Abstract
Ro ribonucleoprotein (RNP) complexes are composed of one molecule of a small noncoding cytoplasmic RNA, termed Y RNA, and the two proteins Ro60 and La. Additional proteins such as hnRNP I, hnRNP K, or nucleolin have recently been shown to be associated with subpopulations of Y RNAs. Ro RNPs appear to be localized in the cytoplasm of all higher eukaryotic cells but their functions have remained elusive. To shed light on possible functions of Ro RNPs, we tested protein components of these complexes for RNA chaperone properties employing two in vitro chaperone assays and additionally an in vivo chaperone assay. In these assays the splicing activity of a group I intron is measured. La showed pronounced RNA chaperone activity in the cis-splicing assay in vitro and also in vivo, whereas no activity was seen in the trans-splicing assay in vitro. Both hnRNP I and hnRNP K exhibited strong chaperone activity in the two in vitro assays, however, proved to be cytotoxic in the in vivo assay. No chaperone activity was observed for Ro60 in vitro and a moderate activity was detected in vivo. In vitro chaperone activities of La and hnRNP I were completely inhibited upon binding of Y RNA. Taken together, these data suggest that the Ro RNP components La, hnRNP K, and hnRNP I possess RNA chaperone activity, while Ro60-Y RNA complexes might function as transporters, bringing other Y RNA binding proteins to their specific targets.
Keywords: La, Ro60, hnRNPs, RNA chaperone, trans-splicing
INTRODUCTION
Ro ribonucleoprotein (RNP) complexes are small cytoplasmic particles of unknown function that were initially identified as major targets of autoantibodies in patients with the rheumatic diseases systemic lupus erythematosus and Sjögren’s syndrome (Youinou et al. 1994). The core structure of human Ro RNPs is composed of one molecule of a small noncoding Y RNA and the two well-characterised proteins, Ro60 and La (for review, see Pruijn et al. 1997). Ro RNPs are presumably present in all higher eukaryotic cells but have not been detected in yeast. Interestingly, however, a Y RNA homolog and a protein related to Ro60 have recently been identified in the eubacterium Deinococcus radiodurans (Chen et al. 2000) but so far not in any other bacterium.
Y RNAs are highly conserved in evolution and their number differs among species, varying from one in the nematode Caenorhabditis elegans to four in humans (O’Brien et al. 1993; Pruijn et al. 1993; Farris et al. 1995). These RNAs fold into an extended stem–loop structure, whose stem is formed by the 5′ and 3′ ends of the RNA except for a short polyuridine tail at the 3′ terminus that, in contrast to other RNA polymerase III transcripts, is not removed during RNA maturation. The Ro60 protein, which appears to be necessary for nuclear export and stabilization of Y RNAs, binds directly to the stem, while La binds to the 3′ oligouridine stretch (Wolin and Steitz 1984; Pruijn et al. 1991; O’Brien et al. 1993; Simons et al. 1996). Studies performed in the last decade have led to the identification of additional proteins that are able to associate with Y RNAs. Thus, interaction of hnRNP I, hnRNP K, and nucleolin with the human (h) Y1 and Y3 RNAs has been demonstrated in vitro and in vivo (Fabini et al. 2001; Fouraux et al. 2002). Interestingly, the internal pyrimidine-rich loop of hY1 RNA was shown to be essential for binding of all three proteins. A similar sequence is also present in hY3 RNA, but not in hY4 and hY5 RNA, which do not seem to be associated with these three proteins.
Although several functions have been ascribed to Y RNA-associated proteins, the role of Ro RNP complexes is still not understood. A primary role of the La protein appears to be stabilization of newly synthesized RNA polymerase III transcripts and to protect them from exonuclease attack (for review, see Wolin and Cedervall 2002). Furthermore, the requirement of La for maturation and efficient folding of pre-tRNAs has been demonstrated (Yoo and Wolin 1997; Fan et al. 1998; Intine et al. 2000; Chakshusmathi et al. 2003). Interestingly, in Saccharomyces cerevisiae La was found to bind also some RNAs transcribed by RNA polymerase II such as U1, U2, U4, and U5 pre-snRNAs (Kufel et al. 2000; Xue et al. 2000). Moreover, association with La favors nuclear retention of the nascent transcripts and/or contributes to assembly of the RNAs into functional RNPs (Simons et al. 1996; Grimm et al. 1997; Pannone et al. 1998; Xue et al. 2000). Besides its roles in the nucleus, a cytoplasmic function for La in the regulation of eukaryotic translation was repeatedly demonstrated (Svitkin et al. 1994; James et al. 1999; Ali et al. 2000). Taken together, there is accumulating evidence that La is necessary for correct folding of many small RNAs, suggesting a general RNA chaperone function for this protein (for review, see Wolin and Cedervall 2002). However, it is completely unknown whether any of the multiple functions of La is dependent on its association with Y RNAs. Moreover, it has to be taken into consideration that La is approximately 50-fold more abundant in the cell than Y RNAs and, therefore, the majority of La molecules are not associated with these RNAs.
While a plethora of data is available for La, relatively little is known about the functions of Ro60. This protein is far less abundant than La and presumably permanently associated with Y RNAs. Disruption of the Ro60 gene in C. elegans and mouse led to a dramatic decrease in Y RNA levels, suggesting a stabilizing role for Ro60 (Labbe et al. 1999; Xue et al. 2003). In Xenopus laevis oocytes, Ro60 was found to bind misfolded 5S rRNAs and murine Ro60 was recently demonstrated to associate with mutated U2 snRNAs (O’Brien and Wolin 1994; Shi et al. 1996; Chen et al. 2003). Interestingly, in both mammalian and bacterial cells Ro60 was shown to contribute to the resistance of cells to UV irradiation (Chen et al. 2000, 2003). Although these observations do not demonstrate a function of Ro RNPs, it has been suggested that they may be involved in some quality control pathway during biogenesis of ribosomes and other RNP particles (O’Brien and Wolin 1994; Shi et al. 1996; Xue et al. 2003).
In contrast to La and Ro60, other Ro RNP components—including nucleolin, hnRNP I, and hnRNP K—are associated with only a subset of Y RNAs (Fabini et al. 2001; Fouraux et al. 2002). HnRNP I is identical to poly-pyrimidine tract binding protein (PTB) isoform 4 and was initially identified as a splicing repressor in mammalian cells (Ghetti et al. 1992; Lin and Patton 1995). Furthermore, hnRNP I was found to be involved in regulation of cap-independent translation, localization of cytoplasmic RNAs, and poly(A) site cleavage (for review, see Wagner and Garcia-Blanco 2001). HnRNP K is also a multifunctional protein and has been shown to act as a transcription factor for c-myc and c-src (Takimoto et al. 1993; Michelotti et al. 1996; Ritchie et al. 2003), to enhance splicing (Expert-Bezancon et al. 2002), to regulate translation of erythroid 15-lipoxygenase mRNA (Ostareck et al. 1997), and to interact with src kinases (Weng et al. 1994; Ostareck-Lederer et al. 2002).
The primarily cytoplasmic localization of Ro RNPs and their conservation throughout evolution suggests a function of these complexes in some fundamental cytoplasmic processes (Wolin and Steitz 1984; O’Brien et al. 1993; Peek et al. 1993; Pellizzoni et al. 1998). In this context it is interesting to note that involvement in cap-independent translation of viral and cellular mRNAs has been demonstrated for La (for review, see Wolin and Cedervall 2002), hnRNP I, hnRNP K, and nucleolin (Hellen et al. 1993; Ali and Siddiqui 1995; Izumi et al. 2001; Evans et al. 2003). It was suggested that proteins interacting with internal ribosome entry site (IRES) sequences induce structural changes of the mRNA to a conformation that permits translation initiation, thereby acting as RNA chaperones (Martinez-Salas et al. 2001; Mitchell et al. 2003; Brown and Jackson 2004; Pickering et al. 2004).
RNA chaperone activity is defined as an activity that either prevents RNAs from misfolding or that helps to open up RNA structures without ATP consumption (Herschlag 1995; Waldsich et al. 2002). For example, in Escherichia coli, the RNA chaperone Hfq was found to enable binding of the small noncoding RNA RhyB to sodB mRNA and as a consequence to block translation of the mRNA (Geissmann and Touati 2004). Since Y RNAs are small noncoding RNAs that are bound by various proteins, we were interested whether individual protein components of Ro RNP complexes possess RNA chaperone activity, which could be essential for a putative role of Ro RNPs in translational control.
To investigate this issue, we tested recombinant La, Ro60, hnRNP K, and hnRNP I in two different RNA chaperone assays in vitro and additionally in a previously developed bacterial chaperone assay in vivo (Clodi et al. 1999). The data obtained demonstrate pronounced RNA chaperone activity for La, hnRNP K, and hnRNP I in at least one of the three different assays. Importantly, binding of Y RNA to these proteins abrogated their chaperone activity in vitro.
RESULTS
La, hnRNP I, and hnRNP K show RNA chaperone activity in vitro in a cis-splicing assay
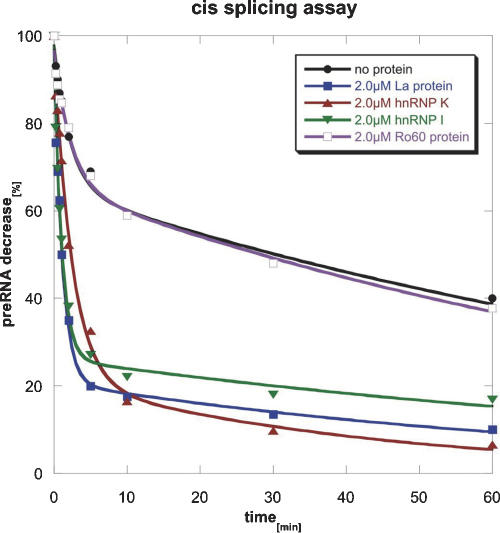
We were interested whether proteins described to bind to hY RNAs possess RNA chaperone activity in vitro. To address this question, the purified proteins were tested in vitro in a cis-splicing assay (Coetzee et al. 1994; Mayer et al. 2002). In this assay, we used a modified construct of the pre-mRNA of the T4 phage thymidylate synthase (td) gene containing a group I intron in which exon sequences were shortened to 27 nt for exon1 and to 2 nt for exon2. This construct splices inefficiently in the absence of chaperones and was shown to depend on StpA for efficient in vitro splicing (Mayer et al. 2002). The minimal td gene was transcribed in vitro in the presence of α-35S-UTP under nonsplicing conditions (see Materials and Methods). Splicing reactions were performed at 37°C and initiated by the addition of GTP (Fig. 1). In the absence of protein, ~30% of the RNA molecules spliced efficiently with a kobs of 0.4 per minute, while the remaining 70% of RNA molecules showed a 50-fold reduced reaction rate (Table 1). This indicates that the majority of RNA molecules are in a folding state that allows only slow reaction rates. Most probably the molecules are partially unfolded or misfolded and refolding into the native state occurs slowly. Upon addition of 2 μM La protein, hnRNP I, or hnRNP K the fraction of fast-reacting molecules increased significantly to 70%–75%. Ro60, on the other hand, did not affect the cis-splicing reaction. To exclude the possibility that Ro60, which in contrast to other proteins was not purified under native conditions, was in an inactive conformation we determined binding of Ro60 to its natural target hY1 RNA using filter-binding and bandshift assays. Since in these assays binding of the Ro60 protein to hY1 RNA was clearly observed, we assumed the protein to be in its native conformation (data not shown).
FIGURE 1.
La, hnRNP K, and hnRNP I increase splicing in a cis-splicing assay. In this assay a modified construct of the pre-mRNA of the td gene of the T4 phage containing a group I intron was used and the decrease of the amount of pre-mRNA with time was measured. In the absence of protein, ~30% of the RNA molecules spliced efficiently (black line). The presence of the proteins La (blue line), hnRNP I (green line), and hnRNP K (red line) significantly increased the fraction of fast-reacting RNA molecules, whereas Ro60 (purple line) had no effect on the splicing reaction.
TABLE 1.
Reaction rates of cis-splicing in the absence and in the presence of hY RNA binding proteins
| Fast reacting (%) | kobs (min−1) | Slow reacting (%) | kobs (min−1) | |
| No protein | 32 ± 3 | 0.44 ± 0.09 | 68 ± 3 | 0.008 ± 0.001 |
| 2.0 μM La protein | 75 ± 3 | 0.9 ± 0.1 | 25 ± 3 | 0.01 ± 0.006 |
| 2.0 μM hnRNP I | 71 ± 2 | 0.9 ± 0.08 | 29 ± 2 | 0.009 ± 0.002 |
| 2.0 μM hnRNP K | 75 ± 4 | 0.4 ± 0.05 | 25 ± 4 | 0.02 ± 0.009 |
| 2.0 μM Ro60 | 31 ± 3 | 0.43 ± 0.1 | 69 ± 3 | 0.009 ± 0.001 |
The percentage of fast-reacting and slow-reacting RNA molecules and the respective rate constants (kobs) are indicated.
hnRNP K and hnRNP I show RNA chaperone activity in vitro in a trans-splicing assay
Next we tested the purified proteins in a previously developed trans-splicing assay. In this assay, the td group I intron is split in two halves (Fig. 2A). The first transcript (H1) contains 549 nt of exon1 and 131 nt of the 5′ part of the intron (I1). The second transcript (H2) contains 147 nt of the 3′ part of the intron (I2) and a truncated exon2 (23 nt). Both RNAs were transcribed individually using α-35S-UTP. Prior to the assay, H1 and H2 were incubated together for 1 min at 95°C and cooled to 55°C or 37°C, respectively. Trans-splicing was initiated by the addition of 32P-GTP so that the spliced product (guanosine-I1) became double labeled (internally with α-35S-UTP and at the 5′ end with 32P-GTP) (Semrad et al. 2004). During two consecutive transesterification reactions, the intron gets excised and the exons are ligated. Finally, the excised intron circularizes (see Fig. 2A) and the formation of 32P-GTP-intron1 (G-I1; Fig. 2B) is used as the main read-out for RNA chaperone activity, because exon ligation may be impaired by the protein storage buffer required to keep the proteins in their native conformation.
FIGURE 2.
hnRNP K and hnRNP I increase splicing in a trans-splicing assay. (A) Scheme for the trans-splicing assay. The two separately transcribed constructs H1 (encoding exon1 + intron1) and H2 (encoding intron2 and exon2) have to fold into a splicing competent structure. The splicing reaction is started upon addition of 32P-labeled GTP. (B) Representative autoradiograph of a 5% polyacrylamide gel. The 5′ part of the intron with the added 32P-guanosine (G-I1) and the ligated exons (E1E2) are indicated, as well as the substrates H1 and H2. At 55°C splicing occurred efficiently (first set of experiments), whereas at 37°C splicing was largely reduced (second set). Trans-splicing was measured at 37°C in the presence of 2 μM La, hnRNP K, hnRNP I, and Ro60. (Prot.b.) Addition of an equivalent volume of protein storage buffer to the splicing reaction (negative control). A slightly different storage buffer was used for Ro60 (“prot.b. (Ro60)”). Note the appearance of the splice product G-I1 in the presence of hnRNP K and hnRNP I and its complete absence in the presence of La and Ro60. (C) Calculated relative splicing rates of the positive control (55°C), the negative control (37°C), and the trans-splicing reaction at 37°C in the presence of hnRNP I or hnRNP K. Relative splicing rates were calculated from (nx − n37)/(n55 − n37), “n55” and “n37” being splicing rates at 55°C or 37°C and “nx” the splicing rate of each evaluated time course.
Trans-splicing is sensitive to low temperatures. At 55°C splicing occurs in the absence of protein, while at 37°C splicing is very low due to the incapability of the constructs to fold into a splicing-competent structure. Therefore, we tested trans-splicing at 37°C in the presence of 2 μM La, Ro60, hnRNP K, or hnRNP I with the splicing reaction at 55°C serving as positive control. A pronounced splicing reaction was observed in the presence of hnRNP K and hnRNP I (Fig. 2B). Thus, intron formation (G-I1) can be seen already after 30 sec and rapidly increases with time; it was, in the case of hnRNP I, even slightly higher than the positive control. Exon ligation (E1E2) is easily seen in the 55°C control, but rather weak in the reactions mediated by hnRNP I and hnRNP K, because the protein storage buffer obviously inhibited this step (see also Fig. 3). In contrast to the hnRNP proteins, neither La nor Ro60 showed a positive effect on splicing (Fig. 2B), either in this assay or when tested at different concentrations (1 μM, 3 μM, 4 μM).
FIGURE 3.
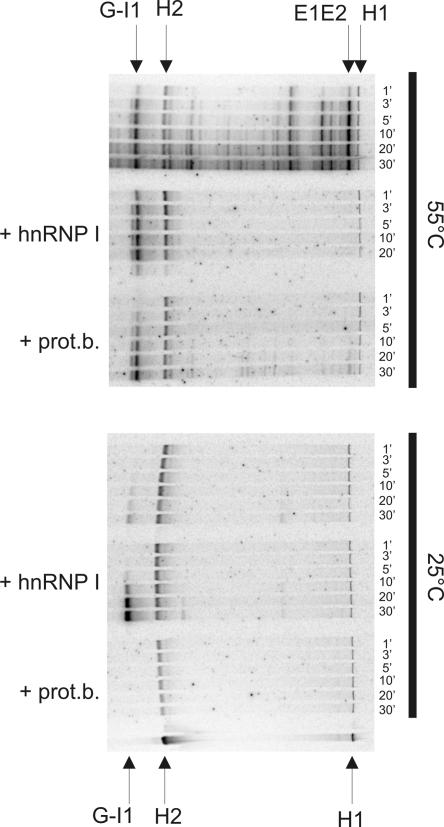
Increased trans-splicing rates upon addition of hnRNP I are observed at low temperatures but not at 55°C. The influence of hnRNP I on trans-splicing was investigated at 55°C and at 25°C. The labeling of the bands is identical to Figure 2 and increase of the splice-product G-I1 was used as read-out for measuring the splicing reaction. At 55°C the presence of hnRNP I did not further stimulate trans-splicing (compare the first set of reactions with the second set). The third set of trans-splicing reactions at 55°C shows splicing in the presence of protein storage buffer (prot. b.), which slightly inhibited the splicing reaction (i.e., formation of G-I1) and almost completely abolished exon ligation. At 25°C splicing was hardly observed but strongly enhanced in the presence of hnRNP I (compare fourth and fifth sets of splicing reactions).
To address whether RNA chaperones may stimulate trans-splicing at 55°C or at a lower temperature (25°C), respectively, activity of hnRNP I was tested also at these temperatures (Fig. 3). At 55°C hnRNP I did not significantly enhance trans-splicing (second set of reactions). The protein storage buffer by itself even slightly decreased the reaction and almost completely inhibited exon ligation (third set of reactions). At 25°C however, the presence of hnRNP I strongly increased the splicing reaction, confirming the data obtained at 37°C (fourth and fifth sets of reactions; Fig. 3).
RNA chaperone activity of La and hnRNP I is inhibited upon association with hY1 RNA
Next we were interested whether binding to Y RNA would affect chaperone activities of the four proteins under investigation. This was particularly important in the case of Ro60, which in contrast to the other proteins is assumed to be permanently associated with Y RNAs. To address this issue we first confirmed stable binding of the four proteins to hY1 RNA in vitro by electrophoretic mobility shift assays (data not shown). We established experimental conditions under which the proteins were almost completely associated with hY1 RNA (described in the Materials and Methods section). These complexes were then tested in both the cis-splicing and the trans-splicing assay (Fig. 4).
FIGURE 4.
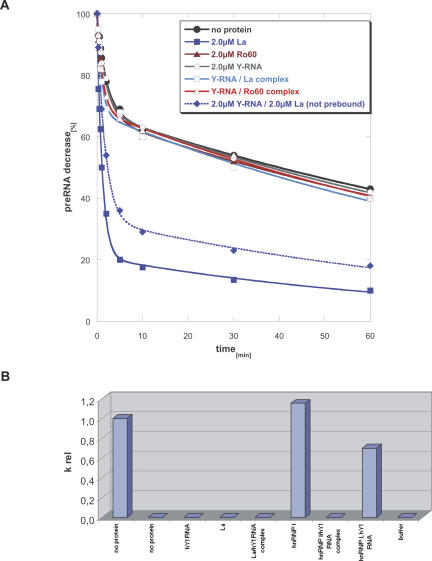
Effect of Y RNA binding to individual components of Ro RNPs on their chaperone activity. (A) Comparison of the cis-splicing activity of free La and Ro60 with La-hY1 RNA and Ro60-hY1 RNA complexes. Two micromolar protein or pre-bound Y RNA–protein complex were tested. As shown previously free La protein increased cis-splicing (filled blue squares); however, when bound to hY1 RNA the RNA chaperone activity was lost (open blue squares). When Y RNA and La were added to the cis-splicing assay without pre-incubation, La could still stimulate the reaction, however to a lesser extent (filled blue diamonds). Ro60 was inactive and also the association of hY1 RNA did not stimulate the reaction (open red triangle); as expected, hY1 RNA by itself did not show any effect on cis-splicing (open circle). (B) Comparison of trans-splicing activity of free hnRNP I and La protein to pre-bound hnRNP I-hY1 RNA and La-hY1 RNA complexes. The bar graph shows relative splicing rates calculated as described in Figure 2 by the formula (nx − n37)/(n55 − n37), where “nx” is the splicing rate of the respective protein or protein–Y RNA complex, “n55” the splicing rate at 55°C in the absence of protein (left bar), and “n37” the splicing rate at 37°C in the absence of protein (second bar from the left). Only free hnRNP I stimulated trans-splicing, whereas the hY1 RNA–hnRNP I complex was completely inactive. A moderate decrease in splicing was seen when hnRNP I and hY1 RNA were added separately (second bar from the right). No activity was observed for La or La-hY1 RNA complexes.
In the cis-splicing assay we compared the activity of La and Ro60 protein–hY1 RNA complexes to the activity of free proteins (Fig. 4A). As observed before, free La stimulated cis-splicing while free Ro60 did not, and also the hY1 RNA complex was completely inactive. However, binding of hY1 RNA to La completely abrogated its RNA chaperoning activity, suggesting that either the domain of La required for chaperoning is bound by Y RNA or that binding of Y RNA could induce a conformational change that impairs the chaperoning activity of La. When La and hY1 RNA were added separately to the same reaction, the La protein could still enhance cis-splicing, although stimulation of splicing was slightly reduced compared to the reaction in the presence of La alone, suggesting binding of the protein to Y RNA occurs during the splicing reaction.
When protein-Y RNA complexes were tested in the trans-splicing assay, similar results were obtained. Here we compared hY1 RNA associated and free hnRNP I and La (Fig. 4B). As demonstrated previously, hnRNP I alone exerted significant splicing activity. Binding of hY1 RNA to hnRNP I completely abrogated its RNA chaperone activity, while a slight reduction in chaperone activity was observed when hnRNP I and hY1 RNA were added separately, in agreement with the result of the cis-splicing assay described above. La did not exert any stimulatory activity, either alone or when complexed to hY1 RNA. Thus, the results obtained in the two in vitro splicing assays demonstrated that Y RNAs are able to inhibit the RNA chaperone activity of Y RNA binding proteins.
La rescues splicing of a stop codon mutant in vivo
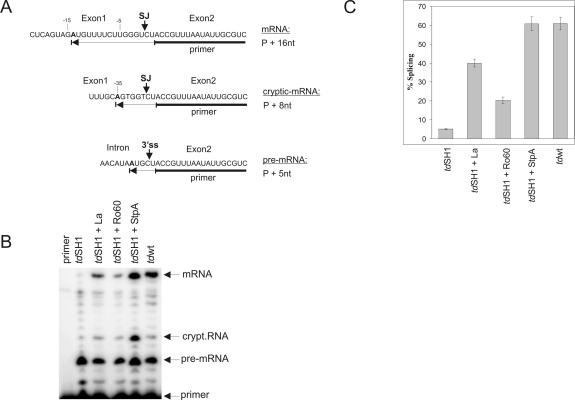
La, Ro60, hnRNP I, and hnRNP K were cloned into the low copy vector pSU20 and each plasmid was cotransformed into E. coli together with a plasmid encoding the td mutant tdSH1 of T4 phage. TdSH1 is a previously described largely splicing deficient double mutant containing a stop codon in exon 1 and the intronic mutation C865U (Pichler and Schroeder 2002). Since overexpression of RNA chaperones was previously shown to alleviate the splicing deficiency of the mutant (Clodi et al. 1999; Pichler and Schroeder 2002), we decided to use this assay to investigate RNA chaperone activity of hY RNA binding proteins in vivo. Expression of the td pre-mRNA and the protein was induced by IPTG and the amount of splicing was visualized using a primer extension assay (see Materials and Methods; Fig. 5A). A representative polyacrylamide gel is shown in Figure 5B, and Figure 5C summarizes the results of three independent experiments. While approximately 60% of wild-type td RNA was spliced, the stop codon mutant cotransformed with the empty vector pSU20 showed only about 5% splicing efficiency. Overexpression of the E. coli protein StpA, known to have pronounced RNA chaperone activity, restored splicing of the stop codon mutant to wild-type levels. Co-expression of La resulted in eightfold stimulation of in vivo splicing and co-expression of Ro60 led to an approximately fourfold increase. Expression of hnRNP I and hnRNP K on the other hand was toxic and, therefore, RNA chaperone activity of both proteins could not be measured in vivo.
FIGURE 5.
Overexpression of La rescues splicing of a splicing deficient mutant in vivo. (A) Reverse transcription reaction analyzing splicing of tdwt and tdSH1 RNA. The DNA primer hybridizes in exon2 and by the addition of dATP, dGTP, dCTP, and ddTTP in the presence of reverse transcriptase either a primer + 5-nt transcript is produced (for pre-mRNA), a primer + 16-nt (for mRNA) or a primer + 8-nt (for cryptic-mRNA). (SJ) Splice junction, (3′ ss) 3′ splice site. (B) Representative autoradiograph of a polyacrylamide gel. Reverse transcription products of mRNA, pre-mRNA, and a cryptic-mRNA are indicated. Cryptic splicing occurs when an alternative fold of the 5′ splice site is formed. The double mutant tdSH1 is significantly reduced in splicing (lane 2). In the presence of La or E. coli protein StpA (lanes 3,5) splicing was significantly increased. In the presence of Ro60 splicing was moderately increased (lane 4). The last lane shows splicing of the wild-type td mRNA. (C) Percentage (mean value ±standard deviation) of splicing [mRNA + cryptic-mRNA] ×100/[mRNA + cryptic-mRNA + pre-mRNA] of three individual experiments.
DISCUSSION
Despite extensive immunological, biochemical, and molecular studies, the exact function(s) of Ro RNPs or Y RNAs, respectively, has remained elusive. Nevertheless, accumulating data indicate that Ro RNP complexes might play a role in a general quality control pathway during biogenesis of small RNAs and/or assembly of RNP complexes as well as in stress resistance (O’Brien and Wolin 1994; Shi et al. 1996; Pellizzoni et al. 1998; Chen et al. 2000, 2003). To shed light on functions of Ro RNPs, we tested RNA chaperone properties of individual components of these complexes by measuring in vitro and in vivo RNA chaperone activities of the human La, Ro60, hnRNP K, and hnRNP I proteins in different chaperone assays based on the self-splicing capacity of group I intron containing RNAs. In the cis-splicing assay, the presence of La, hnRNP K, or hnRNP I significantly increased the fraction of rapidly splicing pre-mRNAs, whereas in the trans-splicing assay, only hnRNP I and hnRNP K showed RNA chaperone activity. It is assumed that RNA chaperones promote cis-splicing by guiding the RNA molecule to correct folding. Contrary to cis-splicing, trans-splicing requires interaction of two RNA molecules that must be correctly folded at the same time. The protein that rescues trans-splicing should assist in correct folding of both RNA molecules and bring them together for proper interaction, which is a more complicated process than the cis-splicing reaction. This suggests that the bimolecular trans-splicing reaction poses higher structural demands on the putative RNA chaperone. Of note, both hnRNP proteins each contain four individual conserved RNA binding motives (four RRMs in hnRNP I, three KH domains and one RGG box in hnRNP K), whereas La contains two RRMs (only one of which is canonical) and Ro60 only one. Thus, it is possible that for promoting a trans-splicing reaction more than two RNA binding motives are required, a hypothesis that will be tested in future experiments.
The observation that La binding to hY RNAs is important for efficient association of hnRNP I and hnRNP K was previously made in our laboratory (Fabini et al. 2001). The demonstration of RNA chaperone activity for La supports the notion that La might guide correct folding of hY RNAs and therefore facilitate efficient assembly of Ro RNPs. This would be consistent with data obtained in yeast, which showed La involvement in snRNP assembly (Pannone et al. 1998; Xue et al. 2000). Recently, in vitro footprinting experiments revealed that the yeast homolog of La promotes folding of the mutated pre-tRNA (Chakshusmathi et al. 2003). In our work we extended the studies on folding activities of La by demonstrating that human La is capable of resolving a misfolded group I intron in vivo using the previously developed in vivo chaperone assay (Clodi et al. 1999). Thus, these data further support a general role of La as a RNA chaperone.
An increasing number of proteins have been identified as interacting with IRES sequences of cellular and/or viral RNAs. These proteins, known as internal initiation transacting factors, include PTB isoforms, members of the poly(rC) family, hnRNP C, La, hnRNP K, unr (upstream of N-ras), nucleolin, and many others (for review, see Stoneley and Willis 2004). It has been suggested by several authors that structural remodeling by such proteins requires RNA chaperone activities (for reviews, see Martinez-Salas et al. 2001; Stoneley and Willis 2004). For example, Mitchell et al. (2003) have shown that neuronal PTB in concert with unr is able to change the structure of the Apaf-1 IRES into the correct conformation accessible for the ribosome. Our data would be in line with such observations confirming RNA chaperone activity of hnRNP I and furthermore clearly demonstrate pronounced RNA chaperone activity also for hnRNP K.
In contrast to the other proteins investigated, Ro60 did not show any chaperone activity in vitro and only moderately increased splicing in vivo. With respect to Ro RNPs, the primary role of Ro60 seems to be Y RNA stabilization (Labbe et al. 1999; Xue et al. 2003) requiring permanent association of this protein. Although interaction of Ro60 with misfolded 5S rRNA (in Xenopus oocytes) and mutated U2 snRNA has been previously demonstrated (O’Brien and Wolin 1994; Shi et al. 1996; Chen et al. 2003), the relevance of these findings is still unclear. It was speculated that Ro60 might resolve the structures of misfolded or mutated RNAs, which would indeed be the task of an RNA chaperone (O’Brien and Wolin 1994; Shi et al. 1996; Chen et al. 2003). Alternatively and more in line with our data, Ro60 might “tag” misfolded RNA species for recognition by the RNA degradation machinery (O’Brien and Wolin 1994; Shi et al. 1996), possibly acting as a quality control checkpoint for RNA misfolding (Labbe et al. 2000), which would not require RNA chaperone activity.
It is still not clear what is the functional relevance of the association of hnRNP I, hnRNP K, and nucleolin with hY1 and hY3 RNAs. It is assumed that these nuclear proteins associate with La-hY1 RNA and La-hY3 RNA complexes during their intranuclear migration to form hY RNPs and, just prior to export into the cytoplasm, Ro60 and other export factor(s) bind (Simons et al. 1996; Pruijn et al. 1997; Rutjes et al. 2001). In the cytoplasm, some Ro RNP components could be released from their Y RNA and eventually get involved in various regulatory processes, such as assembly of snRNP complexes or control of IRES dependent translation. Under certain conditions, like UV irradiation, Ro RNPs may move back to the nucleus, where they might be involved in repair mechanisms (Chen et al. 2003).
Taken together, our experiments suggest that the Y RNA binding proteins La, hnRNP I, and hnRNP K generally possess RNA chaperone activity that is inhibited upon association with Y RNAs. Thus, Ro RNP particles contain multifunctional proteins, which upon binding to Y RNAs may be transported to specific targets where they would be released from the particles to exert functions that require RNA chaperone activity.
MATERIALS AND METHODS
Cloning, expression, and purification of La, Ro60, hnRNP K, and hnRNP I
The cDNAs encoding human La and Ro60 were kindly provided by Ger J. M. Pruijn (Radboud University of Nijmegen, The Netherlands) and the cDNAs encoding hnRNP I and hnRNP K were a gift from Gideon Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania). The cDNAs encoding La and hnRNP I were cloned into the NdeI and SapI cloning sites of the expression vector pTWIN1 (New England Biolabs) resulting in a C-terminal fusion to the intein Mxe GyrA (self-cleveage intein protein). The cDNA encoding human hnRNP K was cloned into the SapI cloning site of the pTWIN1 vector in between two inteins. The fusion proteins were overexpressed in E. coli strain BL21; cells were lysed in a buffer containing 50 mM Tris-HCl (pH 8.5), 500 mM NaCl, 1 mM EDTA, and 20 μM PMSF; and the cell lysates were loaded on chitin columns. High salt concentration was used to reduce unspecific binding of the proteins to RNA. Protein splicing was induced either by increasing the DTT concentration (for the C-terminal fusions) or by a pH and temperature shift (for the N-terminal fusion) according to the manufacturer’s protocol (New England Biolabs, Impact-TWIN-system). Since expression of Ro60 was rather inefficient in this system, the Ro60 cDNA was cloned into the NdeI and XhoI cloning sites of the pET28a vector fused to a His-tag at the N terminus, overexpressed in E. coli strain BL21 codon+, and the extract was purified over a nickel column according to the manufacturer’s protocol (Qiagen). La, hnRNP K, and hnRNP I were stored in a protein storage buffer containing 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 10% glycerol, and Ro60 was stored in a slightly different buffer (50 mM NaH2PO4 at pH 7.5, 300 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 10% glycerol). The purity of the recombinant proteins was verified by SDS-PAGE and Coomassie staining. All proteins migrated as single bands and protein concentration was determined using the Bradford reagent. The RNA binding activity of His-tagged Ro60 was checked by a filter binding assay employing 32P-labeled in vitro transcribed hY1 RNA.
In vitro transcription
For the cis-splicing assay, the pTZ18U-tdΔP6-2 plasmid containing the td group I intron flanked by short exon sequences (exon1: 27 nt, exon2: 2 nt) was linearized with XbaI. Ten micrograms of the linearized plasmid were transcribed under nonsplicing conditions to prevent splicing of the precursor RNA. Transcription was performed by T7 RNA polymerase in 40 mM Tris-HCl (pH 7.5), 2 mM spermidine, 6 mM MgCl2, 10 mM NaCl, 20 mM DTT, 3 mM ATP, 3 mM GTP, 3 mM CTP, 1 mM UTP, and 30 μCi α-35S-UTP. The low Mg2+ concentration and the reaction temperature of 22°C were suboptimal in order to prevent splicing of the precursor RNA during transcription. The reaction was performed overnight and the resulting products were purified on a 5% polyacrylamide gel.
For the trans-splicing assay (see below), the pTZ18U-tdΔP6-2 plasmids called H1 (encoding exon 1 + 5′ part of the td intron) and H2 (encoding 3′ part of the td intron + short version of exon 2) were linearized with SalI (for H1) and BamHI (for H2) (Coetzee et al. 1994). Construct H1 consists of 549 nt of exon 1 and 131 nt of the 5′ part of the intron (total 680 nt). Construct H2 consists of 147 nt of the 3′ half of the intron and 23 nt of exon 2 (total 170 nt). The RNAs H1 and H2 were transcribed by T7 RNA polymerase in 40 mM Tris-HCl (pH 7.0), 26 mM MgCl2, 3 mM spermidine, 10 mM DTT, 5 mM ATP, 5 mM GTP, 5 mM CTP, 2.5 mM UTP, 2.5 mM α-35S-UTP at 37°C for 3 h, followed by 30 min DNase digest and purification of the transcripts over a 5% polyacrylamide gel.
Cis-splicing assay
35S-body-labeled precursor RNA (0.5 pmol) was denatured for 1 min at 95°C and subsequently cooled to 37°C. Splicing buffer containing 50 mM Tris-HCl (pH 7.3), 0.4 mM spermidine, and 5 mM MgCl2 (final concentration) was added followed by either the respective protein (final concentration 2 μM) or the corresponding amount of protein storage buffer. Splicing was induced by addition of 0.5 mM GTP and aliquots were taken after 15 sec, 30 sec, 45 sec, 1 min, 2 min, 5 min, 10 min, 30 min, and 60 min. The reactions were stopped by addition of 40 mM EDTA and 300 μg/mL tRNA (final concentration). Proteins were removed by phenol-CHCl3 extraction and the RNA was precipitated and loaded on 5% polyacrylamide gels. The gels were quantified using a PhosphorImager and the ImageQuant software.
Trans-splicing assay
The trans-splicing assay was performed as described recently (Semrad et al. 2004). Briefly, H1 and H2 transcripts (200 nM each and 20 nM final concentration in the assay) were incubated for 1 min at 95°C and cooled to either 55°C (positive control), or 37°C or 25°C. Next, splicing buffer (40 mM Tris-HCl at pH 7.4, 3 mM MgCl2, 0.4 mM spermidine, 4 mM DTT, final concentration) and 0.33 pmol 32P-GTP were added. Subsequently, either the respective protein was added to a final concentration of 2 μM or an equivalent volume of protein storage buffer. The reactions were incubated at either 55°C (positive control), or 37°C or 25°C and aliquots of the reaction were stopped by adding 40 mM EDTA and 300 μg/mL tRNA (final concentration). Analysis of the reactions was performed essentially as described in the previous section.
In vitro hY RNA binding assay
To prepare protein-Y RNA complexes for the cis- and trans-splicing assay in vitro RNA binding assays were performed with La, Ro60, and hnRNP I. To determine the amount of binding between Y RNA and the respective protein we combined bandshift assays with Coomassie staining. We incubated 2μM 35S-body-labeled hY1 RNA in binding buffer (10 mM Tris-HCl at pH 7.5, 100 mM NaCl, 3 mM MgCl2, 1 mM DTT, final concentration) with different concentrations (1 μM, 2 μM, 3 μM, 4 μM) of the respective protein for 10 min at 37°C. The reactions were applied to native gradient gels (Bio-Rad). Protein complexes and free proteins, respectively, were visualized by Coomassie staining and after destaining, gels were dried and exposed to an X-ray film. Labeled hY1 RNA was visualized by autoradiography. Finally, the binding reaction was performed under conditions where the proteins were almost completely complexed with hY1 RNA and the complexes were immediately employed in the cis-splicing assay or the trans-splicing assay. In some samples, free protein and free hY1 RNA were added separately to the splicing reaction.
In vivo RNA chaperone assay
Plasmids and strains
The thymine deficient (thy−) variant of the E. coli strain C600 [F-, supE44, thi-1, thr-1, leuB6, lacY1, tonA21] was used. Bacterial cells were grown in complete medium TBY-E. Thymine was added to a final concentration of 50 μg/mL. The tdwt and tdSH1 double mutant, which has a stop codon at position −82 in exon 1 and a point mutation in P6 (C865U), were in the context of vector pTZ18-U-tdΔP6-2 (Salvo et al. 1990; Pichler and Schroeder 2002). The E. coli StpA gene was in the context of vector pAZ205, a pSU18 derivative (Zhang et al. 1995). La and Ro60 cDNAs were amplified by PCR using the following primers:
La: 5′-GCCCGCATATGGCTGAAAATGGTGATAATG-3′ and 5′-GCCCGGAATTCCTACTGGTCTCCAGCACC-3′, and
Ro60: 5′-GGTGGTCATATGGAGGAATCTGTAAACC-3′ and 5′-GGTGGTCTCGAGTTAAATCATATCTAATG-3.
The PCR products were digested at NdeI/EcoRI sites (for La) and at NdeI/XhoI sites (for Ro60) and cloned into the corresponding sites of the pSU20 vector.
In vivo RNA chaperoning assay for La and Ro60
E. coli strain C600 (thy−) was transformed with tdwt or tdSH1 together with either the empty vector pSU20 or pSU20 encoding La, or Ro60, or StpA. All transformed cells were washed off the agar plate and used for an overnight culture to average out the possibility of assaying clones with low protein expression. Protein production was induced by addition of IPTG to a final concentration of 1 mM, and cells were grown to an OD600 of 0.2 at 37°C. For isolation of total cellular RNA, the lysozyme freeze–thaw method was used. Splicing of td constructs was analyzed by the poisoned primer reaction as described previously (Clodi et al. 1999). Briefly, primer extension, using γ-32P-ATP end-labeled primer NBS-2, which hybridizes in exon2 close to the 3′ splice site, was performed in the presence of dATP, dGTP, dCTP, ddTTP, resulting in termination of the extension products upon incorporation of the first ddTTP. Extension products were separated on 8% polyacrylamide gel. Bands specifying pre-mRNA and splicing products were quantified with a PhosphorImager using ImageQuant software, and percentage of splicing was calculated from three independent experiments (% splicing = [mRNA + -cryptic-mRNA] ×100/[pre-mRNA + cryptic-mRNA + mRNA]).
Acknowledgments
We thank Ger J.M. Pruijn (Radboud University of Nijmegen, The Netherlands) for providing the cDNAs for La and Ro60 and Gideon Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania) for providing the cDNAs for hnRNP I and hnRNP K. We are grateful to our colleague Andrea Barta for critical reading of the manuscript and her very helpful and enlightening comments. This work was supported by the Austrian Science Foundation (grant SFB 17, project parts F1703 and Z72 to R.S., F1704 to G.S., F1708 to E.W.).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7263905.
REFERENCES
- Ali, N. and Siddiqui, A. 1995. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J. Virol. 69: 6367–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, N., Pruijn, G.J., Kenan, D.J., Keene, J.D., and Siddiqui, A. 2000. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 275: 27531–27540. [DOI] [PubMed] [Google Scholar]
- Brown, E.C. and Jackson, R.J. 2004. All five cold-shock domains of unr (upstream of N-ras) are required for stimulation of human rhinovirus RNA translation. J. Gen. Virol. 85: 2279–2287. [DOI] [PubMed] [Google Scholar]
- Chakshusmathi, G., Kim, S.D., Rubinson, D.A., and Wolin, S.L. 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J. 22: 6562–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Quinn, A.M., and Wolin, S.L. 2000. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultra-violet irradiation. Genes & Dev. 14: 777–782. [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Smith, J.D., Shi, H., Yang, D.D., Flavell, R.A., and Wolin, S.L. 2003. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr. Biol. 13: 2206–2211. [DOI] [PubMed] [Google Scholar]
- Clodi, E., Semrad, K., and Schroeder, R. 1999. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 18: 3776–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee, T., Herschlag, D., and Belfort, M. 1994. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes & Dev. 8: 1575–1588. [DOI] [PubMed] [Google Scholar]
- Evans, J.R., Mitchell, S.A., Spriggs, K.A., Ostrowski, J., Bomsztyk, K., Ostarek, D., and Willis, A.E. 2003. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene 22: 8012–8020. [DOI] [PubMed] [Google Scholar]
- Expert-Bezancon, A., Le Caer, J.P., and Marie, J. 2002. Heterogeneous nuclear ribonucleoprotein (hnRNP) K is a component of an intronic splicing enhancer complex that activates the splicing of the alternative exon 6A from chicken β-tropomyosin pre-mRNA. J. Biol. Chem. 277: 16614–16623. [DOI] [PubMed] [Google Scholar]
- Fabini, G., Raijmakers, R., Hayer, S., Fouraux, M.A., Pruijn, G.J., and Steiner, G. 2001. The heterogeneous nuclear ribonucleoproteins I and K interact with a subset of the Ro ribonucleoprotein-associated Y RNAs in vitro and in vivo. J. Biol. Chem. 276: 20711–20718. [DOI] [PubMed] [Google Scholar]
- Fan, H., Goodier, J.L., Chamberlain, J.R., Engelke, D.R., and Maraia, R.J. 1998. 5′ processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol. Cell. Biol. 18: 3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris, A.D., O’Brien, C.A., and Harley, J.B. 1995. Y3 is the most conserved small RNA component of Ro ribonucleoprotein complexes in vertebrate species. Gene 154: 193–198. [DOI] [PubMed] [Google Scholar]
- Fouraux, M.A., Bouvet, P., Verkaart, S., van Venrooij, W.J., and Pruijn, G.J. 2002. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J. Mol. Biol. 320: 475–488. [DOI] [PubMed] [Google Scholar]
- Geissmann, T.A. and Touati, D. 2004. Hfq, a new chaperoning role: Binding to messenger RNA determines access for small RNA regulator. EMBO J. 23: 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti, A., Pinol-Roma, S., Michael, W.M., Morandi, C., and Dreyfuss, G. 1992. hnRNP I, the polypyrimidine tract-binding protein: Distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 20: 3671–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, C., Lund, E., and Dahlberg, J.E. 1997. In vivo selection of RNAs that localize in the nucleus. EMBO J. 16: 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U., Witherell, G.W., Schmid, M., Shin, S.H., Pestova, T, V., Gil, A., and Wimmer, E. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. 90: 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag, D. 1995. RNA chaperones and the RNA folding problem. J. Biol. Chem. 270: 20871–20874. [DOI] [PubMed] [Google Scholar]
- Intine, R.V., Sakulich, A.L., Koduru, S.B., Huang, Y., Pierstorff, E., Goodier, J.L., Phan, L., and Maraia, R.J. 2000. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol. Cell 6: 339–348. [DOI] [PubMed] [Google Scholar]
- Izumi, R.E., Valdez, B., Banerjee, R., Srivastava, M., and Dasgupta, A. 2001. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res. 76: 17–29. [DOI] [PubMed] [Google Scholar]
- James, M.C., Jeffrey, I.W., Pruijn, G.J., Thijssen, J.P., and Clemens, M.J. 1999. Translational control by the La antigen. Structure requirements for rescue of the double-stranded RNA-mediated inhibition of protein synthesis. Eur. J. Biochem. 266: 151–162. [DOI] [PubMed] [Google Scholar]
- Kufel, J., Allmang, C., Chanfreau, G., Petfalski, E., Lafontaine, D.L., and Tollervey, D. 2000. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20: 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe, J.C., Hekimi, S., and Rokeach, L.A. 1999. The levels of the RoRNP-associated Y RNA are dependent upon the presence of ROP-1, the Caenorhabditis elegans Ro60 protein. Genetics 151: 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe, J.C., Burgess, J., Rokeach, L.A., and Hekimi, S. 2000. ROP-1, an RNA quality-control pathway component, affects Caenorhabditis elegans dauer formation. Proc. Natl. Acad. Sci. 97: 13233–13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.H. and Patton, J.G. 1995. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA 1: 234–245. [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas, E., Ramos, R., Lafuente, E., and Lopez de Quinto, S. 2001. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 82: 973–984. [DOI] [PubMed] [Google Scholar]
- Mayer, O., Waldsich, C., Grossberger, R., and Schroeder, R. 2002. Folding of the td pre-RNA with the help of the RNA chaperone StpA. Biochem. Soc. Trans. 30: 1175–1180. [DOI] [PubMed] [Google Scholar]
- Michelotti, E.F., Michelotti, G.A., Aronsohn, A.I., and Levens, D. 1996. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 16: 2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Coldwell, M.J., Jackson, R.J., and Willis, A.E. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11: 757–771. [DOI] [PubMed] [Google Scholar]
- O’Brien, C.A. and Wolin, S.L. 1994. A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes & Dev. 8: 2891–2903. [DOI] [PubMed] [Google Scholar]
- O’Brien, C.A., Margelot, K., and Wolin, S.L. 1993. Xenopus Ro ribonucleoproteins: Members of an evolutionarily conserved class of cytoplasmic ribonucleoproteins. Proc. Natl. Acad. Sci. 90: 7250–7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck, D.H., Ostareck-Lederer, A., Wilm, M., Thiele, B.J., Mann, M., and Hentze, M.W. 1997. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89: 597–606. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer, A., Ostareck, D.H., Cans, C., Neubauer, G., Bomsztyk, K., Superti-Furga, G., and Hentze, M.W. 2002. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 22: 4535–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone, B.K., Xue, D., and Wolin, S.L. 1998. A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 15: 7442–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek, R., Pruijn, G.J., van der Kemp, A.J., and van Venrooij, W.J. 1993. Subcellular distribution of Ro ribonucleoprotein complexes and their constituents. J. Cell Sci. 106(Pt 3): 929–935. [DOI] [PubMed] [Google Scholar]
- Pellizzoni, L., Lotti, F., Rutjes, S.A., and Pierandrei-Amaldi, P. 1998. Involvement of the Xenopus laevis Ro60 autoantigen in the alternative interaction of La and CNBP proteins with the 5′ UTR of L4 ribosomal protein mRNA. J. Mol. Biol. 281: 593–608. [DOI] [PubMed] [Google Scholar]
- Pichler, A. and Schroeder, R. 2002. Folding problems of the 5′ splice-site containing P1 stem of the group I td intron: Substrate binding inhibition in vitro and mis-splicing in vivo. J. Biol. Chem. 277: 17987–17993. [DOI] [PubMed] [Google Scholar]
- Pickering, B.M., Mitchell, S.A., Spriggs, K.A., Stoneley, M., and Willis, A.E. 2004. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell. Biol. 24: 5595–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijn, G.J., Slobbe, R.L., van Venrooij, W.J. 1991. Analysis of protein–RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res. 19: 5173–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijn, G.J., Wingens, P.A., Peters, S.L., Thijssen, J.P., and van Venrooij, W.J. 1993. Ro RNP associated Y RNAs are highly conserved among mammals. Biochim. Biophys. Acta 1216: 395–401. [DOI] [PubMed] [Google Scholar]
- Pruijn, G.J., Simons, F.H., and van Venrooij, W.J. 1997. Intracellular localization and nucleocytoplasmic transport of Ro RNP components. Eur. J. Cell. Biol. 74: 123–132. [PubMed] [Google Scholar]
- Ritchie, S.A., Pasha, M.K., Batten, D.J., Sharma, R.K., Olson, D.J., Ross, A.R., and Bonham, K. 2003. Identification of the SRC pyrimidine-binding protein (SPy) as hnRNP K: Implications in the regulation of SRC1A transcription. Nucleic Acids Res. 31: 1502–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjes, S.A., Lund, E., van der Heijden, A., Grimm, C., van Venrooij, W.J., and Pruijn, G.J. 2001. Identification of a novel cis-acting RNA element involved in nuclear export of hY RNAs. RNA 7: 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvo, J.L., Coetzee, T., and Belfort, M. 1990. Deletion-tolerance and trans-splicing of the bacteriophage T4 td intron. Analysis of the P6-L6a region. J. Mol. Biol. 211: 537–549. [DOI] [PubMed] [Google Scholar]
- Semrad, K., Green, R., and Schroeder, R. 2004. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA 10: 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., O’Brien, C.A., Van Horn, D.J., and Wolin, S.L. 1996. A misfolded form of 5S rRNA is complexed with the Ro and La autoantigens. RNA 2: 769–784. [PMC free article] [PubMed] [Google Scholar]
- Simons, F.H., Rutjes, S.A., van Venrooij, W.J., and Pruijn, G.J. 1996. The interactions with Ro60 and La differentially affect nuclear export of hY1 RNA. RNA 2: 264–273. [PMC free article] [PubMed] [Google Scholar]
- Stoneley, M. and Willis, A.E. 2004. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene 23: 3200–3207. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Meerovitch, K., Lee, H.S., Dholakia, J.N., Kenan, D.J., Agol, V.I., and Sonenberg, N. 1994. Internal translation initiation on poliovirus RNA: Further characterization of La function in poliovirus translation in vitro. J. Virol. 68: 1544–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto, M., Tomonaga, T., Matunis, M., Avigan, M., Krutzsch, H., Dreyfuss, G., and Levens, D. 1993. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J. Biol. Chem. 268: 18249–18258. [PubMed] [Google Scholar]
- Wagner, E.J. and Garcia-Blanco, M.A. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21: 3281–3288.11313454 [Google Scholar]
- Waldsich, C., Grossberger, R., and Schroeder, R. 2002. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes & Dev. 16: 2300–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, Z., Thomas, S.M., Rickles, R.J., Taylor, J.A., Brauer, A.W., Seidel-Dugan, C., Michael, W.M., Dreyfuss, G., and Brugge, J.S. 1994. Identification of Src, Fyn, and Lyn SH3-binding proteins: Implications for a function of SH3 domains. Mol. Cell. Biol. 14: 4509–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin, S.L. and Cedervall, T. 2002. The La protein. Annu. Rev. Biochem. 71: 375–403. [DOI] [PubMed] [Google Scholar]
- Wolin, S.L. and Steitz, J.A. 1984. The Ro small cytoplasmic ribonucleoproteins: Identification of the antigenic protein and its binding site on the Ro RNAs. Proc. Natl. Acad. Sci. 81: 1996–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, D., Rubinson, D.A., Pannone, B.K., Yoo, C.J., and Wolin, S.L. 2000. U snRNP assembly in yeast involves the La protein. EMBO J. 19: 1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, D., Shi, H., Smith, J.D., Chen, X., Noe, D.A., Cedervall, T., Yang, D.D., Eynon, E., Brash, D.E., Kashgarian, M., et al. 2003. A lupus-like syndrome develops in mice lacking the Ro 60-kDa protein, a major lupus autoantigen. Proc. Natl. Acad. Sci. 100: 7503–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, C.J. and Wolin, S.L. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89: 393–402. [DOI] [PubMed] [Google Scholar]
- Youinou, P., Adler, Y., Muller, S., Lamour, A., Baron, D., and Humbel, R.L. 1994. Anti-Ro(SSA) and anti-La(SSB) antibodies in autoimmune rheumatic diseases. Clin. Rev. Allergy 12: 253–274. [DOI] [PubMed] [Google Scholar]
- Zhang, A., Derbyshire, V., Salvo, J.L., and Belfort, M. 1995. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA 1: 783–793. [PMC free article] [PubMed] [Google Scholar]