Abstract
We have found an extremely large ribonuclease P (RNase P) RNA (RPR1) in the human pathogen Candida glabrata and verified that this molecule is expressed and present in the active enzyme complex of this hemiascomycete yeast. A structural alignment of the C. glabrata sequence with 36 other hemiascomycete RNase P RNAs (abbreviated as P RNAs) allows us to characterize the types of insertions. In addition, 15 P RNA sequences were newly characterized by searching in the recently sequenced genomes Candida albicans, C. glabrata, Debaryomyces hansenii, Eremothecium gossypii, Kluyveromyces lactis, Kluyveromyces waltii, Naumovia castellii, Saccharomyces kudriavzevii, Saccharomyces mikatae, and Yarrowia lipolytica; and by PCR amplification for other Candida species (Candida guilliermondii, Candida krusei, Candida parapsilosis, Candida stellatoidea, and Candida tropicalis). The phylogenetic comparative analysis identifies a hemiascomycete secondary structure consensus that presents a conserved core in all species with variable insertions or deletions. The most significant variability is found in C. glabrata P RNA in which three insertions exceeding in total 700 nt are present in the Specificity domain. This P RNA is more than twice the length of any other homologous P RNAs known in the three domains of life and is eight times the size of the smallest. RNase P RNA, therefore, represents one of the most diversified noncoding RNAs in terms of size variation and structural diversity.
Keywords: RNase P RNA, hemiascomycete, yeast, Candida glabrata, secondary structure
INTRODUCTION
Ribonuclease P (RNase P), required for the 5′-end maturation of transfer RNAs (tRNAs) (for reviews, see Frank and Pace 1998; Altman and Kirsebom 1999), is a ribonucleo-protein particle present in the three domains of life (for review, see Schon 1999). In bacteria and several archaea, the RNA subunit (abbreviated as P RNA) is catalytically active in vitro in the absence of protein components (Guerrier-Takada et al. 1983; Pannucci et al. 1999). However, eukaryal P RNA has never been found to be active on its own.
Comparative sequence analysis classifies bacterial RNase P RNAs into Type A and Type B, while Type A is suggested to be the ancestral form, from which Type B is derived (Haas et al. 1994, 1996). Significant differences of secondary structures are found in these two types, but main elements conspicuous in the tertiary structure are conserved. Indeed, Type A and B structures are each composed of two domains: domain I is the Specificity domain (S domain) recognizing the pre-tRNA substrate and domain II is the Catalytic domain (C domain) (Loria and Pan 1996). Variability exists in domain II, but the P4 pseudoknot region proposed to carry the catalytic function is conserved (Harris and Pace 1995; Frank et al. 1996). A striking difference between the two types lies in the folding of the pre-tRNA recognition responsible elements (in the S domain). For Type A (Krasilnikov et al. 2004), stacked P13 and P14 helices are implicated, whereas for Type B (Krasilnikov et al. 2003), it is the P10.1 helix. In tertiary structures, despite different helical packing, the three strategic points that build the pre-tRNA recognition interface are in close proximity (Krasilnikov et al. 2003, 2004). More analysis of such kind is needed in order to extract rules regarding RNA evolution and RNA architecture. Besides, for structural biology investigations, it was hoped that some species might harbor minimal P RNAs. It was, therefore, decided to analyze the recently sequenced full genomes of yeasts, including some pathogens.
Similar to bacterial P RNA, eukaryotic P RNA is also composed of two domains where almost all of bacteria homologous helices were found (Chen and Pace 1997). However, the specific bacterial four-way junction in the S domain was not exactly identified in eukaryotic RNAs. In fact, the junctions between these helices are more variable in eukaryotes than bacterial ones, thus they were named “eP8” and “eP9” (eP for “eukaryal Paired” region) (Frank et al. 2000) instead of “P8” and “P9” as in bacteria. Apart from structural conservation, five sequences (called CRs for Conserved Regions) are highly conserved even among the three domains of life (Chen and Pace 1997).
We used the conserved core motifs for bacterial and eukaryotic P RNAs to identify more P RNAs in complete yeast genomes. We focused precisely on one class of yeasts: the hemiascomycetes. This class is molecularly as diverse as the entire phylum of chordates (Dujon et al. 2004), and it was hoped that the analysis of yeast P RNAs would help our sequence alignment of eukaryotic P RNAs and facilitate our homogeneous comparison with the known bacterial structural elements. We formulated a conserved core in several hemiascomycete P RNAs by using sequence alignments and found that large variations (insertions or deletions) are minimized and constrained to defined regions. New types of helical insertions in eukaryotes were identified. The most surprising variability was found in the long P RNA (1149 nt) of Candida glabrata, a human pathogen that has become recently an important cause of candidiasis, because of its resistance to antifungal drugs. In this paper, we first describe the search procedure leading to the disclosure of the large RNA, confirm its full-length existence as C. glabrata P RNA, align this long sequence with other hemiascomycete P RNAs to recognize where the insertions in C. glabrata sequence occur, and finally define a yeast hemiascomycete P RNA consensus core to represent P RNA structural conservation and divergence.
RESULTS
RNase P RNA search in yeast genomes
The procedure to search for P RNA from hemiascomycete yeast genomes consisted of two steps. The first step was carried out with the highly conserved pseudoknot P4 and a few nucleotides highly conserved around it. In order to minimize the risk of excluding the correct sequence, only the two strictly conserved motifs among hemiascomycetes were kept: (1) the first motif in the 5′-strand of the P4 helix: 5′-GGGAAAUUCGGUG, (2) the second motif in its 3′-strand: 5′-UACCGAU. These positions were previously identified and called CR I and CR V (Chen and Pace 1997). The first motif is larger than the previously identified CR I conserved region (GNAANNUC), and this extended CR I motif (GGGAAAUUCGGUG) was used in order to increase the specificity of the search. For CR V, we used a shorter segment that is strictly conserved in hemiascomycetes.
Analyses of all the current known RNase P RNAs from different organisms defined the number of nucleotides separating the two motifs to be between 100 and 600 nt (Brown 1999). The RNAMotif program was chosen because this tool allows simultaneous search of two motifs (Macke et al. 2001). We used a descriptor for the RNAMotif program based only on the nature of nucleotides and, thus, bypassed the need to resort to the secondary structure in the first step.
The second step involved manual alignment of the selected sequences with those known helices of yeast RNase P RNAs. This structural alignment was improved to locate most of the homologous helices, in comparison to the previously predicted secondary structures, especially that of Saccharomyces cerevisiae. Thus, each unique result from the RNAMotif search in each genome was aligned with other sequences found by PCR amplification or in databases.
We characterized one sequence of RNase P RNA gene in each of the following genomes (for abbreviations, see Table 1): Candida albicans, Debaryomyces hansenii, Eremothecium gossypii, Kluyveromyces lactis, Kluyveromyces waltii, Naumovia castellii, Saccharomyces kudriavzevii, Saccharomyces mikatae, and Yarrowia lipolytica. For the remaining one, C. glabrata, we could only find a P RNA sequence with the length between the two motifs expanded to 1012 nt. This sequence, which possessed other conserved regions, CR II, CR III, and CR IV, was further aligned with the other P RNAs (see Fig. 4, below) in order to verify that it contains the secondary structure conserved core of a P RNA (see Fig. 3, below). Thus, it allows us to recognize each homologous helix in this long sequence. We also tested other possibilities of degenerated P4 motifs (data not shown), but we could not identify any other sequence as a plausible P RNA gene in the genome of C. glabrata.
TABLE 1.
Abbreviations of yeast species discussed in the text
| Alignment abbreviations | Species |
| Hemiascomycetes | |
| A.tel | Arxiozyma telluris |
| C.alb | Candida albicans |
| C.gla | Candida glabrata |
| Cgui1, Cgui2 | Candida [Pichia] guilliermondii |
| C.kru | Candida krusei |
| C.par | Candida parapsilosis |
| C.ste | Candida stellatoidea |
| C.tro | Candida tropicalis |
| Cl.lu | Clavispora lusitaniae |
| Cl.op | Clavispora opuntiae |
| D.han | Debaryomyces hansenii |
| E.gos | Eremothecium [Ashbya] gossypii |
| K.lac | Kluyveromyces lactis |
| K.pol | Kluyveromyces polysporus |
| K.the | Kluyveromyces thermotolerans |
| K.wal | Kluyveromyces waltii |
| N.cas | Naumovia [Saccharomyces] castellii |
| N.dai | Naumovia dairenensis |
| P.can | Pichia Canadensis |
| P.mis | Pichia mississippiensis |
| P.str | Pichia strasburgensis |
| S.cer | Saccharomyces cerevisiae1 |
| S.glo | Saccharomyces globosus2 |
| S.klu | Saccharomyces kluyveri |
| S.kud | Saccharomyces kudriavzevii |
| S.mik | Saccharomyces mikatae |
| S.pas | Saccharomyces pastorianus3 |
| S.ser | Saccharomyces servazzii |
| S.uni | Saccharomyces unisporus |
| Sp.fi | Saccharomycopsis fibuligera |
| T.del | Torulaspora delbrueckii |
| W.flu | Wickerhamia fluorescens |
| Y.lip | Yarrowia lipolytica |
| Z.bai | Zygosaccharomyces bailii |
| Z.flo | Zygosaccharomyces florentinus |
| Z.rou | Zygosaccharomyces rouxii |
| Archiascomycete | |
| S.pom | Schizosaccharomyces pombe |
The brackets indicate abbreviations used. The square brackets indicate other synonym genus names (Kurtzman 2003). The numbers indicate that several species, where P RNA was found, are synonyms (Kurtzman and Fell 1998): 1S. cerevisiae and S. diastaticus;2S. bayanus, S. globosus, and S. uvarum; 3S. carlsbergensis and S. pastorianus.
FIGURE 4.
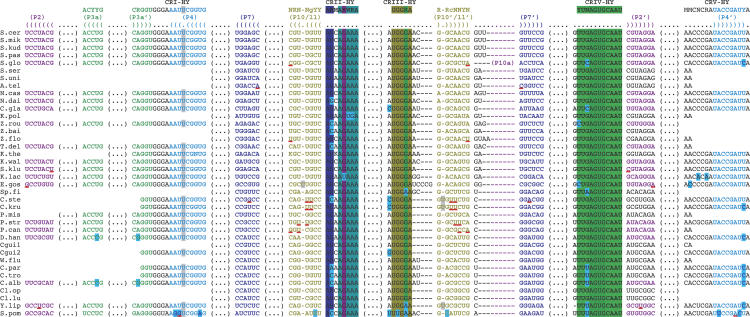
Hemiascomycete sequence alignment of the conserved segments of the RNase P RNA structure (helices with conserved lengths and the conserved regions CR I-HY to CR V-HY as shown in Fig. 3). The complete alignment can be obtained from the authors. The names and the conserved sequences are shown on the first two lines. On the next two lines are displayed the names of helices with the bracket notation (a dot indicates a single-stranded nucleotide and brackets, open for the 5′-end and closed for the 3′-end, indicate helices). For the other lines, sequences of each species are aligned in a phylogenetic order (Kurtzman and Robnett 1998, 2003; Dujon et al. 2004). The first column indicates species names (for abbreviations, see Table 1). The first sequence of C. guilliermondii (Cgui1) was found in the RNase P database with a synonymous name Pichia guilliermondii. The second sequence (Cgui2) was found by PCR amplification and has a few sequence variations, especially in CR III-HY. The archiascomycete S. pombe sequence is indicated here as an external reference, and the consensus does not include it. In each sequence, a dash sign (—) indicates a 1-nt gap and a gap of several nucleotides is noted by (. . .). Red underlined pairs of nucleotides indicate that they do not form either a Watson-Crick or a GoU wobble pair. Bulges are highlighted in light gray and terminal loops in dark gray. Some nucleotides are highlighted in light blue to emphasize the variations that occur in only few hemiascomycetes. P RNA sequences of other species were found in the RNase P database (Brown 1999), but were omitted here because identity percentage with the aligned sequences are higher than 95%. Sequences of Saccharomyces diastaticus has 99.64% identity (id.) with S. cerevisiae, Saccharomyces bayanus has 100% id. with Saccharomyces uvarum, S. bayanus has 99.27% id. with S. cerevisiae, and Saccharomyces carlsbergensis has 97.49% id. with Saccharomyces pastorianus (this last difference occurs only in the P1 helix). Furthermore, S. diastaticus is another name for the S. cerevisiae species; S. carlsbergensis for the S. pastorianus species; and S. uvarum and S. bayanus for the S. globosus species (Kurtzman and Fell 1998). However, paradoxically, the S. globosus sequence is quite different from the S. bayanus sequence (100% id. with S. uvarum), which is almost identical to the S. cerevisiae sequence. Moreover, the S. globosus sequence presents a P10a insertion, while neither S. bayanus nor S. cerevisiae presents such an insertion. The C. stellatoidea sequence has 98.71% identity with that of C. krusei.
FIGURE 3.
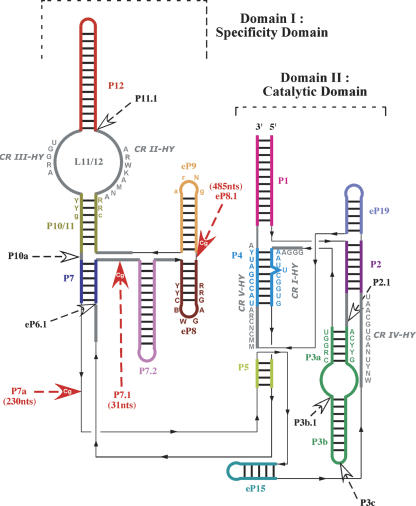
Secondary structure consensus of the conserved core of hemiascomycete RNase P RNAs. Domain I is the Specificity domain and domain II the Catalytic domain. The colored lines represent helices. The color code corresponds to that in the alignment (Fig. 4). Only five helices (P2, P3a, P4, P7, P10/11) have a constant length in the hemiascomycetes. Gray thick lines represent single strands. Black thin lines indicate junctions between two segments containing no nucleotide and necessary for the drawing. The black thin line arrows indicate the 5′ → 3′ direction. Letters beside the lines indicate conserved nucleotides among the hemiascomycete species. CR I (Conserved Region) to CR V are five well-conserved sequences among the three domains of life (Chen and Pace 1997). Because the consensus sequences are longer, we note them CR I-HY to CR V-HY (HY for Hemiascomycete Yeasts). The consensus nomenclature follows that of Cornish-Bowden (1985) with an uppercase letter when it concerns 100% of sequences and a lowercase when there is a difference in less than three sequences among the 37 sequences of the alignment. The nucleotide abbreviations are (N) any nucleotide; (R) A or G; (Y) U or C; (B) C, G, or U; (D) A, G, or U; (M) A or C; (W) A or U. Large arrows indicate sequence insertions in at least one hemiascomycete species. Red large arrows indicate sequence insertions in C. glabrata and sometimes in other species. Helix nomenclature comes from the homologous helix nomenclature of bacteria (Haas et al. 1994). Helices with unsettled homology were named eP, for “eukaryal Paired” region (Frank et al. 2000). (See Table 2 for variation types found in P RNAs of each species.)
The putative RNA is expressed in C. glabrata
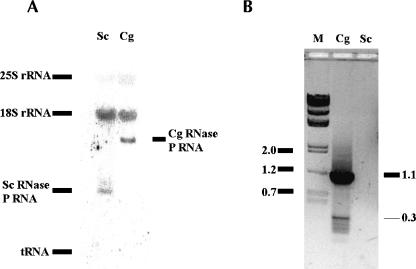
To determine whether this unusually large RNA is expressed in C. glabrata cells, we performed Northern blot and RT-PCR analysis. The oligonucleotide probe complementary to the conserved P4 helix region was used to probe total RNAs isolated from S. cerevisiae (Sc) and C. glabrata (Cg) cells. Figure 1A demonstrates that a relatively small RNA was expressed in S. cerevisiae (as expected, its length was at 369 nt according to Lee et al. 1991). In contrast, C. glabrata cells expressed a much larger RNA, ~1100 nt long. To reveal the identity of this long RNA, we performed a RT-PCR from C. glabrata total RNAs with oligonucleotides specific to the predicted RNA. A DNA fragment ~1100 nt long (Fig. 1B) was amplified and subjected to DNA sequencing. We confirmed that the amplified product is a full-length DNA of the RNase P RNA subunit from C. glabrata (Dujon et al. 2004). Therefore, this RNase P RNA subunit is expressed in C. glabrata with no apparent post-transcriptional event reducing its length.
FIGURE 1.
The putative RNA expressed in C. glabrata. (A) Northern blot. An oligonucleotide complementary to the conserved P4 helix region was used to probe the total RNA of S. cerevisiae (Sc) and C. glabrata (Cg). The position of tRNAs and 18S and 25S rRNA is shown by nonspecific binding or ethidium bromide staining. (B) RT-PCR. The total RNA of C. glabrata or S. cerevisiae was reverse-transcribed with CAGL3 primer, and the DNA was amplified by PCR with CAGL5 and CAGL3 as primers (see Materials and Methods). The DNA markers (M) were shown as 2.0, 1.2, and 0.7 kb, and so on. The 1.1-kb product was sequenced to be the right DNA coding for the putative whole-length P RNA. The small band (~300 bp) was determined to be a nonspecific product from an mRNA (CAGL0L06116g, putative protein similar to YGL185c of S. cerevisiae).
The long RNA is in the active RNase P enzyme complex
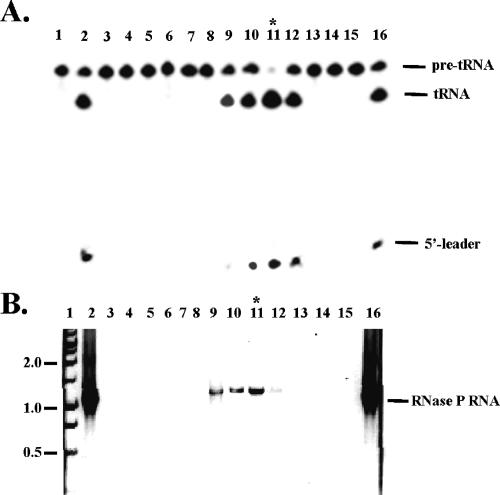
To further investigate whether this RNA is in the active enzyme complex, a small preparation of nuclear RNase P enzyme from C. glabrata was made. The nuclear cell extract was fractionated by ion exchange chromatography, and both the RNase P enzyme activity and the presence of the P RNA were determined. Figure 2 demonstrates that the RNA followed the enzyme activity, and both the peak of enzyme and the long RNA appeared in the same fraction. Considering that the RNA contains all the conserved regions and the homologous helices found in the other yeast RNase P RNAs, and that it follows the enzymatic activity in our enzyme preparation, we are confident that this large RNA (1149 nt) is the RNA subunit of the RNase P enzyme in C. glabrata.
FIGURE 2.
The putative RNA is in the RNase P enzyme complex. The nuclear RNase P enzyme from C. glabrata was purified by an ion exchange column. (Lane 2) The initial sample; (lanes 3–15) fractions eluted from the column; (lanes 1,16) controls or markers. (A) The enzyme activity assay of the fractions. The enzyme complex was eluted by ~350 mM KCl from anion exchange column, similar to that of S. cerevisiae enzyme and human enzyme. (Lane 1) Control of the pre-tRNA alone; (lane 16) the pre-tRNA processed by Escherichia coli enzyme. The products of #11 and #16 had the exact same length when separated by a denaturing 12% high-resolution gel (data not shown). (B) RT-PCR reactions of the fractions. (Lane 1) The DNA markers are shown as 2.0, 1.0, and 0.5 kb, and so on; (lane 16) total RNA of C. glabrata.
A conserved core with supplementary helices in the RNase P RNA
One can define a conserved core of RNase P RNA from hemiascomycetes that consists of two main domains (equivalent to the Specificity and Catalytic domains of bacterial species) by sequence comparison (Fig. 3). This conserved core is similar to the conserved minimum structure of bacterial P RNA. The representations of the secondary structures in Figure 3 are based on the modeling of the architecture of the two types of bacterial P RNAs (Massire et al. 1998). Recent crystal structures of bacterial Specificity domains (Krasilnikov et al. 2003, 2004) display a good agreement with the modeled structures. The diagrams of the secondary structures emphasize the stackings of the helices and the central positioning of the P4 pseudoknot, which is a key element for catalysis. Figure 3 represents the conserved core specific for hemiascomycetes and do not represent the minimal consensus structure.
During the alignments, we systematically attempted to present the insertions between the main helices common to both bacterial and eukaryotic structures. Figure 4 illustrates the results. Seventeen known P RNAs of hemiascomycetes (including D. hansenii, K. waltii, N. castellii, S. kudriavzevii, S. mikatae, and Y. lipolytica) are only composed of the conserved core without any insertion sequences (Fig. 3; Table 2); whereas, for other species (including C. albicans, C. glabrata, Candida krusei, Candida parapsilosis, Candida tropicalis, Candida stellatoidea, E. gossypii, and K. lactis), there are variable helices inserted or deleted in the constant core. Three insertions (P2.1, P3b.1, and P7a) occur in the E. gossypii P RNA, while the P7.2 helix is absent in the P RNA of C. albicans, C. parapsilosis, and C. tropicalis. Two of the insertions in the P RNA of E. gossypii occur also in C. glabrata, C. krusei, and C. stellatoidea (P7a), and in K. lactis (P3b.1). In K. lactis, a long variation occurs in P3b helix (133 nt), called P3c. Another helix (P7.1) is found in several sequences (C. glabrata, C. krusei, C. stellatoidea, and K. lactis). Most remarkably, C. glabrata P RNA presents an extreme variability: two insertions within the Specificity domain (P7.1 of 31 nt and a surprising eP8.1 of 485 nt) and a third one positioned after P7 (P7a of 230 nt).
TABLE 2.
Types of variations found in the conserved core of hemiascomycete RNase P RNAs
| Domain I | Domain II | |||||||||
| Deletion | Insertion | |||||||||
| Species | P7.2 | eP6.1 | P7.1 | eP8.1 | P10a | P11.1 | P2.1 | P3b.1 | P3c | P7a |
| Arxiozyma telluris | — | 60 | — | — | 12 | — | — | — | — | |
| Saccharomyces globosus | — | — | — | 20 | — | — | — | — | — | |
| Naumovia dairenensis | X | 66 | — | — | — | — | — | — | — | — |
| Candida glabrata | — | 31 | 485 | — | — | — | — | — | 230 | |
| Kluyveromyces polysporus | 20 | — | — | — | — | — | — | — | — | |
| Zygosaccharomyces rouxii | — | — | — | — | — | — | 15 | — | — | |
| Zygosaccharomyces florentinus | — | 33 | — | — | — | — | — | — | — | |
| Torulaspora delbrueckii | — | 25 | — | — | — | — | — | — | — | |
| Kluyveromyces lactis | — | 28 | — | — | — | — | 17 | 133 | — | |
| Eremothecium gossypii | — | — | — | — | — | 19 | 59 | — | 41 | |
| Saccharomycopsis fibuligera | — | 22 | — | — | — | — | — | — | — | |
| Candida stellatoidea | — | 30 | — | — | — | — | — | — | 14 | |
| Candida krusei | — | 30 | — | — | — | — | — | — | 14 | |
| Pichia mississippiensis | X | — | — | — | — | — | — | — | — | — |
| Pichia strasburgensis | X | — | — | — | — | — | — | — | — | — |
| Pichia canadensis | — | 29 | — | — | — | — | — | — | — | |
| Candida parapsilosis | X | — | — | — | — | — | — | — | — | — |
| Candida tropicalis | X | — | — | — | — | — | — | — | — | — |
| Candida albicans | X | — | — | — | — | — | — | — | — | — |
The variations were deduced from phylogenetic comparative analysis. The helices names inserted or deleted in the conserved core (Fig. 3) are indicated horizontally and vertically, the species names in which these variations were observed. The order respects the phylogenetic order in the alignment (Fig. 4). The numbers indicate the length (in nucleotides) of each insertion. The first line indicates in which domain the variations occur. The domain of P7a is not indicated since this helix is still undefined structurally. The new types of helical insertions are bold. The 17 other species (Candida guilliermondii, Clavispora lusitaniae, Clavispora opuntiae, Debaryomyces hansenii, Kluyveromyces thermotolerans, Kluyveromyces waltii, Naumovia castellii, Saccharomyces cerevisiae, Saccharomyces kluyveri, Saccharomyces kudriavzevii, Saccharomyces mikatae, Saccharomyces pastorianus, Saccharomyces servazzii, Saccharomyces unisporus, Wickerhamia fluorescens, Yarrowia lipolytica, Zygosaccharomyces bailii) without variations and composed only of the conserved core are not in the table.
Besides secondary structure changes, sequence variations occur also. The yeast-specific BUGA eP8 terminal loop should be extended to BWGA (see E. gossypii P RNA). The terminal loop of eP9 is generally GNRA, with several exceptions (P RNAs of C. glabrata, Clavispora lusitaniae, Clavispora opuntiae, E. gossypii, K. lactis, Pichia canadensis, and Saccharomyces kluyveri). Variations are also located in the CRs regions. Concerning CR II, the yeast-specific sequence is GAAA except in K. lactis, where U is present instead of G, and, more remarkably, GUGA is found in Kluyveromyces polysporus, as in Bacillus subtilis, a bacterium with Type B P RNA. Concerning CR V, in eukaryotes, the consensus is ACYCRAUNCNGAYUA (Frank et al. 2000), which after inclusion of the hemiascomycete sequences becomes less stringent, MCNCRAUNCNGAYYA. Moreover, regarding the universal consensus (ACNNRANNNNGNNUA), a change occurs also, since the penultimate nucleotide (the last nucleotide of P4) is U for the three domains of life except in C. albicans, C. krusei, C. parapsilosis, C. tropicalis, D. hansenii, Saccharomyces globosus, and Y. lipolytica, which have a C instead of a U. Thus, the universal consensus of CR V is now MCNNRANNNNGNNYA.
DISCUSSION
All RNase P RNAs disclosed so far have basic similarly conserved secondary architecture. The subtle differences between them can be used to divide the living world into three domains (bacteria, archaea, and eukarya) (Li and Altman 2004). Efficient methods have been developed to search the RNase P RNA sequence from genomes of bacteria and archaea with considerable success (Li and Altman 2004; Griffiths-Jones et al. 2005). However, the same method has difficulty in finding P RNAs from eukaryotic genomes, mainly because of less conserved sequence motifs, larger genome sizes, and fewer complete genomic sequences available.
The present phylogenetic comparative analysis led to a secondary structure consensus specific to the class of hemi-ascomycete yeasts (Fig. 3). This consensus is composed of a conserved core with variable insertions or deletions depending on the species. Despite the extreme divergence of the hemiascomycete P RNA sequences (see the complete alignment), some regions are strictly conserved.
Specific structural elements that occur only in hemiascomycete P RNAs can also be detected. First, three helices have a strictly conserved length: P2 and P10/11 with each 7 bp and P7 with 6 bp. Secondly, a hemiascomycete-specific helix can be defined (eP15). Indeed, this helix exists in all hemiascomycete sequences and is absent in the archiascomycete Schizosaccharomyces pombe yeast and in higher eukaryotes. Similarly, the P7.2 helix could also be considered as a hemiascomycete-specific helix, but exceptions occur in some species close to S. pombe, where the helix is absent (Table 2). However, it is not clear whether helix P5 exists despite some covariations, but the 5′- and 3′-strands appear to decrease in length for sequences close to S. pombe and disappear in higher eukaryotes. Finally, we can notice that the eP8 helix has a specific hemiascomycete terminal loop (BWGA) and three strictly Watson-Crick base pairs preceding the loop.
Interestingly, insertion sequences occur, for almost each hemiascomycete (K. lactis is an exception), either in the Specificity domain or in the Catalytic domain, but not in both domains for the same species (Table 2). Most of the insertions found in eukaryotes occur also in the hemiascomycete yeasts. However, four new helical insertions have been found in the newly characterized P RNAs of this class of yeast: (1) P2.1 (after P2 helix) in E. gossypii P RNA; (2) P3c of K. lactis; (3) P7a (after P7 helix) in C. glabrata, C. krusei, C. stellatoidea, and E. gossypii P RNAs; (4) eP8.1 (after eP8) for C. glabrata. The latter insertion (485 nt) can be folded as a single very long helix, which seems stable with more than 200 bp and few bulges. However, we have no supporting experimental evidence. Besides these new secondary structure variations, less stringent eukaryotic and universal primary structure consensus could be deduced.
The most significant variability is found in C. glabrata P RNA. Three lines of evidence support the conclusion that the large RNA found in C. glabrata is, indeed, the RNase P RNA of C. glabrata. First, it possesses all five conserved regions (CR I to CR V) found in every P RNA in three domains of life, as well as all the minimal eukaryotic homologous helices. Second, this RNA is expressed with full length (1149 nt) in C. glabrata cells. Third, this RNA is in the active RNase P enzyme complex as it follows the enzymatic activity. Like other eukaryal P RNAs, the in vitro synthesized P RNA of C. glabrata is not active on its own even with high concentration of salts and magnesium (data not shown).
The disclosure of the P RNA in C. glabrata with such an unusual size is remarkable, since until now the longest one among eukaryotic cells has a size of 465 nt (Naumovia dairenensis)with an average of 300–350 nt. Previously, all the RNase P RNAs from bacteria, archaea, and eukaryotic nuclei and chloroplasts ranged in size from 276 (of Mycoplasma fermentans) (Siegel et al. 1996) to 465 nt (N. dairenensis) (Brown 1999; Li and Altman 2004). Mitochondrial RNase P RNA has variable size (Seif et al. 2003), with the respective lengths of predicted mitochondrial P RNA being 423 nt for S. cerevisiae (Stribinskis et al. 1996), 227 nt for C. glabrata (Shu et al. 1991), and as short as 140 nt for Saccharomycopsis fibuligera (Wise and Martin 1991). However, no RNase P RNA has ever been found to be longer than 500 nt (Brown 1999; Li and Altman 2004). The size of the P RNA of C. glabrata is thus at least twice as long as its homolog in all other organisms in the three domains (eukarya, bacteria, and archaea), and it is about eight times that of the smallest RNase P RNA.
There are only four classes of noncoding RNAs present in every cell of bacteria, archaea, and eukarya. They are rRNAs, SRP RNAs, tRNAs, and RNase P RNAs. Structural variation has been observed in others, but none matches the observed variation for RNase P RNA (ranging from 140 nt to 1149 nt, eight times different). The sizes of rRNAs in bacteria and archaea are about 16S, 5S, and 23S, and about 18S, 5.8S, and 25S/28S in eukaryotes, which translates into a factor of <2 in size variation. For the size of SRP RNAs, the range is approximately from 77 nt (of Mycoplasma mycoides) to 330 (of the archaeon Methanococcus jannaschii), about four times variation (data taken from the Signal Recognition Particle Database; Rosenblad et al. 2003). The tRNAs range from 70 to 85 nt with the exception of some mammalian mitochondrial ones, which are ~10 nt shorter (Sprinzl et al. 1998). Therefore, it appears that the P RNA is the most diversified molecule in terms of size variation relative to all other noncoding RNAs existing in every cell of bacteria, archaea, and eukarya.
The structural diversity of bacterial P RNA has been illuminated by the recent X-ray crystal structure study comparing the Specificity domains from Types A and B RNase P RNAs (Krasilnikov et al. 2003, 2004). Both Specificity domains form a similar substrate recognition interface with quite different secondary structures. The variable helices (P13 and P14 in Type A, and P10.1 in Type B) are suggested to be auxiliary elements that serve to stabilize the invariant three-dimensional core. The present results show that most of the variability in the secondary structures of P RNAs of hemiascomycetes can be accommodated around a conserved core very similar to that of bacterial P RNAs. However, because of the structural diversity and lack of conservation, it is rather unlikely that the two insertion elements (eP8.1 and P7a) in P RNA of C. glabrata play similar roles in RNA folding. The predicted secondary structures of eP8.1 and P7a apparently share no similarity with any known RNA motifs. The extremely diversified structure of this P RNA indicates that it may possess other unknown functions, for example, in the binding of protein cofactors. Such a hypothesis is currently being investigated. The origins of the inserted sequences, especially in C. glabrata, although still unknown and under investigation, might be useful as a diagnostic tool. In addition, there is an interesting analogy between the very long helix of C. glabrata and the Rickettsia Palindromic Elements (RPE) found either in protein genes or in RNA genes in the Rickettsia bacteria (Ogata et al. 2000). In the human pathogen Rickettsia conorii, the P RNA contains a 172-nt insertion in the P12 helix (Ogata et al. 2002), which, as in the present case, should not disturb the three-dimensional folding and P RNA function. The molecular mechanisms for the insertions of those palindromic gene segments in the Rickettsia bacteria are still debated.
MATERIALS AND METHODS
Bioinformatics
P RNA sequences were searched with the RNAMotif program version 3.0.0 (Macke et al. 2001). This search was done in 10 hemiascomycete genomes: C. glabrata, D. hansenii, K. lactis, Y. lipolytica (Dujon et al. 2004), C. albicans (Jones et al. 2004), E. gossypii (Dietrich et al. 2004), K. waltii (Kellis et al. 2004), N. castellii, S. kudriavzevii, and S. mikatae (Cliften et al. 2003; for S. mikatae see also Kellis et al. 2004). The output sequences from RNAMotif were aligned manually, and some helices were recognized using the folding program Mfold server with constraints version 3.1 (Zuker 2003). Reference sequences and structures were found in the RNase P database (Brown 1999) and the Entrez Nucleotides database (http://www.ncbi.nlm.nih.gov/). The resulting alignments of P RNAs can be obtained from the authors.
Strains, media, and reagents
Yeast cells were cultured in rich medium including 1% Bacto-yeast extract, 2% Bacto-peptone, and 2% glucose. Solid medium for plates included 2% Bacto-agar. Culture medium reagents were Fisher Scientific or Difco. The yeast strains used in this study were S. cerevisiae strain YMW1 (Zieler et al. 1995) and C. glabrata strain Q (Zhou et al. 1992).
Northern blot and RT-PCR
Total RNA was isolated from S. cerevisiae and C. glabrata by hot phenol extraction (Kohrer and Domdey 1991). Forty micrograms of total RNA was separated on a 1.5% formaldehyde agarose gel. After electrophoresis, the RNA was transferred onto a Zeta-Probe membrane (Bio-Rad), and then probed with the CAGLP4 primer (5′-CTGTAATCCACCGAATTTCCC, complementary to the P4 helix region of RNase P RNA of S. cerevisiae and C. glabrata) labeled with [γ-32P]ATP by T4 polynucleotide kinase. Ten micrograms of RNA from C. glabrata was reverse-transcribed with the CAGL3 oligo (5′-TTGGACAGCAACCTCAGGTAA, complementary to the last 21 nt of the putative DNA) by M-MLV reverse transcriptase (Promega), and the cDNA was subjected to a PCR reaction with CAGL5 (5′-CTGGACAGCAACCGGGATCCT, the first 21 nt of the putative DNA) and CAGL3 as primers. The PCR procedure included 94°C for 2 min, and 30 cycles (94°C for 2 min, 50°C for 1 min, 72°C for 1.5 min), and 72°C for 10 min. The PCR products were separated on a 1.0% agarose gel and cloned into pCR2.1 vector (Invitrogen) before DNA sequencing was performed.
Small preparation of the enzyme
The cell extract was obtained by disrupting 8 g of C. glabrata cells as described (Evans and Engelke 1990). After the ammonium precipitation, about one-third of the extract was resuspended in buffer A (20 mM HEPES at pH 7.9, 10 mM MgCl2, 10 mM KCl, 5% glycerol) (fraction #2), and loaded onto a Bio-Rad Econo-Pac High Q anion exchange column (5 mL; flowthrough, fraction #3) powered by a Pharmacia FPLC system. The chromatography was performed with an initial wash (fraction #4) with buffer A with 100 mM KCl, followed by a gradient of buffer A with 100 mM to 1.0 M KCl (fractions #5–#30). A yeast precursor tRNA (a precursor to serine tRNA) internally labeled with [α-32P]GTP was used as a substrate to assay the enzyme activities (Guerrier-Takada et al. 1983). (Fractions #2–#15 are shown in Fig. 2. No enzyme activities in fractions #16–#30 were detected.) To examine the RNA existence in the samples, an aliquot (0.5 mL) of each fraction was extracted by phenol/chloroform and precipitated by ethanol. About one-tenth of the pellets were used as the template for the RT-PCR reactions as described above.
PCR amplification of partial P RNA genes from other Candida species
Genomic DNAs from five other Candida species were used as templates for PCR reactions to amplify their partial P RNA genes. The primers were designed to anneal to the highly conserved P4 helix and its flanking sequence (P4For: 5′-GGT GGGAAATTCGGTG; P4Rev: 5′-CTRATCGGTATCGGGA). The strains (Candida guilliermondii strain 2309[H], C. krusei 351, C. parapsilosis CDC60, C. tropicalis 350, C. stellatoidea 44) were acquired from Paul L. Fidel Jr. (Department of Microbiology, Immunology, and Parasitology, Louisiana State University Medical Center, New Orleans, LA). The PCR products were cloned into the pGEM-T vector (Promega), respectively, and subsequently subjected to DNA sequencing. The resulting DNA sequences were assessed to encode the P RNA by motif retrieving (CR II, CR III, and CR IV) and sequence alignment.
Acknowledgments
This work was supported in part by the Génolevures-2 sequencing consortium GDR CNRS 2354. R.K. and E.W. are especially thankful to Bernard Dujon and Jean-Luc Souciet for numerous discussions. Y.L. is grateful to Steve Ellis and Robert Gray for their help, and to Paul L. Fidel Jr. of Louisiana State University for supplying Candida strains.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2130705.
REFERENCES
- Altman, S. and Kirsebom, L. 1999. Ribonuclease P. In The RNA world (eds. R.F. Gesteland et al.), pp. 351–380. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Brown, J.W. 1999. The Ribonuclease P database. Nucleic Acids Res. 27: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.L. and Pace, N.R. 1997. Identification of the universally conserved core of ribonuclease P RNA. RNA 3: 557–560. [PMC free article] [PubMed] [Google Scholar]
- Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden, A. 1985. Nomenclature for incompletely specified bases in nucleic acid sequences: Recommendations 1984. Nucleic Acids Res. 13: 3021–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, F.S., Voegeli, S., Brachat, S., Lerch, A., Gates, K., Steiner, S., Mohr, C., Pohlmann, R., Luedi, P., Choi, S., et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304: 304–307. [DOI] [PubMed] [Google Scholar]
- Dujon, B., Sherman, D., Fischer, G., Durrens, P., Casaregola, S., Lafontaine, I., De Montigny, J., Marck, C., Neuveglise, C., Talla, E., et al. 2004. Genome evolution in yeasts. Nature 430: 35–44. [DOI] [PubMed] [Google Scholar]
- Evans, C.F. and Engelke, D.R. 1990. Yeast extracts for transfer RNA gene transcription and processing. Methods Enzymol. 181: 439–450. [DOI] [PubMed] [Google Scholar]
- Frank, D.N. and Pace, N.R. 1998. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67: 153–180. [DOI] [PubMed] [Google Scholar]
- Frank, D.N., Ellington, A.E., and Pace, N.R. 1996. In vitro selection of RNase P RNA reveals optimized catalytic activity in a highly conserved structural domain. RNA 2: 1179–1188. [PMC free article] [PubMed] [Google Scholar]
- Frank, D.N., Adamidi, C., Ehringer, M.A., Pitulle, C., and Pace, N.R. 2000. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA 6: 1895–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Moxon, S., Marshall, M., Khanna, A., Eddy, S.R., and Bateman, A. 2005. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33: D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N., and Altman, S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. [DOI] [PubMed] [Google Scholar]
- Haas, E.S., Brown, J.W., Pitulle, C., and Pace, N.R. 1994. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc. Natl. Acad. Sci. 91: 2527–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, E.S., Banta, A.B., Harris, J.K., Pace, N.R., and Brown, J.W. 1996. Structure and evolution of ribonuclease P RNA in Gram-positive bacteria. Nucleic Acids Res. 24: 4775–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, M.E. and Pace, N.R. 1995. Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA 1: 210–218. [PMC free article] [PubMed] [Google Scholar]
- Jones, T., Federspiel, N.A., Chibana, H., Dungan, J., Kalman, S., Magee, B.B., Newport, G., Thorstenson, Y.R., Agabian, N., Magee, P.T., et al. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. 101: 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., Birren, B.W., and Lander, E.S. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624. [DOI] [PubMed] [Google Scholar]
- Kohrer, K. and Domdey, H. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194: 398–405. [DOI] [PubMed] [Google Scholar]
- Krasilnikov, A.S., Yang, X., Pan, T., and Mondragon, A. 2003. Crystal structure of the specificity domain of ribonuclease P. Nature 421: 760–764. [DOI] [PubMed] [Google Scholar]
- Krasilnikov, A.S., Xiao, Y., Pan, T., and Mondragon, A. 2004. Basis for structural diversity in homologous RNAs. Science 306: 104–107. [DOI] [PubMed] [Google Scholar]
- Kurtzman, C.P. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 4: 233–245. [DOI] [PubMed] [Google Scholar]
- Kurtzman, C.P. and Fell, J.W. 1998. The yeasts, a taxonomic study. Elsevier Science BV, Amsterdam.
- Kurtzman, C.P. and Robnett, C.J. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73: 331–371. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 3: 417–432. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Rohlman, C.E., Molony, L.A., and Engelke, D.R. 1991. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell. Biol. 11: 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. and Altman, S. 2004. In search of RNase P RNA from microbial genomes. RNA 10: 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria, A. and Pan, T. 1996. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA 2: 551–563. [PMC free article] [PubMed] [Google Scholar]
- Macke, T.J., Ecker, D.J., Gutell, R.R., Gautheret, D., Case, D.A., and Sampath, R. 2001. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 29: 4724–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massire, C., Jaeger, L., and Westhof, E. 1998. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 279: 773–793. [DOI] [PubMed] [Google Scholar]
- Ogata, H., Audic, S., Barbe, V., Artiguenave, F., Fournier, P.E., Raoult, D., and Claverie, J.M. 2000. Selfish DNA in protein-coding genes of Rickettsia. Science 290: 347–350. [DOI] [PubMed] [Google Scholar]
- Ogata, H., Audic, S., Abergel, C., Fournier, P.E., and Claverie, J.M. 2002. Protein coding palindromes are a unique but recurrent feature in Rickettsia. Genome Res. 12: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannucci, J.A., Haas, E.S., Hall, T.A., Harris, J.K., and Brown, J.W. 1999. RNase P RNAs from some Archaea are catalytically active. Proc. Natl. Acad. Sci. 96: 7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad, M.A., Gorodkin, J., Knudsen, B., Zwieb, C., and Samuelsson, T. 2003. SRPDB: Signal Recognition Particle Database. Nucleic Acids Res. 31: 363–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon, A. 1999. Ribonuclease P: The diversity of a ubiquitous RNA processing enzyme. FEMS Microbiol. Rev. 23: 391–406. [DOI] [PubMed] [Google Scholar]
- Seif, E.R., Forget, L., Martin, N.C., and Lang, B.F. 2003. Mitochondrial RNase P RNAs in ascomycete fungi: Lineage-specific variations in RNA secondary structure. RNA 9: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, H.H., Wise, C.A., Clark-Walker, G.D., and Martin, N.C. 1991. A gene required for RNase P activity in Candida (Torulopsis) glabrata mitochondria codes for a 227-nucleotide RNA with homology to bacterial RNase P RNA. Mol. Cell. Biol. 11: 1662–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.W., Banta, A.B., Haas, E.S., Brown, J.W., and Pace, N.R. 1996. Mycoplasma fermentans simplifies our view of the catalytic core of ribonuclease P RNA. RNA 2: 452–462. [PMC free article] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribinskis, V., Gao, G.J., Sulo, P., Dang, Y.L., and Martin, N.C. 1996. Yeast mitochondrial RNase P RNA synthesis is altered in an RNase P protein subunit mutant: Insights into the biogenesis of a mitochondrial RNA-processing enzyme. Mol. Cell. Biol. 16: 3429–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, C.A. and Martin, N.C. 1991. Dramatic size variation of yeast mitochondrial RNAs suggests that RNase P RNAs can be quite small. J. Biol. Chem. 266: 19154–19157. [PubMed] [Google Scholar]
- Zhou, P., Szczypka, M.S., Sosinowski, T., and Thiele, D.J. 1992. Expression of a yeast metallothionein gene family is activated by a single metalloregulatory transcription factor. Mol. Cell. Biol. 12: 3766–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieler, H.A., Walberg, M., and Berg, P. 1995. Suppression of mutations in two Saccharomyces cerevisiae genes by the adenovirus E1A protein. Mol. Cell. Biol. 15: 3227–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]