltransferase enzymes belong to two structural classes (class I and IV) among the five AdoMet dependent Mtase structural families described so far (Schubert et al. 2003) for review. The class I Mtase family regroups a tremendous variety of Mtases, including the great majority of RNA Mtases. These enzymes all belong to the superclass of Rossman-fold enzymes (Lo Conte et al. 2000). They share a common structure with seven-stranded β-sheets where the last one is inserted in an antiparallel conformation between the fifth and the sixth β-sheet. AdoMet is bound in a deep cleft in the center of the molecule. The class IV structure consists solely of the SPOUT (SpoU/ TrmD) family of RNA Mtases (Anantharaman et al. 2002). Their shared structural characteristic is a deep trefoil knot in a core structure including six-stranded parallel β-sheets, of which the first three form half of a Rossman fold, flanked by seven α-helices. AdoMet is bound above the “knot” in a bent conformation (Michel et al. 2002; Nureki et al. 2002, 2004; Elkins et al. 2003; Lim et al. 2003; Zarembinski et al. 2003).
In many instances, archaea use C/D box guide RNPs to mediate the 2′-O-methylation in tRNAs (as is the case for eukaryotic and archaeal rRNAs). The C/D box guide particle includes a box C/D guide RNA, characterized by a bipartite structure with conserved C (RUGAUGA) and D (CUGA) box motifs near the 5′ and 3′ ends, and related C′ and D′ box motifs near the center of the molecule (Cavaille and Bachellerie 1998; Kiss 2001). Previous biochemical studies have shown that box C and D are the sites of RNP assembly (Szewczak et al. 2002; Watkins et al. 2002; Bortolin et al. 2003; Decatur and Fournier 2003; Tran et al. 2003). Archaeal C/D RNPs contain three proteins with a symmetric distribution: the archaeal fibrillarin (aFib), the ribosomal protein L7Ae and Nop5p (Kuhn et al. 2002; Omer et al. 2002; Watkins et al. 2002; Aittaleb et al. 2003; Bortolin et al. 2003; Ziesche et al. 2004). The box C/D sequences generate a characteristic RNA secondary structure motif, the kink-turn (K-turn) which is directly bound by L7Ae and, together with aFib and Nop5p, form the core RNP complex. Guide sequence complementarity to an RNA target located upstream of box D establish a site of 2′-O-methylation within the paired region, located five nucleotides upstream of box D (Cavaille and Bachellerie 1996; Kiss-Laszlo et al. 1996). Methylation is catalyzed by aFib, a class I Mtase. In a few archaeal species, several C/D sRNAs were predicted to target more than 23 tRNA positions for 2′-O-methylation (Clouet d’Orval et al. 2001; Dennis et al. 2001; Tang et al. 2002; Omer et al. 2003). Archaeal C/D RNP-guided tRNA 2′-O-methylations were recently demonstrated in vitro, on the Haloferax volcanii pre-tRNATrp for C34 and U39 2′-O-methylation (Clouet d’Orval et al. 2001; Bortolin et al. 2003) and on the S. solfataricus tRNAGln for G18 methylation in an unique tRNA (Ziesche et al. 2004). The absence of archaeal tRNA 2′-O-Mtases up until now raised the following question: Are all the tRNA 2′-O-methylations in archaea carried out by C/D box guide sRNPs?
The tRNA Gm18 modification is found in eubacteria and eukaryotes and in plant organella (McCloskey and Crain 1998; Rozenski et al. 1999). So far it has not been found in sequenced archaeal tRNAs (Auffinger and Westhof 1998), even if a unique S. solfataricus tRNAGln was recently proposed to be methylated at this position by a C/D sRNP (Ziesche et al. 2004). However, a recent search to define methyl-transferase superfamilies by conserved structural elements highlighted a relationship between the eukaryotic and bacterial 2′-O-methylases specific to position G18 (Trm3 and SpoU, respectively) and several uncharacterized archaeal proteins, suggesting their common evolutionary origin. All of these proteins belong to the SPOUT superfamilies (Anantharaman et al. 2002), and constitute the class IV of Mtases. We postulated that these archaeal enzymes may be responsible for another species-specific tRNA methylation. We focused our attention on one of these proteins, belonging to a Cluster of Orthologous Groups (COG; Tatusov et al. 2003) exclusively found in archaea, encoded by the PAB 1040 gene. We cloned the gene, expressed and purified the corresponding P. abyssi protein, and demonstrated that in vitro, it specifically catalyzes the AdoMet-dependent formation of the Cm at position 56 (Cm56) in tRNA transcripts. The gene for this enzyme (named “aTrm56” for archaeal tRNA-methylase for position 56) is the first archaeal tRNA 2′-O-methylase identified, and is present in all the archaeal genomes sequenced up to now, except in Pyrobaculum aerophilum. In this crenarchaeon, we show that the C56 2′-O-methylase activity is provided by a C/D guide sRNP.
RESULTS
COG 1303 proteins are only found in Archaea
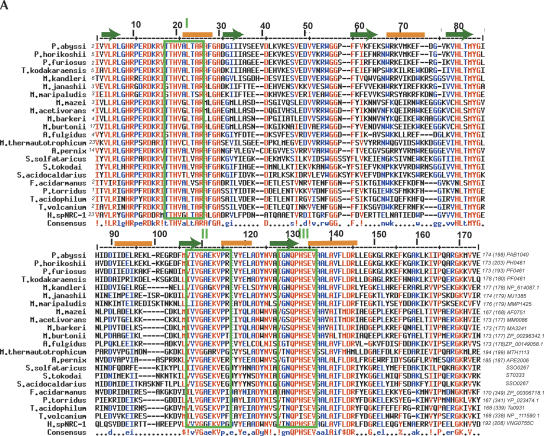
A search of the nonredundant protein database from NCBI using the PSI-BLAST tool (Altschul et al. 1997) for the P. abyssi 1040 gene revealed a set of 22 full-length closely related archaeal proteins (hits with E-values below 10−26 and sequence identity above 47%, Fig. 1A) which constitute the COG 1303 (http://www.ncbi.nlm.nih.gov/COG/; Tatusov et al. 2003). No close homologs (E values worse than the threshold of 0.01) from eukaryota or bacteria were found in the second or third PSI-BLAST iteration. Of the totally or partially completed 23 archaeal genomes analyzed, only a single crenarchaeon, Pyrobaculum aerophilum, does not possess a gene belonging to this family. Nevertheless, the gene is present in the four other sequenced crenarchaeon genomes (Fig. 1C) (Aeropyrum pernix, Sulfolobus solfataricus, Sulfolobus acidocaldarius and Sulfolobus tokodaï; Fig. 1A). All of these proteins present conserved domains related to the three motifs of the SpoU Mtase family (Koonin and Rudd 1993; Gustafsson et al. 1996; Fig. 1A), in which only a few amino-acids are conserved as compared to the eubacterial and eukaryal enzymes of the same family (Fig. 1B). Additional conserved domains are specific to the COG 1303. The secondary structure predicted by the JPRED software (Cuff et al. 1998) proposes six β-sheets and five α-helices characteristic of the catalytic fold with a deep trefoil knot of the SPOUT family (or class IV) Mtases (Fig. 1A; for review, see Schubert et al. 2003).
FIGURE 1.
(A) Sequence alignments of the COG 1303 proteins from the archaea genomes available in the public databases (http://www.ncbi.nlm.nih.gov/) by PSI-BLAST searches (Altschul et al. 1997). Multiple alignments were performed by Multalin (http://prodes.toulouse.inra.fr/multalin/multalin.html). Secondary structure elements of the P. abyssi sequence encoded by the PAB1040 gene were predicted using the JPRED software (Cuff et al. 1998) (http://barton.ebi.ac.uk/servers/jpred.html). Putative β-sheets and α-helices are represented by green arrows, and orange stretches respectively. Amino acid numbering refers to the P. abyssi sequence. Motifs I, II, and III corresponding to conserved domains of the SpoU-like enzymes (Gustafsson et al. 1996) are framed in green. Organism names and first and last amino acids are indicated for each sequence, as well as the total number (in brackets) of amino acids, and the gene bank identifiers. Strictly conserved residues are in red, and residues with > 70% identity or strictly of the same type are in blue. The symbols in the consensus are: !, IV; $, LM; %, FY; h, hydrophobic; -, gap;. , any amino acid. (B) Multiple sequence alignment of SPOUT proteins. Amino acid sequences of five eubacterial and eukaryotal proteins are compared. The consensus determined from the SpoU-like enzymes (Gustafsson et al. 1996) are reported on the first line. In the alignment, color, and symbol codes for residues conservation is as in (A). Reported in the last line are the consensus sequences of the COG 1303 (part A). (C) Phylogenetic tree of the archaeal genomes derived from 16S rRNA genes (Marck and Grosjean 2002).
The P. abyssi PAB1040 ORF encodes a Mtase (aTrm56) involved in the methylation of the invariant C56 in tRNAs
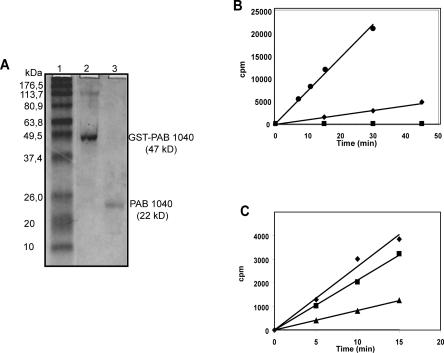
The cloned P. abyssi PAB1040 ORF expressed as a N-terminal GST-tagged protein in E. coli was purified to quasi-homogeneity and showed an apparent molecular mass of 22 kD when subjected to SDS-PAGE (Fig. 2A). To test the RNA Mtase activity, the purified recombinant protein was assayed for its ability to catalyze transfer of the labeled methyl group from [3H]AdoMet to different RNA substrates. No methyl incorporation was observed with the C/D guide sR47 or HIV-TAR RNAs (Fig. 2B). However, the bulk yeast tRNAs showed specific kinetics of 3H methyl incorporation at 37°C and 50°C (Fig. 2B). Similar methylation kinetics were obtained with unfractionated E. coli tRNAs as substrates (data not shown). In vitro transcribed E. coli tRNALeu2, yeast tRNAPhe, and P. abyssi tRNALeu/CAA tRNAs were methylated with significant initial rates at 50°C (Fig. 2C). In conclusion, tRNAs are suitable substrates of this enzyme. The Km measured for one of its substrates (yeast tRNAPhe) at 50°C in the presence of 6 μM AdoMet is 80 nM for a Vmax of 0.4 μM methyl/mg.h, similar to values found for another class IV Mtase, the Thermus thermophilus TrmH (Watanabe et al. 2005). The RNA Mtase activity is thermostable (80% of the initial activity persists after incubating 2 h at 95°C, data not shown).
FIGURE 2.
(A) SDS-PAGE analysis of the purified recombinant P. abyssi Mtase. SDS-PAGE was performed on a 10% polyacrylamide gel, in Laemmli buffer. The gel was stained with Coomassie Brillant blue. Lane 1, MW marker (Sigma); Lane 2, GST-tagged protein eluted from the Glutathione-Sepharose beads (1/30 of the protein retained on Glutathion-Sepharose beads); Lane 3, purified P. abyssi Mtase (0.5 μg) after Thrombin cleavage. (B) The purified P. abyssi protein is a tRNA methyl-transferase. Kinetics of 3H(Met) incorporation in 1 μM total yeast tRNAs, incubated at 37°C, (♦), or 50°C, (•), or in 1 μM TAR or SR47 RNAs, incubated at 50°C (•) in the presence of (3H)AdoMet and purified enzyme. (C) Individual specific tRNA methylation by the P. abyssi enzyme. Kinetics of 3H(Met) incorporation in individual tRNA transcripts, yeast tRNAPhe (♦), E. coli tRNALeu2 (▪), P. abyssi tRNALeu/CAA (▴), 0.5μM each incubated at 50°C in the same conditions as in (B).
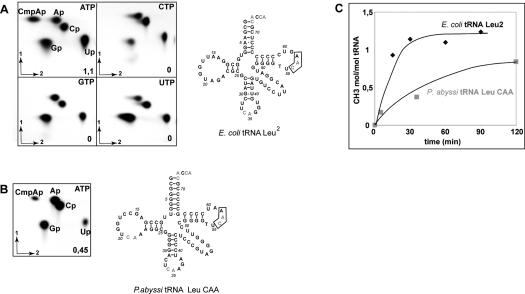
To characterize the substrate specificity of the enzyme, we predicted that the product of the PAB1040 gene could catalyze the 2′-O-methylation of C56 since this conserved position is invariably methylated in archaeal tRNAs and is never modified in bacterial or eukaryotic tRNAs (see http://www.staff.uni-bayreuth.de/~btc914/search/index.html) (Auffinger and Westhof 1998; Sprinzl et al. 1998). To identify the position methylated by the archaeal enzyme, the E. coli tRNALeu2 transcript was uniformly labeled either with [α-32P]-ATP, GTP, CTP, or UTP for a nearest neighbors analysis (Grosjean and Benne 1998). It was then subjected to methylation in the presence of AdoMet and the purified P. abyssi protein, followed by RNase T2 digestion to generate 3′ monophosphate nucleosides. 2′-O-methylation inhibits RNase T2 hydrolysis, and generates a dinucleoside labeled by the adjacent 3′[32P]phosphate. The TLC analysis shows a single spot corresponding to the dinucleoside CmpAp for the E. coli tRNALeu2 transcript labeled with [α-32P]-ATP (Fig. 3A). In the tRNALeu2 sequence, the only CmpAp that can be labeled by the neighbor [α-32P]- A is Cm56A57 located in the T-loop. Quantification of the relative amount of [32P] in the different radioactive spots on the TLC plates revealed that ~1 mol of Cm56 is formed per mole of E. coli tRNALeu2 after 1 h incubation at 50°C (Fig. 3A). Similar results were obtained with the yeast tRNAPhe (not shown), whereas 0.45-mol Cm56/mol tRNA were formed with P. abyssi tRNALeuCAA (Fig. 3B) Kinetic analyses performed with E. coli tRNALeu2 and P. abyssi tRNALeu/CAA revealed that the E. coli substrate is more suitable than that of P. abyssi at this temperature (Fig. 3C). Altogether, these data demonstrate that in vitro, the P. abyssi protein catalyzes the 2′-O-methylation of C at position 56 in the T-loop of tRNAs. Indeed, we named this enzyme “aTrm56” for “archaeal tRNA methylase for position 56.”
FIGURE 3.
Position 56 is specifically methylated by the P. abyssi enzyme. T7 in vitro transcripts of E. coli wild type tRNALeu2 and P. abyssi tRNALeu/CAA, uniformly labeled by incorporation of [α-32P]-NTP were incubated for 1 h at 50°C in the presence of the purified P. abyssi protein. (A) 2D TLC in system A+C of RNase T2 digests obtained from E. coli tRNALeu2 labeled with ATP, CTP, GTP, or UTP as indicated. (B) 2D TLC of RNase T2 digest of P. abyssi tRNALeuCAA labeled with ATP. The yield of methylation is indicated on each chromatogram. A cloverleaf representation of the corresponding tRNAs analyzed is shown on the right with CA dinucleosides marked in gray. (C) Comparative kinetic analysis of aTrm56-catalyzed methylation of two tRNA substrates, E. coli tRNALeu2, and P. abyssi tRNALeu/CAA. tRNAs uniformly labeled by incorporation of [α-32P]-ATP were incubated for different period of times as described above. The yield of methylation was determined by quantitative analysis of 2D-TLC.
The aTrm56 enzymatic activity absolutely requires a C at position 56, and recognizes the T-loop structure
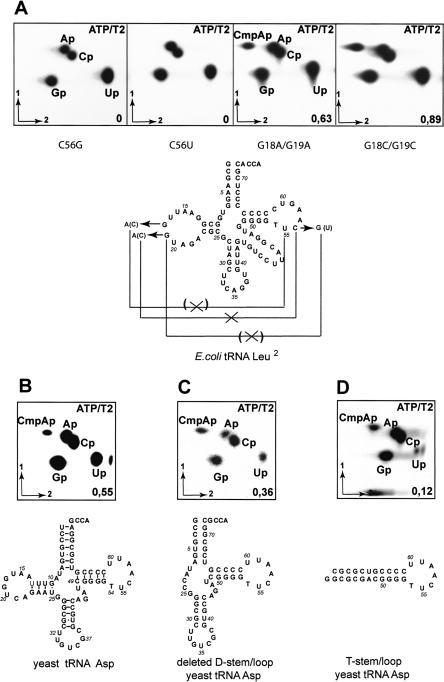
To determine the substrate specificity of the aTrm56 protein, mutants of the E. coli tRNALeu2 transcript were tested. Mutations of C at position 56 either by U or G in tRNA abolish methyl incorporation (Fig. 4A, left), implying a strict substrate requirement with a C at position 56. We then tested the influence of the 3D architecture of the tRNA on aTrm56 activity. Two double mutants of the E. coli tRNALeu2 transcript, G18A/G19A and G18C/G19C in which the G18-Ψ55, and G19-C56 interactions responsible for the compact 3D L-architecture of the tRNA are disrupted, are efficiently methylated at 37°C (Fig. 4A, right). We tested a tRNA mini-substrate corresponding to the yeast tRNAAsp transcript missing the D stem-loop (Fig. 4C), and found that it was methylated in vitro at a slightly lower efficiency than the wild-type tRNAAsp (0.36-mol Cm56 formed per mol tRNA against 0.55 for the wild-type tRNAAsp, Fig. 4B). A second minisubstrate consisting of the T-loop of tRNAAsp closed by a synthetic stem (“T stem-loop or TSL”, Fig. 4D) was also tested. This TSL minisubstrate is still methylated in vitro even though the reaction is four- to fivefold less efficient than with the wild type substrate. We conclude that the complete 3D architecture of the tRNA substrate is not required for aTrm56 catalysis. A T stem-loop structure alone is thus a minimal substrate recognized by aTrm56.
FIGURE 4.
aTrm56 specifically recognizes T-loop structure. Mutant T7 transcripts of E. coli tRNALeu2 (A) or yeast tRNAAsp (B,C,D) uniformly labeled by incorporation of [α-32P]-ATP were incubated for 1 h at 50°C with aTrm56 and treated as described in Figure 3. The yield of methylation is indicated and cloverleaf representations of each mutant show the effect of the mutation on the D-T loops’ interactions. the T and D loops of tRNA have no effect on the efficiency
A C/D sRNA in Pyrobaculum aerophilum responsible for C56 methylation
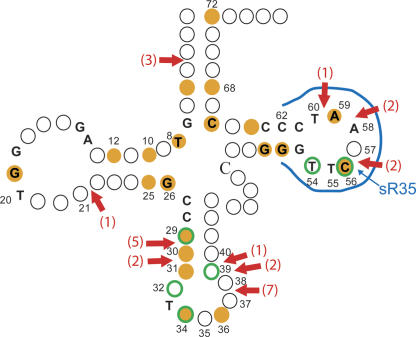
A gene orthologous to PAB 1040 is present in all the sequenced archaeal genomes except P. aerophilum (Fig. 1A). We hypothesized that in this archaeal species, a C/D guide machinery could catalyze the C56 methylation. A previous global analysis of C/D box guide sRNAs in Archaeal genomes revealed that many of them harbor antisense elements matching a total of 23 different sites in tRNAs (Dennis et al. 2001). Interestingly, the genome of the crenarchaeon P. aerophilum appears to have the largest number of genes for sRNAs targeting tRNA positions. It implies that this archaeon may have developed a greater number of RNA guides involved in tRNA modification. To fully identify these sRNAs, we performed a systematic computer search for the potential sRNA/tRNA duplexes. We used the 51 intergenic regions annotated as being sRNAs in the genomic sequence of P. aerophilum available at the NCBI site, and the archaeal tDNA database (Marck and Grosjean 2003) to assign a tRNA target to each of the P. aerophilum sRNAs (assembled in http//atg.toulouse.inra.fr/~rnaworld/PAE.html). Dennis et al. (2001) found 16 different sRNAs, targeting 13 tRNA positions (Table 1, second column; Fig. 5), and we found in addition the sRNAs (sR39, sR23, sR32, sR41, sR37, sR49, sR46, sR48) with antisense elements complementary to tRNAs, for methylation at positions 5, 8, 10, 25, 29, 31, 34, 64, and 66 (Table 1, third column). For the sRNAs already identified as targeting tRNAs (Dennis et al. 2001), we found additional potential tRNA targets (Table 1, third column), as sR35 (Table 1, cf. second and third columns). SR35 presents a 11–-13 nt antisense element upstream of its D box, complementary to the T-loop sequence of all 46 tRNAs of P. aerophilum, where the fifth position in the RNA/RNA duplex corresponds to C56 (Fig. 6A). A Northern blot hybridization performed on total RNAs extracted from P. aerophilum confirmed the presence of this sRNA (Fig. 6B). Thus sR35 may contribute to the C56 methylation of its tRNAs, in the absence of a gene coding an aTrm56 homolog.
TABLE 1.
sRNAs from Pyrobaculum aerophilum showing complementarities with tRNAs
| tRNA targeted position | sRNAs [Dennis et al. 2001] | sRNAs [this study] |
| 5 | sR39: D (6tRNAs) | |
| 8 | sR23: D (4tRNAs) | |
| 10 | sR32: D (5tRNAs) | |
| sR34: D (8tRNAs) | sR34: D (9tRNAs) | |
| sR48: D (2tRNAs) | sR48: D (15tRNAs) | |
| sR5: D’ (6tRNAs) | sR5: D’ (4tRNAs) | |
| 12 | sR46: D (2tRNAs) | sR46: D (3tRNAs) |
| 19 | sR11: D (3tRNAs) | |
| 25 | sR41: D (10tRNAs) | |
| 26 | sR51: D’ (3tRNAs) | sR51: D’ (1tRNA) |
| 29 | sR39: D’ (9tRNAs) | |
| sR37: D (5tRNAs) | ||
| 30 | sR49: D (1tRNA) | |
| 31 | sR49: D’ (1tRNA) | |
| 34 | sR27: D (1tRNA) | sR27: D (1tRNA) |
| sR46: D’ (3tRNAs) | ||
| sR51: D (1tRNA) | sR51: D (1tRNA) | |
| sR45: D (2tRNAs) | sR45: D (2tRNAs) | |
| sR26: D (3tRNAs) | sR26: D (1tRNA) | |
| 36 | sR36: D (1tRNA) | |
| 51 | sR29: D’ (3tRNAs) | sR29: D’ (10tRNAs) |
| 52 | sR37: D’ (11tRNAs) | sR37: D’ (5tRNAs) |
| 56 | sR35: D (14tRNAs) | sR35: D (32tRNAs) |
| 59 | sR4: D’ (1tRNA) | sR4: D’ (14tRNAs) |
| 64 | sR48: D’ (6tRNAs) | |
| 66 | sR23: D (8tRNAs) | |
| 68 | sR39: D’ (1tRNA) | sR39: D’ (2tRNAs) |
| 72 | sR37: D (3tRNAs) | sR37: D (4tRNAs) |
Potential tRNA modified positions are indicated in the first column from the left; sRNAs targeting these positions found by Dennis et al. (2001) are in the second column, and additional sRNAs found in this study are in the third column with either new or more tRNA position targets. In each case, the nature of antisense element targeting the indicated tRNA position (upstream D or D’ box) is given. For example, (in bold) sR39 targets three tRNAs for position 68 with the antisense element upstream its box D’, and six tRNAs for position 5 with the antisense element upstream its box D. The particular case of sR35 (in bold) was previously annotated as targeting 14 tRNAs (Dennis et al. 2001) and was found to target 46 tRNAs (http://atg.toulouse.inra.fr/~rnaworld/PAE.html).
FIGURE 5.
Predicted sRNAs targeting tRNA sites in Pyrobaculum aerophilum. The structure of a tRNA is depicted in the standard cloverleaf configuration. Conserved positions among the 46 P. aerophilum tRNAs are indicated by letters, positions known to be methylated in archaeal tRNAs are circled in green (Auffinger and Westhof 1998); positions predicted to be methylated based on C/D sRNA guide sequence complementarities predicted by Dennis et al. (2001) and from this study are in orange (see Table 1). Intron insertion sites are indicated by red arrows, and the number of tDNAs bearing these introns is indicated (Marck and Grosjean 2003). The hybridization site for the antisense element of sR35 targeting position C56 is shown (in blue).
FIGURE 6.
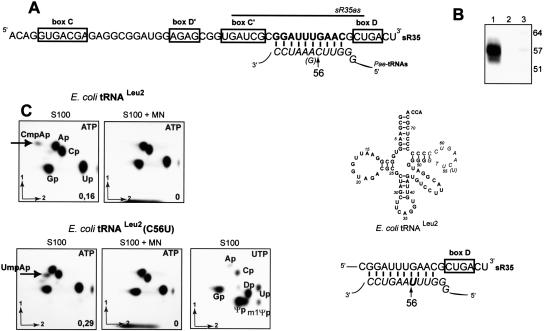
The P. aerophilum sR35 C/D sRNA. (A) Sequence of the C/D sRNA sR35, the consensus boxes C, D, C′ D′ are framed. The antisense element (AE) complementary to all of the P. aerophilum tRNA T-loop sequences (in italic) is indicated in bold letters. Position 56 within the complementary sequence is indicated, as well as the oligonucleotide SR35as used for Northern hybridization. (B) Detection of the predicted C/D sRNA sR35 by Northern hybridization: Aliquots of total cellular RNA before (lane 1) and after micrococcal nuclease (MN) treatment (lane 2) were analyzed. Hybridization was performed using [32P]-labeled oligonucleotide SR35as (partA); lane 3, size marker. (C) T7 transcript of E. coli wild type tRNALeu2 (which TΨC stem-loop can duplex with the antisense element of C/D guide sR35) uniformly labeled by [α-32P]-ATP incorporation was incubated for 2 h at 70°C, in the presence of P. aerophilum S100 extract, not treated (S100), or treated (S100 + MN) with micrococcal nuclease (MN). Analysis was performed as in Figure 3. The labeled nucleotide is indicated, as well as the yield of methylation. T7 transcript of E. coli mutant tRNALeu2 C56U was tested in similar conditions: in the presence of P. aerophilum S100 extract, not treated (S100), or MN-treated (S100 + MN). T7 transcript of wild type E. coli tRNALeu2 uniformly labeled by incorporation of [α-32P]-UTP was treated as described above. The U modifications performed by the P. aerophilum extract are indicated.
We looked in vitro for evidence of C56 2′-O-ribose methylation activity in a P. aerophilum S100 extract using tRNA as a substrate. The E. coli tRNALeu2 transcript (whose TΨC stem-loop is able to form a duplex with the antisense element of C/D guide sR35) was used as a substrate. A labeled [α-32P]ATP transcript incubated with S100 extract and analyzed by T2 digestion and 2D-TLC gives rise to the dinucleoside Cmp56Ap57 (Fig. 6C; S100). To prove that Cm56 depends on C/D guide machinary present in the S100 extract, we treated the S100 extract with micrococcal nuclease (MN), prior to reaction. Degradation of sR35 was verified by Northern blot (Fig. 6B). Under these conditions, the Cmp56Ap57 spot was not observed (Fig. 6C, S100 + MN), suggesting that the machinery for Cm56 is RNA-dependent in P. aerophilum. A similar experiment performed with the P abyssi, tRNALeuCAA, gave the same results (data not shown). The MN treatment was also controlled to be without effect on the P. abyssi aTrm56 enzymatic activity (data not shown). An E. coli tRNALeu2 C56U mutant transcript (whose RNA duplex with the antisense element of C/D guide sR35 involves a U-G pair at the fifth position instead of a C-G) was labeled by [α-32P]ATP incorporation and incubated with the P. aerophilum extract as described above. A spot corresponding to dinucleoside Ump56Ap57 was observed that was no more present when the extract was MN treated (Fig. 6D; S100 & S100 +MN). We showed previously that this E. coli tRNALeu2 C56U mutant was not methylated by the purified aTrm56 enzyme (Fig. 4A), supporting our identification of an sRNA-dependent 2′-O-methylation activity targeting tRNA position 56 in P. aerophilum extracts. U56 does not exist in the archaeal tRNA genes identified until now; all possess a C at this position (Marck and Grosjean 2002).
In conclusion, our observations strongly suggest that in the absence of an enzyme orthologous to the P. abyssi aTrm56, the 2′-O-methylation of tRNAs at their 56th position is performed by a sRNP-guided system in the crenarchaeon P. aerophilum. The presence of other tRNA modification activities in the P. aerophilum extract is evident in the TLC of Figure 6C (S100[U]), were an E. coli tRNALeu2 transcript labeled with [α-32P]UTP was treated as described above: Spots corresponding to Ψ, m1Ψ, and D are clearly identified.
DISCUSSION
aTrm56, a new archaeal class IV RNA Mtase
In this study, we identify a new RNA Mtase, aTrm56, which is the first archaeal tRNA ribose 2′-O-Mtase to be characterized. This protein which catalyzes the archaeal-specific methylation of the conserved position C56 in the TΨC loop of tRNAs, belongs to the class IV Mtases (Schubert et al. 2003) that groups the enzymes of the SPOUT (“SpoU/TrmD”) family (Anantharaman et al. 2002). Only three tRNA ribose Mtases have been included in this class: the bacterial SpoU, its yeast homolog Trm3, catalyzing G18 2′-O-ribose methylation, and the aTrm56 Mtase. It is noticeable that both positions involved in the tertiary interaction which anchors the acceptor side to the anticodon side in the L-shape of tRNA (G18 in eukarya and bacteria and C 56 in archaeal tRNAs (Limbach et al. 1994; Bjork 1995; McCloskey and Crain 1998; Sprinzl et al. 1998) are methylated by the only tRNA ribose methyl-transferases belonging to structural class IV, suggesting a common origin for these enzymes.
aTrm56 belongs to the COG 1303 that contains 22 archaeal proteins (Fig. 1A), that all present partially conserved domains characteristic of the SpoU-like Mtases (Fig. 1A,B; Gustafsson et al. 1996). In addition, all of the proteins contain conserved domains in the N-terminal region and around positions 56, 80, 123, and 170 (P. abyssi protein numbering, Fig. 1A) that may contribute to substrate specificity. In the SPOUT enzymes crystallized to date (Ahn et al. 2003; Elkins et al. 2003; Lim et al. 2003; Nureki et al. 2004), conserved residues belonging to motifs I, II, and III were demonstrated to form the characteristic knotted catalytic pocket for AdoMet binding. These amino-acid residues are only partially conserved in aTrm56, indicating that a similar trefoil knot structure providing different interactions with AdoMet is formed. It will be of great interest to identify whether similar interactions occur in crystals of the aTrm56 protein, as these differences may reflect the distinct evolution of archaeal enzymes from a common ancestor of the SPOUT enzymes.
C56 2′-O-methylation involves conformational change of the tRNA
aTrm56 in vitro methylates C56 in various tRNA substrates. This modification at the 56th position of tRNAs, (together with the modifications at positions 54, 55, 18, or 15), is located in the “corner” formed by the tertiary interactions of the canonical L-shape, where bases are less accessible to modification enzymes. Despite the fact that aTrm56 was demonstrated to be highly thermostable, we fixed the incubation temperature with heterologous tRNA transcripts at 50°C, taking into account the melting of unmodified T7 transcripts of yeast and E. coli tRNAs which begins at 55–60°C (Sampson and Uhlenbeck 1988; Sampson et al. 1990). In these conditions we observed a rapid and complete methylation of the E. coli tRNALeu2, when the P. abyssi tRNALeuCAA (Tm> 60°C) was slowly and incompletely modified (Fig. 3C). This suggests that at least a partial melting of the substrate, leading to the disruption of the tertiary structure of the tRNA, is required for efficient methylation. We observed also that mutations disrupting the association of of Cm56 formation in vitro, and that the TSL minihelix is modified by aTrm56 at the correct site, assuming that the 11bp T-stem of the TSL minihelix persists at a temperature of 50°C. This implies that the global or tertiary structure of tRNA is not the feature recognized by aTrm56 and that the T-loop is the minimal recognition element. Formation of other modifications in the T-loop also occured efficiently in vitro on TSL minihelices incubated with an extract of Haloferax volcanii (Constantinesco et al. 1999b), and with yeast or E.coli enzymes catalyzing tRNAs T54 and Ψ55 (Becker et al. 1997; Gu et al. 1998), strongly suggesting that enzymes modifying T-loop positions require a conformational change of the tRNA that disrupts the D-T loops’ interactions. Indeed, in the complex formed between T-loop and E. coli TruB (catalyzing the formation of Ψ55) U55 is flipped out, and highly accessible for modification (Hoang and Ferre-D’Amare 2001). Modifications occuring on the D-loop also require a conformational change of the tRNA L-shape (Ishitani et al. 2003; Nureki et al. 2004), as observed when the enzyme archaeosin tRNA transglycosylase (ArcTGT) catalyzes the formation of the precursor for archaeosin at position 15 of archaeal tRNAs (Bai et al. 2000). When complexed with its tRNA substrate, ArcTGT stabilizes a tRNA λ-form in which the D-stem and the canonical core are completely disrupted, releasing the T-loop for access to other modification enzymes. In this conformation of the tRNA, the T-loop conserves its local structure, particularly the reverse Hoogsteen pair U54:A58 (Romby et al. 1987) demonstrated to be an important identity determinant for substrate recognition by the enzymes modifying positions 54 and 55 (Becker et al. 1997; Gu et al. 1998; Hoang and Ferre-D’Amare 2001). We suggest that tRNA modification by aTrm56 involves an L to λ shape transition that might be rate-limiting for the reaction. Binding of ArcTGT may initiate this transition.
A large number of C/D guide RNAs with antisense elements to tRNAs in Pyrobaculum aerophilum
The crenarchaeota thermoproteale Pyrobaculum aerophilum is the only archaea (among the 23 archaea genomes totally or partially sequenced that we could analyze) that has no gene coding an aTrm56 Mtase. Instead, it contains the sR35 sRNA that potentially targets the TΨC loop of all the 46 tRNAs for the Cm56 modification (Fig. 5; Table 1; http://atg.toulouse.inra.fr/~rnaworld/PAE.html). C/D guide-RNA directed tRNA 2′-O-methylation is specific to archaea, as there is no evidence to date that any eukaryotic tRNA modifications are C/D guide-RNA directed. Nine archaeal species contain sRNAs for which one or both D and D′ box guide regions exhibit complementarities to tRNAs (Omer et al. 2000; Clouet d’Orval et al. 2001; Dennis et al. 2001; this study). Among the archaea studied (listed at http://rna.wustl.edu/snoRNAdb/; Dennis et al. 2001; this study), P. aerophilum displays the highest number of sRNA targeting tRNA positions: altogether, 19 sRNAs for 20 tRNA positions (Table 1; Fig. 5). Another pecularity of P. aerophilum is the presence of a high number of introns in its tRNA genes (26 introns in 23 iso-acceptors) and a high diversity of locations of these introns (Marck and Grosjean 2003; Fig. 5). Usually, most tRNA introns are found between positions 37–38 of the anticodon. In the crenarchaeon P. aerophilum, 19 of the 26 introns are located elsewhere. For their excision, the archaeal splicing machinery does not require the three dimensional structure of the pre-tRNA and cleaves introns at variable positions in pre-tRNAs within a bulge–helix– bulge motif such as BHB (Arn and Abelson 1996; Trotta et al. 1997; Li et al. 1998). Five P. aerophilum pre-tRNAs contain an intron in their T-loop (Fig. 5; Marck and Grosjean 2003). Since sR35 base-pairs to the mature sequence of this loop in the 46 tRNAs, guided-2′-O-methylation should arise after intron excision in these tRNAs. Thus, sR35 may have a chaperone function, its base-pairing with the mature tRNA directing the efficient local folding of the tRNA after intron processing and preventing the formation of nonproductive structures that may block or delay the folding process. This mechanism is different from that of the intronic C/D guide in the euryarchaeal pretRNA Trp which is predicted to act at positions 34 and 39 of the unspliced tRNA precursor, through a pair of guide RNA duplexes spanning exon–intron junctions, directing transition from a splicing-competent structure to a methylation-competent RNA conformer (Clouet d’Orval et al. 2001, 2005; Singh et al. 2004). Considering the high number of introns in P. aerophilum tRNA genes, the numerous C/D sRNAs targeting 20 tRNA positions (Fig. 5; Table 1) may be the result of a selective advantage provided by the chaperone function of the duplexes formed between guide and tRNAs to insure a correct folding of the tRNAs after intron splicing. P. aerophilum growing at an optimal temperature of 100°C may also require numerous tRNA positions 2′-O-methylated for stabilization of the tRNA structures (Davis 1998). Nevertheless, other archaea (like Pyrococcus) growing at similar temperatures, do not have as many sRNAs.
Site-specific Mtase or C/D sRNP: Which came first?
In this study, we demonstrated the invention of two independent systems to catalyze the same tRNA modification within the archaeal kingdom: 1) a class IV enzyme, present in most of the archaeal species, and 2) a sRNP (the methylation being catalyzed by aFib, a class I Mtase) present in one crenarchaeal species among the five crenarchaea genomes sequenced to date. In the crenarchaea P. aerophilum, the sRNP is the only system able to catalyze 2′-O-methylation at position C56. To determine if this feature may be specific to the crenarchaeota, as no other sequenced genomes of Pyrobaculum are yet available, we carefully checked if a sR35-like sRNP could be present in the other sequenced crenarchaea. The database of archaeal sRNA (http://lowelab.ucsc.edu/snoRNAdb/#archaea) was examined for selecting known candidate sRNA able to target C56 position in tRNAs. sRNA candidates of A. pernix, S. solfataricus, and S. acidocaldarius were examined. No such sRNA was found in the genomes concerned. Alltogether, these results do not exclude that both systems (site-specific enzyme and sRNP) may be present in a same archaea. At this step, the knowledge of both C56 methylation systems represents a new case of two independent inventions of enzymes to catalyze the same reaction.
The existence of two modification systems acting on a same site was reported once for snRNA modification in eukaryotes. Pseudouridylation at position 34 of the U2 snRNA is modified by an H/ACA snoRNP in X. laevis, and by the Pus7p enzyme in S. cerevisiae, (Behm-Ansmant et al. 2003). Another situation was recently described in yeast, concerning the 2′-O-methylation of positions U2921 and G2922 in the large ribosomal subunit (LSU). In a wildtype context, Um2921 is RNA-guided by a C/D snoRNA (snR52), whereas the neighbor Gm2922 is enzymatically modified by Sbp1 at a later stage of 60S maturation. In a context of snR52 depletion, however, Um2921 may also be performed by Sbp1 described as a “region” specific enzyme (Lapeyre and Purushothaman 2004).
From an evolutionary point of view, it is hard to say which RNA modification system was created first, as the ancestral RNP complex was recently proposed to have a very ancient origin derived from the primitive translation apparatus (Tran et al. 2004). The absence of RNA-guided nucleotide-modification complexes in bacteria does not necessarily indicate that RNPs arose in the archaeal and eukaryotic branches after divergence from bacteria. Both systems—the RNP-guided nucleotide modification system and the site-specific enzymes system—may have evolved in parallel. As a consequence, in most cases, the enzyme responsible for the RNA 2′-O-methylation is a Class I methylase (site-specific Mtase or the C/D RNP Mtase of the Fibrillarin family), and in some cases (aTrm56), the site specific Mtase is a class IV Mtase that could have derived from the class I. Altogether, these observations show how diverse are the modification systems in the three domains of life. In the case of archaea, the 2′-O-methylation at the conserved tRNA position C56 is the first example of the coexistence of two different mechanisms to modify the same nucleoside in tRNAs within the same kingdom.
MATERIALS AND METHODS
Unless otherwise noted, all techniques for cloning and manipulating nucleic acids were performed according to standard protocols (Sambrook 1989). The following oligodeoxynucleotides were used:
PAB1040(5′): TACCGGATCCATAGTCGTCCTTAGACTAGG;
PAB1040(3′): ACGCAAGCTTTTAGCTAGGTAACTGGCCGT;
TSL(3′): TAATACGACTCACTAT;
TSL(5′): GmCGCGACGGGGAATTGAACCCCGTCGCGCCATA GTGAGTCGTATTA;
EC Leu2(5′): TAATACGACTCACTATAGCCGAGGTGGTGGAA TTGGT;
EC Leu2(3′): TGGTACCGAGGACGGGACTT;
EC Leu2 U56(3′): TGGTACCGAGGACGGGACTTAAACCCGTA AG;
EC Leu2G56(3′): TGGTACCGAGGACGGGACTTGAACCCGTAAG;
EC Leu2 C18/C19(3′): TAATACGACTCACTATAGCCGAGGT GGTGGAATTCCTAGACACG;
EC Leu2 A18/A19(3′): TAATACGACTCACTATAGCCGAGGTG GTGGAATTAATAGACACG; and
SR35as: AGCGTTCAAATCCGCGATC.
Strains, media, growth conditions, and general procedures
Transfer RNAs from both yeast and E. coli were obtained from Sigma.
P. abyssi genomic DNA was a gift from B. Clouet d’Orval (LBME Toulouse), P. aerophilum cells were from I. Schroeder (UCLA, Los Angeles).
Amplification and cloning of the PAB 1040 gene
The PAB 1040 ORF was amplified from P. abyssi genomic DNA using Pfu DNA polymerase (Promega) and the primer pairs PAB1040(5′) and PAB1040(3′) containing the EcoRI (5′) and XhoI (3′) restriction sites (Table 1). After cloning the PCR product in PKS vector (BluescriptII KS+, Stratagene), the EcoRI/XhoI fragment was extracted and cloned in pGEX 2TK-p vector, generating MH1x plasmid. Plasmid pGEX 2TK-p was derived from pGEX 2TK plasmid (Amersham) by insertion of the sequence: AATTCTAGACTCCATGGGTCGACTCGAGCTCAAGCTT at the EcoRI site allowing bacterial expression of the N-terminal-GST tagged P. abyssi protein by IPTG induction of the tac promoter.
Expression and purification of the GST–P. abyssi recombinant protein
The GST-P. abyssi protein was expressed in E. coli strain BL21 (DE3). Freshly transformed cells were grown to an OD600 of 0.5–0.6, at 37°C in 200 mL Luria broth with antibiotics. IPTG (isopropyl-β-D-thiogalactopyranoside; 1mM) was then added to induce recombinant protein expression (37°C, overnight). Cells were harvested, rinced with 1× PBS, and the pellet resuspended in 4 mL of the same buffer complemented with protease inhibitors (complete Mini, EDTA-free protease inhibitor, Roche) prior to lysis by sonication. The lysate was cleared by centrifugation (14,000g for 20 min) and applied to a GST-sepharose column (Pharmacia Biotech) previously equilibrated with 1× PBS. The column was washed with the same buffer, and the absorbed material was directly cleaved by incubation of the beads with thrombin (10 u) during 2 h at room temperature. The eluted fractions were then pooled, heated 10 min at 70°C, centrifuged 10 min at 15,000g, to eliminate the denatured GST and thrombin, and concentrated by centrifugation on YM-10 centricon. Aliquots of the purification steps were analyzed by 10% SDS-PAGE.
Northern hybridization
The SR35as oligonucleotide was end-labeled with [γ-32P]ATP and T4 polynucleotide kinase for 1 h at 37°C, then purified by filtration through Sephadex G25. Total RNA from P. aerophilum was isolated by TRIZOL (Invitrogen). Aliquots (24 μg) were separated on 6% denaturing polyacrylamide gel (8 M urea, 1× TBE buffer) and electro-transferred onto a nylon membrane (HybondN+, Amersham). After crosslinking (Stratagene crosslinker), prehybridization was performed 1 h at 50°C in 5× SSPE, 1× Denhardt, 1% SDS with calf thymus DNA. Hybridization was carried out at 50°C for 4 h in 5× SSPE, SDS 0.1% buffer. After three washes at room temperature in 0.1× SSPE, 0.1% SDS, membrane was exposed to Kodak MS-1 film for 8 h.
Preparation of P. aerophilum S100 extract; micrococcal nuclease treatment
A S100 extract of P. aerophilum was prepared as follows: About 1 g of frozen cells was resuspended in 4 mL PBS in the presence of half a tablet of complete Mini, EDTA-free protease inhibitor (Roche) and sonicated. The lysate was then centrifuged at 100,000g for 20 min and the resulting supernatant was dispatched in 100μL aliquots and stored at −80°C. For the micrococcal nuclease treatment, 20 μL of P. aerophilum S100 extract were incubated 15 min at 37°C, in Tris-HCl 10 mM (pH 8.8), 1 mM CaCl2, in the presence (or not) of 50 units of micrococcal nuclease (USB): The nuclease reaction was stopped by addition of 10 mM EGTA.
Reaction inhibition was controlled by addition of EGTA prior to the addition of micrococcal nuclease. The effect of micrococcal nuclease on purified aTrm56 enzyme was controlled by incubation of 10 μL of the purified enzyme under conditions described above.
In vitro T7 transcription of tRNA genes
Generation of in vitro T7 transcripts from tRNA genes is based on the method described in Reyes and Abelson (1987). Transcription was performed from cloned DNA, oligonucleotides, or PCR fragments with 15 U of T7 RNA polymerase (Promega). We used the plasmids carrying the sequences for the yeast tRNAPhe, tRNAAsp (wild type and deleted for stem-loop D), the P. abyssi tRNALeu/CAA, sR47, and the HIV TAR RNA cloned behind the T7 promoter to produce the corresponding RNA transcripts. The TSL minisubstrate was transcribed from the oligonucleotides TSL(3′) and TSL(5′) (1 pmol of each oligonucleotide introduced in the transcription mixture). The E. coli tRNAleu2, wild type and C56U, C56G, G18C/G19C or G18A/G19A mutants were transcribed from PCR fragments obtained by amplification of E. coli genomic DNA with the oligonucleotide pairs formed with the EC Leu2(5′) and one of the EC Leu2 (3′) oligonucleotides. Radioactive (32P) transcripts were generated using [α-32P]ATP, [α-32P]GTP, [α-32P]CTP, or [α-32P]UTP (20–50 μCi, 400–800 Ci/mM) (Amersham) in the transcription mix. Cold or radioactive transcripts were purified by 8% polyacrylamide/urea gel electrophoresis, eluted in 500 mM NH4-acetate, 1 mM EDTA, and ethanol-precipitated in the presence of polyU.
In vitro assays for tRNA methyltransferase activity
Two types of tRNA Mtase assays were used. The first measured the amount of 3H transferred to total E. coli or yeast tRNA or in vitro tRNA transcripts, using [methyl-3H]AdoMet (Amersham Biosciences) as the methyl donor. Reactions were at 37, 50, or 70°C in 80 μL containing 100 mM Tris-HCl (pH 8.0), 100 mM ammonium acetate, 5 mM MgCL2, 2 mM DTT, 0.1 mM EDTA (methylation buffer 1× as described in Motorin and Grosjean (1999), 8 μM AdoMet (5000cpm/pM), and 1–2 μM tRNA transcript (T7 tRNA in vitro transcripts) or 10 μM bulk yeast or E. coli tRNA mix (Sigma). The reaction was initiated by addition of purified enzyme (30 nM); aliquots (15 μL) were removed at regular time intervals to cold 5% trichloroacetic acid and the precipitate was collected by filtration through a GF/C filter (Whatman); after two washes with cold 5% TCA, and one with EtOH, filters were dried and radioactivity measured by liquid scintillation counting.
The second tRNA Mtase assay used in vitro transcribed uniformly [32P]-labeled tRNAs as substrates. Incubation was performed in the buffer described above, in the presence of 20 μM AdoMet, 1–2 fmol of radiolabeled tRNA, and purified enzyme (30 nM) or P. aerophilum S100 extract (15 μL) and incubated 1 h at 50 or 70°C. AdoMet stock solutions were made at 2 mM concentration in sterile water, and stored at −80°C. Prior to the reaction, the solutions of tRNA substrates (in sterile water) were denatured by a 1-min incubation at 90°C, transferred on ice, and renatured, after addition of the reaction buffer, at 50°C for 10 min.
Analysis of modified nucleosides
Modified nucleoside(s) were analyzed as described (Jiang et al. 1997). Each modified tRNA was purifed on 8% acrylamide/urea gel and completely hydrolyzed to 3′-nucleoside monophosphates by RNAse T2 (nearest neighbor analysis). Digestion was in 10 μL of 50 mM Na-acetate (pH 5.2) with 0.05 U/ml RNase T2, for 24 h at 37°C. Digestion products were analyzed by 2D-thin layer chromatography (TLC) on cellulose plates (10×10 cm, Merck) using chromatographic solvents A and C (Grosjean et al. 2004). The first dimension developed with solvent A (isobutyric acid/concentrated NH4OH/H2O (66:1:33; v/v/v) ; second dimension developed with solvent C: ter-butanol/HCl/H2O (70:15:15; v/v/v) or 2-propanol/HCl/H2O (68:18:14). Radiolabeled spots were identified by autoradiography by comparison with standard 2D-TLC maps (Grosjean et al. 2004). The yield of modified nucleotides in mol/mol was evaluated by quantification of 2D-TLC spot intensities by a Fuji-F-3000 imager, taking into account the nucleotide composition of the tRNA transcripts.
Search for Pyrobaculum aerophylum C/D sRNA targeting tDNAs
We used the 51 intergenic regions annotated as being sRNAs in the genomic sequence of P. aerophilum available at the NCBI site, and the archaeal tDNA database (Marck and Grosjean 2003) to assign tRNA targets to the P. aerophilum sRNAs. For each sRNA we searched a minimal ten-base pairing in a tDNA sequence with or without intron, allowing at most one G-U pair, and one mismatch. All the potential duplexes that can be formed may be computed and visualized at the http://atg.toulouse.inra.fr/~rnaworld/PAE.html Web site.
Acknowledgments
We gratefully acknowledge H. Grosjean for helpful discussions and critical reading of the manuscript, and for the plasmid carrying the yeast tRNAAsp deleted for stem-loop D. We thank R. Giégé for the plasmid carrying the yeast tRNAAsp, O.C. Uhlenbeck for the plasmid carrying the yeast tRNAPhe sequence, S. Auxilien for the oligonucleotides used for transcription of the TSL minisubstrate, B. Clouet d’Orval for the plasmids carrying P. abyssi tRNALeu/CAA and SR47. I Schroeder is thanked for the gift of P. aerophilum frozen cells, and Y. de Préval for oligodeoxynucleotide synthesis. Thanks to Y. Henri, M. Ferrer, Leonora Poljak, and B. Clouet d’Orval for critical reading of the manuscript. This work was supported by laboratory funds from the Centre National de la Recherche Scientifique and the Institut National de la Recherche Agronomique.
Abbreviations
Mtase, methyltransferase
AdoMet, S-adenosyl-methionine
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2110805.
REFERENCES
- Agris, P.F. 1996. The importance of being modified: Roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 53: 79–129. [DOI] [PubMed] [Google Scholar]
- Agris, P.F., Koh, H., and Soll, D. 1973. The effect of growth temperatures on the in vivo ribose methylation of Bacillus stearothermophilus transfer RNA. Arch. Biochem. Biophys. 154: 277–282. [DOI] [PubMed] [Google Scholar]
- Ahn, H.J., Kim, H.W., Yoon, H.J., Lee, B.I., Suh, S.W., and Yang, J.K. 2003. Crystal structure of tRNA(m1G37)methyltransferase: Insights into tRNA recognition. EMBO J. 22: 2593–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aittaleb, M., Rashid, R., Chen, Q., Palmer, J.R., Daniels, C.J., and Li, H. 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 10: 256–263. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman, V., Koonin, E.V., and Aravind, L. 2002. SPOUT: A class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 4: 71–75. [PubMed] [Google Scholar]
- Armengaud, J., Urbonavicius, J., Fernandez, B., Chaussinand, G., Bujnicki, J.M., and Grosjean, H. 2004. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain containing, AdoMet-dependent methyltransferase, conserved in archaea and eukaryota. J. Biol. Chem. 279: 37142–37152. [DOI] [PubMed] [Google Scholar]
- Arn, E.A. and Abelson, J.N. 1996. The 2′-5′ RNA ligase of Escherichia coli. Purification, cloning, and genomic disruption. J. Biol. Chem. 271: 31145–31153. [DOI] [PubMed] [Google Scholar]
- Auffinger, P. and Westhof, E. 1998. Location and distribution of modified nucleotides in tRNA. In Modification and editing of RNA (H. Grosjean and R. Benne, eds.), pp. 569–576. ASM Press, Washington, D.C.
- Bai, Y., Fox, D.T., Lacy, J.A., Van Lanen, S.G., and Iwata-Reuyl, D. 2000. Hypermodification of tRNA in Thermophilic archaea. Cloning, overexpression, and characterization of tRNA-guanine transglycosylase from Methanococcus jannaschii. J. Biol. Chem. 275: 28731–28738. [DOI] [PubMed] [Google Scholar]
- Becker, H.F., Motorin, Y., Sissler, M., Florentz, C., and Grosjean, H. 1997. Major identity determinants for enzymatic formation of ribothymidine and pseudouridine in the T ψ-loop of yeast tRNAs. J. Mol. Biol. 274: 505–518. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant, I., Urban, A., Ma, X., Yu, Y.T., Motorin, Y., and Branlant, C. 2003. The Saccharomyces cerevisiae U2 snRNA: pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Ψ-synthase also acting on tRNAs. RNA 9: 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork, G.R. 1995. tRNA: Structure, biosynthesis, and function. ASM Press, Washington, DC.
- Bjork, G.R., Jacobsson, K., Nilsson, K., Johansson, M.J., Bystrom, A.S., and Persson, O.P. 2001. A primordial tRNA modification required for the evolution of life? EMBO J. 20: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin, M.L., Bachellerie, J.P., and Clouet-d’Orval, B. 2003. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp). Nucleic Acids Res. 31: 6524–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille, J. and Bachellerie, J.P. 1996. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: An exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie 78: 443–456. [DOI] [PubMed] [Google Scholar]
- ———. 1998. SnoRNA-guided ribose methylation of rRNA: Structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 26: 1576–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille, J., Chetouani, F., and Bachellerie, J.P. 1999. The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-Oribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA 5: 66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet d’Orval, B., Bortolin, M.L., Gaspin, C., and Bachellerie, J.P. 2001. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribosemethylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 29: 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet d’Orval, B., Gaspin, C., and Mougin, A. 2005. Two different mechanisms for tRNA ribose methylation in Archaea: A short survey. Biochimie (in press). [DOI] [PubMed]
- Constantinesco, F., Motorin, Y., and Grosjean, H. 1999a. Characterisation and enzymatic properties of tRNA(guanine 26, N (2), N (2))-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J. Mol. Biol. 291: 375–392. [DOI] [PubMed] [Google Scholar]
- ———. 1999b. Transfer RNA modification enzymes from Pyrococcus furiosus: Detection of the enzymatic activities in vitro. Nucleic Acids Res. 27: 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff, J.A., Clamp, M.E., Siddiqui, A.S., Finlay, M., and Barton, G.J. 1998. JPred: A consensus secondary structure prediction server. Bioinformatics 14: 892–893. [DOI] [PubMed] [Google Scholar]
- Davis, D.R. 1998. Biophysical and conformational properties of modified nucleosides in RNA (nuclear magnetic resonance studies). In Modification and editing of RNA (H. Grosjea and R. Benne, eds.), pp. 85–102. ASM Press, Washington, DC.
- Decatur, W.A. and Fournier, M.J. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278: 695–698. [DOI] [PubMed] [Google Scholar]
- Dennis, P.P., Omer, A., and Lowe, T. 2001. A guided tour: Small RNA function in Archaea. Mol. Microbiol. 40: 509–519. [DOI] [PubMed] [Google Scholar]
- Droogmans, L., Haumont, E., de Henau, S., and Grosjean, H. 1986. Enzymatic 2′-O-methylation of the wobble nucleoside of eukaryotic tRNAPhe: Specificity depends on structural elements outside the anticodon loop. EMBO J. 5: 1105–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins, P.A., Watts, J.M., Zalacain, M., van Thiel, A., Vitazka, P.R., Redlak, M., Andraos-Selim, C., Rastinejad, F., and Holmes, W.M. 2003. Insights into catalysis by a knotted TrmD tRNA methyl-transferase. J. Mol. Biol. 333: 931–949. [DOI] [PubMed] [Google Scholar]
- Grosjean, H. and Benne, R. 1998. Modification and editing of RNA. ASM Press, Washington, DC.
- Grosjean, H., Sprinzl, M., and Steinberg, S. 1995. Posttranscriptionally modified nucleosides in transfer RNA: Their locations and frequencies. Biochimie 77: 139–141. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Keith, G., and Droogmans, L. 2004. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Humana Press, Inc., Totowa, NJ. [DOI] [PubMed]
- Gu, X., Yu, M., Ivanetich, K.M., and Santi, D.V. 1998. Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry 37: 339–343. [DOI] [PubMed] [Google Scholar]
- Gustafsson, C., Reid, R., Greene, P.J., and Santi, D.V. 1996. Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res. 24: 3756–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, C. and Ferre-D’Amare, A.R. 2001. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell 107: 929–939. [DOI] [PubMed] [Google Scholar]
- Hori, H., Yamazaki, N., Matsumoto, T., Watanabe, Y., Ueda, T., Nishikawa, K., Kumagai, I., and Watanabe, K. 1998. Substrate recognition of tRNA (Guanosine-2′-)-methyltransferase from Thermus thermophilus HB27. J. Biol. Chem. 273: 25721–25727. [DOI] [PubMed] [Google Scholar]
- Ishitani, R., Nureki, O., Nameki, N., Okada, N., Nishimura, S., and Yokoyama, S. 2003. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 113: 383–394. [DOI] [PubMed] [Google Scholar]
- Jiang, H.Q., Motorin, Y., Jin, Y.X., and Grosjean, H. 1997. Pleiotropic effects of intron removal on base modification pattern of yeast tRNAPhe: An in vitro study. Nucleic Acids Res. 25: 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20: 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M., and Kiss, T. 1996. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85: 1077– 1088. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V. and Rudd, K.E. 1993. SpoU protein of Escherichia coli belongs to a new family of putative rRNA methylases. Nucleic Acids Res. 21: 5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak, J.A., Dalluge, J.J., McCloskey, J.A., and Stetter, K.O. 1994. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33: 7869– 7876. [DOI] [PubMed] [Google Scholar]
- Kuhn, J.F., Tran, E.J., and Maxwell, E.S. 2002. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/ Snu13p snoRNP core protein. Nucleic Acids Res. 30: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre, B. and Purushothaman, S.K. 2004. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell 16: 663–669. [DOI] [PubMed] [Google Scholar]
- Li, H., Trotta, C.R., and Abelson, J. 1998. Crystal structure and evolution of a transfer RNA splicing enzyme. Science 280: 279–284. [DOI] [PubMed] [Google Scholar]
- Lim, K., Zhang, H., Tempczyk, A., Krajewski, W., Bonander, N., Toedt, J., Howard, A., Eisenstein, E., and Herzberg, O. 2003. Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): A cofactor bound at a site formed by a knot. Proteins 51: 56–67. [DOI] [PubMed] [Google Scholar]
- Limbach, P.A., Crain, P.F., and McCloskey, J.A. 1994. Summary: The modified nucleosides of RNA. Nucleic Acids Res. 22: 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte, L., Ailey, B., Hubbard, T.J., Brenner, S.E., Murzin, A.G., and Chothia, C. 2000. SCOP: A structural classification of proteins database. Nucleic Acids Res. 28: 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck, C. and Grosjean, H. 2002. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anti-codon-sparing strategies and domain-specific features. RNA 8: 1189–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: Evolutionary implications. RNA 9: 1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey, J.A. and Crain, P.F. 1998. The RNA modification database—1998. Nucleic Acids Res. 26: 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey, J.A., Graham, D.E., Zhou, S., Crain, P.F., Ibba, M., Konisky, J., Soll, D., and Olsen, G.J. 2001. Post-transcriptional modification in archaeal tRNAs: Identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 29: 4699–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, G., Sauve, V., Larocque, R., Li, Y., Matte, A., and Cygler, M. 2002. The structure of the RlmB 23S rRNA methyltransferase reveals a new methyltransferase fold with a unique knot. Structure (Camb) 10: 1303–1315. [DOI] [PubMed] [Google Scholar]
- Motorin, Y. and Grosjean, H. 1998. Chemical structures and classification of postranscriptionally modified nucleosides in RNA. In Modification and editing of RNA (H. Grosjean and R. Benne, eds.), pp. 543–549. ASM Press, Washington, DC.
- ———. 1999. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA 5: 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon, K.R., Guymon, R., Crain, P.F., McCloskey, J.A., Thomm, M., Lim, J., and Cavicchioli, R. 2003. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23°C) and Stetteria hydrogenophila (Topt, 95°C). J. Bacteriol. 185: 5483–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nureki, O., Shirouzu, M., Hashimoto, K., Ishitani, R., Terada, T., Tamakoshi, M., Oshima, T., Chijimatsu,M., Takio, K.,Vassylyev,D.G., et al. 2002. An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallogr. D Biol. Crystallogr. 58: 1129–1137. [DOI] [PubMed] [Google Scholar]
- Nureki, O., Watanabe, K., Fukai, S., Ishii, R., Endo, Y., Hori, H., and Yokoyama, S. 2004. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure (Camb) 12: 593–602. [DOI] [PubMed] [Google Scholar]
- Omer, A.D., Lowe, T.M., Russell, A.G., Ebhardt, H., Eddy, S.R., and Dennis, P.P. 2000. Homologs of small nucleolar RNAs in Archaea. Science 288: 517–522. [DOI] [PubMed] [Google Scholar]
- Omer, A.D., Ziesche, S., Ebhardt, H., and Dennis, P.P. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribo-nucleoprotein complex. Proc. Natl. Acad. Sci. 99: 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer, A.D., Ziesche, S., Decatur, W.A., Fournier, M.J., and Dennis, P.P. 2003. RNA-modifying machines in archaea. Mol. Microbiol. 48: 617–629. [DOI] [PubMed] [Google Scholar]
- Pierrel, F., Hernandez, H.L., Johnson,M.K., Fontecave, M., and Atta,M. 2003. MiaB protein from Thermotoga maritima. Characterization of an extremely thermophilic tRNA-methylthiotransferase. J. Biol. Chem. 278: 29515–29524. [DOI] [PubMed] [Google Scholar]
- Pintard, L., Lecointe, F., Bujnicki, J.M., Bonnerot, C., Grosjean, H., and Lapeyre, B. 2002. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 21: 1811–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, V.M. and Abelson, J. 1987. A synthetic substrate for tRNA splicing. Anal. Biochem. 166: 90–106. [DOI] [PubMed] [Google Scholar]
- Romby, P., Carbon, P., Westhof, E., Ehresmann, C., Ebel, J.P., Ehresmann, B., and Giege, R. 1987. Importance of conserved residues for the conformation of the T-loop in tRNAs. J. Biomol. Struct. Dyn. 5: 669–687. [DOI] [PubMed] [Google Scholar]
- Roovers, M., Wouters, J., Bujnicki, J.M., Tricot, C., Stalon, V., Grosjean, H., and Droogmans, L. 2004. A primordial RNA modification enzyme: The case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 32: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenski, J., Crain, P.F., and McCloskey, J.A. 1999. The RNA modification database: 1999 update. Nucleic Acids Res. 27: 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sampson, J.R. and Uhlenbeck, O.C. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 85: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, J.R., DiRenzo, A.B., Behlen, L.S., and Uhlenbeck, O.C. 1990. Role of the tertiary nucleotides in the interaction of yeast phenylalanine tRNA with its cognate synthetase. Biochemistry 29: 2523–2532. [DOI] [PubMed] [Google Scholar]
- Schubert, H.L., Blumenthal, R.M., and Cheng, X. 2003. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem Sci. 28: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.K., Gurha, P., Tran, E.J., Maxwell, E.S., and Gupta, R. 2004. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribo-nucleoprotein of pre-tRNA. J. Biol. Chem. 279: 47661–47671. [DOI] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczak, L.B., DeGregorio, S.J., Strobel, S.A., and Steitz, J.A. 2002. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9: 1095–1107. [DOI] [PubMed] [Google Scholar]
- Tang, T.H., Bachellerie, J.P., Rozhdestvensky, T., Bortolin, M.L., Huber, H., Drungowski, M., Elge, T., Brosius, J., and Huttenhofer, A. 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. 99: 7536–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Fedorova, N.D., Jackson, J.D., Jacobs, A.R., Kiryutin, B., Koonin, E.V., Krylov, D.M., Mazumder, R., Mekhedov, S.L., Nikolskaya, A.N., et al. 2003. The COG database: An updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, E.J., Zhang, X., and Maxwell, E.S. 2003. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 22: 3930–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, E., Brown, J., and Maxwell, E.S. 2004. Evolutionary origins of the RNA-guided nucleotide-modification complexes: From the primitive translation apparatus? Trends Biochem. Sci. 29: 343–350. [DOI] [PubMed] [Google Scholar]
- Trotta, C.R., Miao, F., Arn, E.A., Stevens, S.W., Ho, C.K., Rauhut, R., and Abelson, J.N. 1997. The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89: 849–858. [DOI] [PubMed] [Google Scholar]
- Watanabe, K., Nureki, O., Fukai, S., Ishii, R., Okamoto, H., Yokoyama, S., Endo, Y., and Hori, H. 2005. Roles of conserved amino acid sequence motifs in the SpoU (TrmH) RNA methyltransferase family. J. Biol. Chem. 280: 10368–10377. [DOI] [PubMed] [Google Scholar]
- Watkins, N.J., Dickmanns, A., and Luhrmann, R. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5 K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22: 8342–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarembinski, T.I., Kim, Y., Peterson, K., Christendat, D., Dharamsi, A., Arrowsmith, C.H., Edwards, A.M., and Joachimiak, A. 2003. Deep trefoil knot implicated in RNA binding found in an archaebacterial protein. Proteins 50: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziesche, S.M., Omer, A.D., and Dennis, P.P. 2004. RNA-guided nucleotide modification of ribosomal and non-ribosomal RNAs in Archaea. Mol. Microbiol. 54: 980–993. [DOI] [PubMed] [Google Scholar]