Abstract
Cytoplasmic polyadenylation is important in the control of mRNA stability and translation, and for early animal development and synaptic plasticity. Here, we focus on vertebrate poly(A) polymerases that are members of the recently described GLD2 family. We identify and characterize two closely related GLD2 proteins in Xenopus oocytes, and show that they possess PAP activity in vivo and in vitro and that they bind known polyadenylation factors and mRNAs known to receive poly(A) during development. We propose that at least two distinct polyadenylation complexes exist in Xenopus oocytes, one of which contains GLD2; the other, maskin and Pumilio. GLD2 protein interacts with the polyadenylation factor, CPEB, in a conserved manner. mRNAs that encode GLD2 in mammals are expressed in many tissues. In the brain, mouse, and human GLD2 mRNAs are abundant in anatomical regions necessary for long-term cognitive and emotional learning. In the hippocampus, mouse GLD2 mRNA colocalizes with CPEB1 and Pumilio1 mRNAs, both of which are likely involved in synaptic plasticity. We suggest that mammalian GLD2 poly(A) polymerases are important in synaptic translation, and in polyadenylation throughout the soma.
Keywords: poly(A) polymerase, GLD2, translational control
INTRODUCTION
Control of the movement, translation and stability of mRNAs determines when, where, and how much protein an mRNA generates (for reviews, see Sonenberg 1994; Hentze 1995; Wickens et al. 2002; Kuersten and Goodwin 2003). A tract of adenosines at the 3′ end of the mRNA—the poly(A) tail—plays a key role in these events. Cytoplasmic lengthening of the tail can activate translation and stabilize the mRNA, while poly(A) removal can cause translational repression and mRNA decay. Regulated changes in the length of the poly(A) tail are critical during early development; for example, they control the meiotic and mitotic cell cycles of Xenopus oocytes and embryos by regulating specific mRNAs (for reviews, see Mendez and Richter 2001; Wickens et al. 2002). In the nervous system, repeated stimulation of synapses activates polyadenylation and local translation (Wu et al. 1998; Huang et al. 2002; Si et al. 2003a; Theis et al. 2003). These polyadenylation events are thought to be important in long-term potentiation (LTP) and learning (Wu et al. 1998; Alarcon et al. 2004).
Cytoplasmic polyadenylation in frog oocytes requires multiple protein components, including Cytoplasmic Polyadenylation Element Binding Protein (CPEB) and Cleavage and Polyadenylation Specificity Factor (CPSF) (for reviews, see Mendez and Richter 2001). CPEB binds directly to specific sequences in the 3′UTRs of target mRNAs (Hake et al. 1998). CPSF, a multiprotein complex, binds the sequence AAUAAA and is necessary for both nuclear and cytoplasmic polyadenylation (Bilger et al. 1994). CPEB binds CPSF, which is thought then to recruit the enzyme that adds the poly(A), a cytoplasmic poly(A) polymerase (PAP) (Mendez et al. 2000; Dickson et al. 2001). Although oocytes contain cytoplasmic PAPs related to the nuclear enzyme (Ballantyne et al. 1995; Gebauer and Richter 1995), their role in cytoplasmic polyadenylation is unclear.
GLD-2, a divergent cytoplasmic PAP, was identified in Caenorhabditis elegans (Wang et al. 2002), and is related to the Cid1 and Cid13 PAPs of Schizosaccharomyces pombe (Read et al. 2002; Saitoh et al. 2002). GLD-2 polymerization activity is stimulated by interaction with an RNA binding protein, GLD-3 (Wang et al. 2002). Together GLD-2 and GLD-3 are thought to form a novel heterodimeric PAP, in which the RNA binding component, GLD-3, recruits the catalytic subunit, GLD-2, to specific mRNAs (Wang et al. 2002; Kwak et al. 2004). Homologs of GLD-2 that possess polyadenylation activity recently were identified in mice and humans (Kwak et al. 2004). Similarly, a Xenopus protein related to GLD-2 was identified by virtue of its association with CPEB and shown to participate in cytoplasmic polyadenylation in oocytes (Barnard et al. 2004).
Repression of specific mRNAs in oocytes and embryos involves multiple RNA binding proteins. In Xenopus oocytes, maskin, Pumilio, and Nanos (Xcat-2) all appear to be bound to repressed RNAs and involved in repression (Stebbins-Boaz et al. 1999; Nakahata et al. 2001, 2003; for review, see Richter 2000). Maskin binds CPEB on the 3′UTR, and sequesters eIF4E to repress translation (Stebbins-Boaz et al. 1999; for review, see Richter 2000). Similarly, Nanos and PUF (e.g., Pumilio) proteins interact physically and assemble on specific sequences in the 3′UTR (Kraemer et al. 1999; Sonoda and Wharton 1999; Nakahata et al. 2001; for review, see Wickens et al. 2002). These multiprotein complexes are required for repression. Release from repression is accompanied by cytoplasmic polyadenylation.
Translational regulation of dendritic mRNAs is important in synaptic plasticity. Stimulation of synapses results in locally increased protein synthesis, which requires cytoplasmic polyadenylation and CPEB (Si et al. 2003a). This local translation is required for the late phase of LTP, an electro-physiological, cellular correlate of memory (Nguyen et al. 1994; Frey et al. 1988; Liu and Schwartz 2003; for reviews, see Wells et al. 2000; Richter 2001; Tang and Schuman 2002). Four isoforms of CPEB are found in the hippocampus (Wu et al. 1998; Theis et al. 2003). Knockout mice lacking one of these, mCPEB1, exhibit a modest deficit in LTP (Alarcon et al. 2004). After LTP induction, cytoplasmic polyadenylation regulates the translation of proteins enriched in synaptic spines, including αCaMKII (Wu et al. 1998; Miller et al. 2002; Otmakhov et al. 2004), cytoskeletal actin (Fukazawa et al. 2003; Liu and Schwartz 2003; Matsuzaki et al. 2004), Erg1 (Simon et al. 2004), and tissue plasminogen activator (TPA) (Shin et al. 2004). Both Erg1 and TPA are necessary for LTP and long-term memory formation (Jones et al. 2001; Pawlak et al. 2002; Malkani et al. 2004; Pang et al. 2004).
In this paper, we focus on GLD2 in vertebrates. We identify two Xenopus GLD-2 enzymes and analyze their interaction with known polyadenylation factors, confirming and extending the work of Barnard et al. (2004). We demonstrate that the mammalian enzymes associate with Xenopus polyadenylation factors, and that they are expressed in regions of the hippocampus associated with learning and memory. Their expression pattern in the hippocampus parallels that of CPEB1 and Pumilio1, both of which are implicated in synaptic plasticity. Based on these findings. we suggest the existence of two distinct translational control complexes in oocytes, and propose that the GLD2 PAP participates in translational activation at synapses.
RESULTS
Vertebrate GLD2 homologs: proteins and mRNAs
We recently identified C. elegans, murine, and human homologs of C. elegans GLD-2 (Wang et al. 2002; Kwak et al. 2004). To identify the enzyme that catalyzes cytoplasmic polyadenylation in frog oocytes, we designed degenerate primers directed against regions conserved among GLD2 homologs, and performed PCR to identify GLD2-related cDNAs in a Xenopus oocyte cDNA library. We determined the sequences of two independent isolates of Xenopus GLD-2 cDNA that comprised the entire ORF, and six cDNAs from the NIBB (Japan) and the IMAGE consortium. These sequences were consolidated with those of 40 fragmentary ESTs (see Materials and Methods).
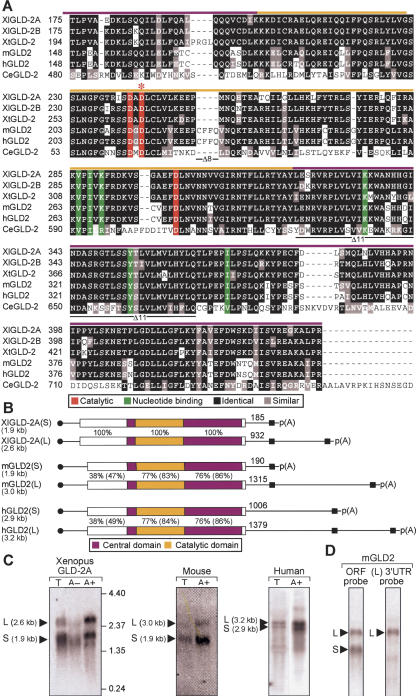
Two different isoforms of Xenopus leaevis GLD-2, termed XlGLD-2A and XlGLD-2B, were identified. The two predicted proteins are 88% identical and differ in nucleotide sequence at multiple locations throughout their length. All cDNAs and ESTs belonged to one group or the other, suggesting that XlGLD-2A and XlGLD-2B are different genes. XlGLD-2B corresponds to the protein recently shown to be involved in cytoplasmic polyadenylation (Barnard et al. 2004). XlGLD-2A and XlGLD-2B each are 62% identical to human and mouse GLD-2 (hGLD2 and mGLD2) (Kwak et al. 2004) and 42% identical to catalytic and central domains of the C. elegans protein. The predicted Xenopus proteins possess the hallmarks of the β-nucleotidyl transferase superfamily, including specific amino acids that participate in catalysis and bind the nucleotide (Fig. 1A, red and green residues; Aravind and Koonin 1998; Martin et al. 2004).
FIGURE 1.
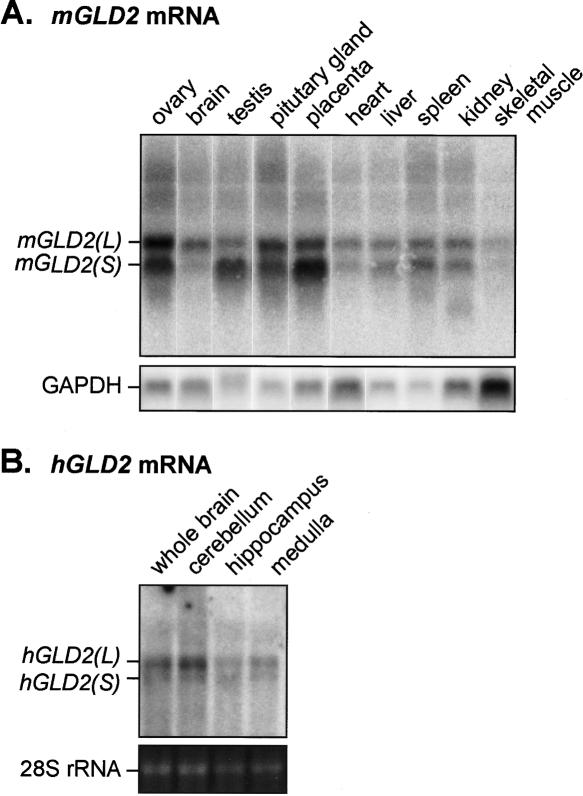
XlGLD-2, mGLD2, and hGLD2 mRNAs and proteins. (A) Predicted protein sequences in the catalytic and central domains of GLD2 homologs. The sequences from five animal different species (X. laevis, X. tropicalis, mouse, human, and C. elegans) are presented. Black indicates amino acid identity; red, three carboxylate side-chains required for catalysis; green, six residues that position the nucleotide (Martin et al. 2000; Wang et al. 2002); and red asterisk, location of the active site mutation, D242A. Colored bars above the sequence indicate the central (purple) or catalytic (yellow) domains. Black lines below the sequence indicate the regions missing in Δ8 and Δ11 forms of the human protein. (B) Vertebrate GLD2 mRNAs. Approximate lengths of mRNAs, as calculated by cDNA sequencing and confirmed by Northern blotting, are shown to the left of each panel. Purple indicates PAP central domain; yellow, PAP catalytic domail (corresponding to Fig 1A). Percentage of amino acid sequence identity relative to XlGLD-2A is given (percentage similarity is in parentheses). Black circles and boxes indicate 5′ cap and 3′ cleavage and polyadenylation signal (AAUAAA and AAUACA). Distances from the termination codon to the poly(A) tail are indicated. (C) Two mRNA forms. RNAs from Xenopus oocytes, mouse 3T3 cells, and human spleen were analyzed by Northern blotting. T indicates total RNA; A+, RNA retained by oligo(dT) cellulose; and A-, RNA not retained by oligo (dT). Amounts of RNA and hybidization probes are described in Materials and Methods. (D) mGLD2(L) and mGLD2(S) mRNAs differ by their 3′UTRs. Total RNA from spleen was analyzed by using either a probe complementary to the entire ORF (left) or just the 3′UTR of the long form of mRNA, mGLD2(L) (right).
We analyzed by Northern blotting the mRNA produced from the XlGLD-2 genes, and compared it to the mouse and human GLD2 mRNAs. In all three species, two mRNA forms were detected, which we term GLD2(L) and GLD2(S) (Fig. 1B,C). Each apparent 3′ end was deduced from the location of poly(A) on multiple cDNAs and ESTs, and was preceded by a polyadenylation signal (Fig. 1B). The two mRNAs produced from one gene in each species are identical in the protein coding region but differ in the length of their 3′UTRs. This was deduced from multiple cDNAs and corroborated by using a probe that was predicted to detect only mGLD2(L) form (Fig 1D). In Xenopus, XlGLD-2A produces two forms; XlGLD-2B expresses one mRNA with a long 3′UTR (data not shown). Human cDNAs representing alternatively spliced variants that lack exon 8 or exon 11 were detected among ESTs (Δ8 and Δ11; indicated in Fig. 1A). Throughout the protein coding region, each of the vertebrate genes display identical exonintron organizations (data not shown).
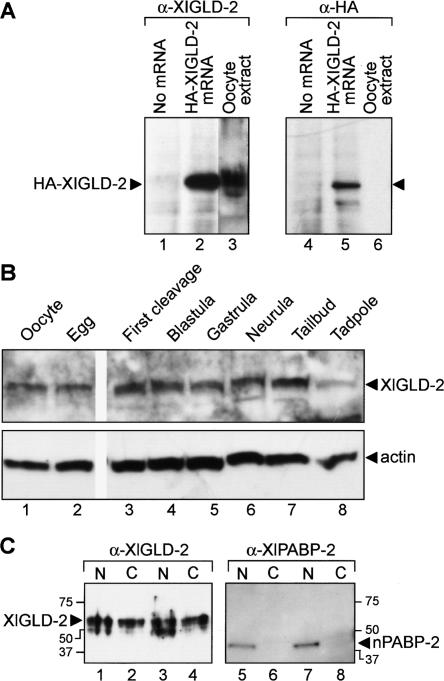
We next focused on the activity and biological role of GLD2 protein in Xenopus oocytes. Antibodies raised against XlGLD-2A were used to examine the abundance and subcellular distribution of endogenous XlGLD-2 protein. The antibodies were specific: In vitro translation of an mRNA encoding HA-tagged GLD-2A yielded a single protein of −60 kDa that was detected by α-XlGLD-2 and α-HA antibodies (Fig. 2A, lanes 2,5). This protein comigrates with the endogenous oocyte protein (Fig. 2, lanes 2,3). The antibody recognizes both XlGLD-2A and XlGLD-2B proteins (data not shown). A single prominent polypeptide was detected throughout oogenesis, oocyte maturation, and post-fertilization development (Fig. 2B; data not shown).
FIGURE 2.
XlGLD-2 is present throughout embryogenesis and is in the cytoplasm. (A) Antibody specificity. mRNA encoding HA-XlGLD-2A was translated in vitro, and the products were analyzed by Western blotting using α-XlGLD-2 antibodies. A predominant protein of 60 kDa is detected by α-XlGLD-2 (lanes 1–3) and α-HA antibodies (lanes 4–6). The in vitro translated protein (lane 2) roughly comigrates with endogenous XlGLD-2 protein in oocyte extracts (lane 3). (B) XlGLD-2 protein abundance. Proteins extracted from the indicated stages were analyzed by Western blotting using either α-XlGLD-2 or α-actin. (C) XlGLD-2 subcellular distribution. Nuclear and cytoplasmic fractions prepared manually from stage VI oocytes were analyzed by Western blotting with α-XlGLD-2 (lanes 1–4). α-XlPABP-2 (lanes 5–8), which detects a nuclear poly(A) binding protein (Good et al. 2004), was used to control for proper enucleation and a lack of leakage into the cytoplasm. N and C indicate nucleus and cytoplasm, respectively.
The factors needed to catalyze regulated polyadenylation during oocyte maturation are cytoplasmic (Fox et al. 1989). XlGLD-2 protein was detected in the oocyte cytoplasm, as well as the nucleus (Fig. 2C). α-XlPABP2 antibodies confirmed that the manual separation of nuclei and cytoplasms was successful, since nPABP2 was predominantly nuclear (Good et al. 2004).
XLGLD-2 is a PAP
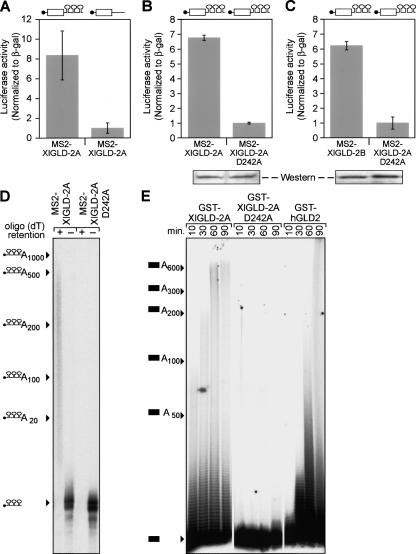
To test whether the Xenopus GLD2 proteins possessed PAP activity, we tethered XlGLD-2A to an mRNA reporter using MS2 coat protein. Oocytes first were injected with mRNAs that direct the synthesis of MS2-XlGLD-2A fusion proteins. After allowing time for protein to accumulate, the same oocytes were injected with luciferase reporter mRNAs containing MS2 sites. A β-galactosidase reporter mRNA lacking MS2 sites was coinjected as a control. MS2-XlGLD-2A and MS2-XlGLD-2B proteins stimulated translation of mRNAs containing MS2 sites but did not affect translation of mRNAs lacking them (Fig. 3A–C). A putative active site mutation, D242A, abolished translational stimulation but accumulated to the same level as did the wild-type protein (Fig. 3B). To analyze polyadenylation directly, we injected 32P-labeled RNAs bearing MS2 sites. The labeled RNA was polyadenylated in oocytes containing wild-type MS2-XlGLD-2A, but not the D242A, form of fusion protein (Fig. 3D). Furthermore, we purified recombinant, full-length human GLD2 and XlGLD-2A proteins from bacteria. These enzymes added long tails to 32P-labeled RNAs in vitro, while a D242A mutant form of XlGLD-2A protein did not (Fig. 3E). The added tails were composed of poly-adenosine, as they were removed by oligo(dT)/RNAse treatment (data not shown).
FIGURE 3.
Vertebrate GLD2 homologs are poly(A) polymerases in vivo and in vitro. (A) Translational stimulation in vivo. Oocytes containing MS2/XlGLD-2A protein increase the expression of a luciferase reporter mRNA that contains MS2 binding sites in its 3′UTR (left), but does not increase expression with an mRNA reporter that lacks MS2 binding sites (right). Luciferase activity is normalized to that of β-galactosidase, generated from a coinjected mRNA that lacks MS2 binding sites. (B) An active site mutation abolishes translational stimulation. Oocytes containing MS2-XlGLD-2A protein with an active site mutation, D242A, do not stimulate translation of the luciferase reporter mRNA that contains MS2 sites. The wild-type protein was used as a control. Western blotting with α-HA antibodies demonstrates the wild-type and mutant proteins are comparable in abundance. (C) XlGLD-2B is active in vivo. Oocytes containing MS2-XlGLD-2B protein increase the expression of a luciferase reporter mRNA that contains MS2 binding sites in its 3′UTR (left). The point mutant protein was used as a control and is inactive. Western blotting with α~HA antibodies demonstrated the wild-type and mutant proteins are comparable in abundance (shown below the histogram). (D) Poly-adenylation in vivo. A 32P-labeled RNA containing three MS2 sites was injected into oocytes expressing either MS2-XlGLD-2 or MS2-XlGLD-2A (D242A). Oocytes were collected after 16 h, and the RNA was extracted and fractionated using biotinylated oligo(dT). + indicates RNAs that bound the resin; −, RNAs that did not bind the resin. Positions of RNAs carrying various lengths of poly(A) (determined by using markers) are shown to the left. (E) Poly-adenylation in vitro. Recombinant GST-XlGLD-2A, GST-XlGLD-2A(D242A), and GST-hGLD2 proteins, purified from Escherichia coli, were incubated at 24°C with L1 RNA (see Materials and Methods) and a mixture of all four ribonucleoside triphosphates at 1 mM. Samples were collected at the indicated time points and analyzed by denaturing gel electrophoresis. Positions of RNAs carrying various lengths of poly(A) (determined by using markers) are shown to the left.
We conclude that the two Xenopus proteins, XlGLD-2A and XlGLD-2B, as well as human GLD2, possess PAP activity. In the following experiments, we focus first on the interaction of XlGLD-2A with proteins involved in cytoplasmic polyadenylation and translational control, and then turn to mouse and human GLD2.
XlGLD-2A interacts with polyadenylation factors and target mRNAs
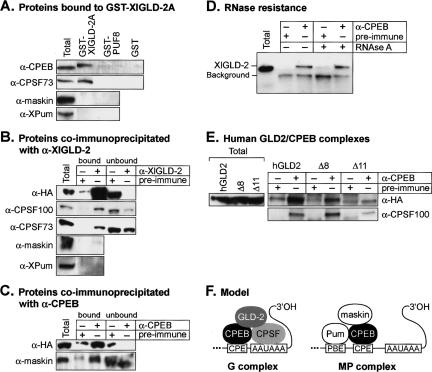
To test whether XlGLD-2 interacted with CPEB or CPSF, recombinant GST-XlGLD-2A was attached to Sepharose-glutathione beads and incubated with crude lysates of stage VI oocytes. The beads were washed with buffer, and bound proteins were eluted and analyzed by Western blotting (Fig. 4A). CPEB bound to GST-XlGLD-2A but not to either GST-PUF-8 or GST alone, used as controls (Fig. 4A). A subunit of CPSF, CPSF73, also bound specifically to GST-XlGLD-2A.
FIGURE 4.
XlGLD-2 interacts with polyadenylation factors. (A) Proteins bound to GST-XlGLD-2A. Stage VI oocyte lysate was inubated with purified GST-XlGLD-2A, GST-PUF-8, or GST, bound to glutathione-Sepharose. Proteins retained on beads after three washes were analyzed by Western blotting with antibodies specific to CPEB, CPSF-73, maskin, and XlPu-milio. Total indicates unfractionated lysate (fourfold less starting material than in the other lanes). (B) Proteins coimmunoprecipitated with α-XlGLD-2. Crude lysates were prepared from oocytes that were expressing HA/XlGLD-2. Lysates were incubated with α-XlGLD-2 (top panel) or α-CPEB (bottom panel), or with respective preimmune sera, as indicated above each lane. Proteins were detected by Western blot analysis using α-CPSF100, α-CPSF73, α-maskin, and α-XlPumilio, as indicated. Western analysis using α-HA as probe demonstrated that the α-XlGLD-2 antibody efficiently immunoprecipitates the endogenous protein. Bound indicates proteins that bound to the beads; unbound, proteins that did not bind to the beads. Total indicates unfractionated lysate (threefold less starting material than in bound, and equal starting material to unbound). (C) Proteins coimmunoprecipitated with α-CPEB. Oocytes were injected with mRNAs that encode HA-XlGLD-2. Oocyte lysates were incubated with α-CPEB, or with preimmune serum. Both XlGLD-2 and maskin are associated with α-CPEB. Total indicates unfractionated lysate (threefold less starting material than in bound, and equal starting material to unbound). (D) Ribonuclease-resistance of the CPEB/GLD-2 interaction. Xenopus oocyte extracts, prepared as in B, were treated with RNAseA. Gel electrophoresis demonstrated that the RNA had been degraded (data not shown). Coimmunoprecipitations were performed with α-CPEB or preimmune sera, on X. laevis oocyte extracts as in B. The two proteins continue to coimmunoprecipitate after RNAse treatment. (E) Human GLD2/CPEB complexes. Immunoprecipitations using either α-CPEB or preimmune antibodies were incubated with X. laevis oocyte extracts prepared from oocytes expressing either HA-hGLD2, HA-hGLD2Δ8, or HA-hGLD2Δ11. Total indicates crude lysate. (F) Model of polyadenylation complexes in resting oocytes. See text for details. CPSF and CPEB are shown interacting in the resting oocyte (Mendez et al. 2000; Dickson et al. 2001). However, CPEB phosphorylation increases binding affinity to CPSF (Mendez et al. 2000). Symplekin is likely to be in the G complex, as inferred from the data of Barnard et al. (2004).
Maskin and Pumilio proteins, both of which are involved in translational repression, physically interact with CPEB in resting oocytes (Stebbins-Boaz et al. 1999; Nakahata et al. 2003). In contrast, GST-XlGLD-2A binds CPEB, but it did not bind either maskin or Pumilio proteins (Fig. 4A).
To test whether endogenous XlGLD-2 interacts with CPSF, we performed coimmunoprecipitation experiments (Fig. 4B). α-XlGLD-2 antibodies were incubated with crude oocyte lysates, and the bound proteins were analyzed by Western blotting. Oocytes first were injected with mRNA encoding HA-XlGLD-2A. As expected, HA-tagged XlGLD-2A was efficiently immunoprecipitated by the α-XlGLD-2 antibodies (Fig 4B, “α-HA”). Endogenous CPSF100 and CPSF73 were immunoprecipitated by α-XlGLD-2; maskin and Pumilio were not (Fig. 4B).
In a reciprocal experiment, we used α-CPEB antibodies to immunoprecipitate complexes from oocyte lysates (Fig. 4C). α-CPEB antibodies efficiently precipitated HA-XlGLD-2A from the extracts, confirming that CPEB and XlGLD-2 interact (Fig. 4C). The coimmunoprecipitation of the two proteins was resistant to digestion with RNa-seA that was sufficient to degrade the endogenous RNA (Fig. 4D; data not shown). Thus the coimmunoprecipitation of CPEB and XlGLD-2 was not due to co-occupancy of a single RNA. α-CPEB also immunoprecipitated CPSF and maskin, as reported previously (Fig. 4C; Dickson et al. 1999; Stebbins-Boaz et al. 1999; data not shown). Together, the interaction data demonstrate that XlGLD-2 binds CPSF and CPEB, consistent with the results of Barnard et al. (2004). We conclude that XlGLD-2 interacts with CPEB and CPSF, but does not interact with maskin or Pumilio.
To test whether these interactions between GLD-2 and Xenopus polyadenylation components were conserved, we expressed wild-type human GLD2, and the Δ8 and Δ11 forms of human GLD2, in oocytes. The wild-type and Δ8 forms of hGLD2 were immunoprecipitated by α-CPEB; the Δ11 form was not (Fig. 4E). Similarly, the Δ8 form was active in polyadenylation assays, while Δ11 was inactive (data not shown).
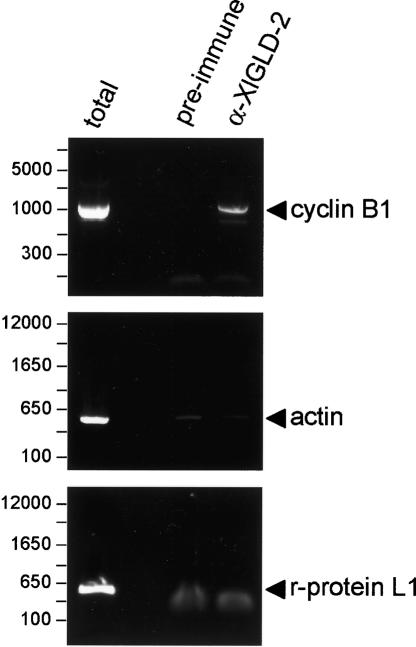
If XlGLD-2 catalyzes cytoplasmic polyadenylation, then it must associate with mRNAs that receive poly(A). We asked whether cyclin B1 mRNA, an mRNA that is polyadenylated during oocyte maturation, was bound to XlGLD-2 in stage VI oocyte (Fig. 5). Oocyte lysates were incubated with α-XlGLD-2 antibodies, and the immunoprecipitated RNAs were extracted and analyzed by RT-PCR (Fig. 5). Cyclin B1 mRNA was readily detected in immunoprecipitates obtained with α-XlGLD-2, but not with control, preimmune antibodies (Fig. 5). Actin and riboso-mal protein L1 mRNAs were not specifically immunoprecipitated by α-GLD2, as they were present at equivalent, low levels in the α-XlGLD-2 and preimmune immunoprecipitates (Fig. 5). c-mos mRNA, which also receives poly(A) during maturation, was associated with XlGLD-2 as well (data not shown).
FIGURE 5.
XlGLD-2 interacts with target, but not nontarget, mRNAs. Oocyte extracts were incubated with α-XlGLD-2 or preimmune guinea pig serum and bound to Protein A–Sepharose. Bound material was eluted and the RNA recovered. mRNAs were detected by semiquantitative PCR, using oligo(dT)-primed reverse transcription reactions, followed by PCR using gene-specific primers. Cyclin B1 mRNA receives poly(A) during oocyte maturation, and is immuno-precipitated by α-XlGLD-2. Cytoskeletal actin and ribosomal protein L1 mRNAs do not receive poly(A) during maturation and are not immunoprecipitated.
Taken together, our findings imply that at least two complexes exist in resting, stage VI oocytes (Fig. 4F). One, which we term the G complex (for GLD-2), contains GLD-2 and CPEB. The other complex, which we term MP (for maskin and Pumilio), contains CPEB, maskin, and Pumilio. CPSF is present in one or both complexes. The finding that XlGLD-2 associates with symplekin, and is stimulated by that protein in combination with CPSF, implies that symplekin and CPSF are in the G complex (Barnard et al. 2004).
Expression of mammalian GLD2 homologs
Our findings that mouse and human GLD2 are PAPs and that they interact with polyadenylation factors in the Xenopus oocyte cytoplasm, suggested that the mammalian proteins were involved in regulated polyadenylation. We examined the tissue distribution of GLD2 mRNAs in mouse and human tissues by Northern blotting (Fig. 6). GLD2 mRNAs are expressed in a wide range of tissues, including the ovary, brain, and testes. Both mGLD2(S) and mGLD2(L) were detected in each tissue. Their relative abundance varied: for example, in the mouse brain, mGLD2(L) mRNA was much more abundant than mGLD2(S), while the opposite is true in placenta. GLD2(L) mRNAs also were detected in RNA from human brain and in RNAs prepared from isolated human cerebellum, hippocampus, and medulla (Fig. 6B). Again, the GLD2(L) form predominated (Fig. 6B). Subcellular fractionation studies suggested that mGLD2 mRNA was enriched in synaptic fractions (data not shown).
FIGURE 6.
Analysis of mGLD2 and hGLD2 mRNAs. (A) Tissue distribution of mGLD2 mRNA; 10 μg of total RNA from each of the indicated tissues was analyzed by Northern blotting, using a probe directed against the entire ORF region of mGLD2. GAPDH mRNA served as a loading control. (B) Distribution of hGLD2 mRNAs within human brain; 2 μg of RNA prepared from each of the indicated regions of the brain (Ambion) was analyzed by hybridization with a probe directed against a portion of the hGLD2 ORF. 28S rRNA stained with ethidium bromide served as a loading control.
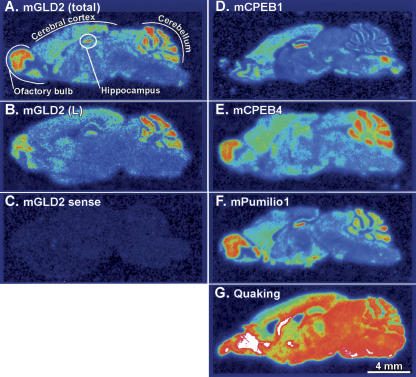
To determine more precisely the distribution of mGLD2 mRNA within mouse brain, we performed in situ hybridization on sagittal sections by using 35S-labeled RNA probes (Fig. 7A). An antisense probe that detects both mGLD2 mRNA isoforms yielded signal throughout the brain in a pattern consistent with the presence of mGLD2 transcripts within neurons. The distribution of mGLD2(L) mRNA, detected by using an L-form–specific probe, was identical (Fig. 7, cf. A and B). As expected, a sense-strand probe yielded no significant signal (Fig. 7C). The abundance of mGLD2 mRNA was highest in the cerebral cortex, cerebellum, hippocampus, and olfactory bulb. mGLD2 mRNA abundance paralleled neuronal density. In the hippocampus and cerebellum, for example, intense mGLD2 mRNA expression was evident in the cell-dense, granule cell layers of the dentate gyrus and cerebellar cortex. We analyzed several other mRNAs in parallel sections. The distribution of mGLD2 mRNA was almost identical to the distribution of mouse Pumilio1 mRNA and was very similar to mCPEB1 in the hippocampus and to mCPEB4 in the cerebellum and olfactory bulb (Fig. 7D–F). The distribution of Quaking mRNA, used as a control, differs substantially, and is abundant in many regions of the brain (Fig. 7G).
FIGURE 7.
The pattern of mGLD2 mRNA abundance in neuronal cell populations in the mouse brain is similar to those of mCPEB1 and mPumilio1 mRNAs. (A) In situ hybridization on sagittal mouse brain sections using an 35S-labeled mGLD2 RNA probe that detects total mGLD2 mRNA (i.e., mGLD2(L) plus mGLD2(S)). The detected pattern of mGLD2 mRNA is consistent with expression in the major neuronal cell layers in the olfactory bulb, cerebral cortex, cerebellum, and hippocampus, each of which is labeled. (A–G) Bar, 4 mm. (B) Same as A, but using a probe specific to the unique region of the 3′UTR of mGLD2(L). (C) Same as A, but hybridized to a probe with the same sense as mGLD2 mRNA. (D–G) Adjacent sections hybridized with the indicated probes to detect mCPEB1 (D), mCPEB4 (E); Pumilio1 (F) and Quaking (G) mRNAs.
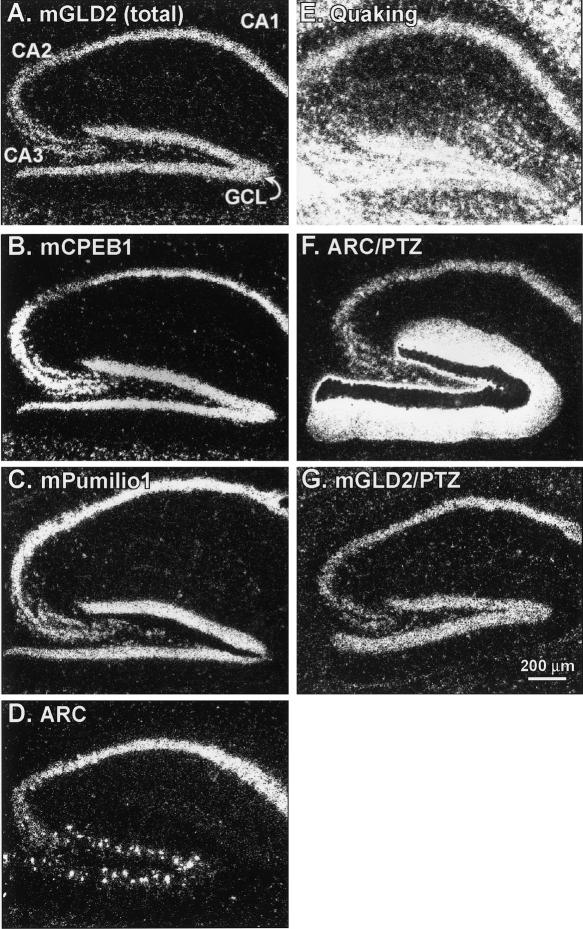
Since the hippocampus mediates certain types of long-term memory formation (Scoville and Milner 1957; for review, see Eichenbaum 2004), we examined the distribution of GLD2 transcripts in the hippocampus at higher resolution. After hybridization, sections were treated with radiographic emulsion and visualized by darkfield microscopy (Fig. 8). mGLD2 mRNA was specifically detected in the granule cell layer of the dentate gyrus (GCL) and the pyramidal cell layer of the hippocampus (CA1, CA2, and CA3) (Fig. 8A). The distribution of mGLD2 mRNA in the hippocampus was nearly identical to that of mCPEB1 and Pumilio1 mRNAs, which were analyzed in parallel sections (Fig. 8B,C). The distribution of mRNA for the activity-regulated protein, ARC, was also similar to mGLD2 except that fewer cells in the granule cell layer expressed ARC mRNA, compared with mGLD2 and Pumilio1 mRNAs (Fig. 8D). In contrast, the distribution of mRNA for the RNA binding protein, Quaking (Ebersole et al. 1996), differed dramatically from the other mRNAs analyzed (Fig. 8E). Induction of seizures using pentylenetetrazole (PTZ) did not affect either the abundance or distribution of mGLD2 mRNA, but dramatically increased the abundance of ARC mRNA, as expected (Fig. 8F,G; Steward and Worley 2001).
FIGURE 8.
The distribution of mGLD2 in hippocampus is nearly identical to those of mCPEB1, mPumilio1, and ARC. Darkfield microscopy of 35S-labeled RNA probes on radiographic emulsion-coated sections. (A–G) Bar, 200 μm. (A–E) mGLD2 mRNA was detected in the pyramidal cell layers (CA1 through CA3) of the hippocampus and granule cell layers (GCL) of the dentate gyrus (A). Nearly identical distributions were detected in adjacent sections hybridized with probes complementary to mCPEB1 (B), Pumilio1 (C), or ARC (D). The mRNA for the RNA binding protein Quaking, which is expressed primarily in glial cells, was more abundant and distributed differently than mGLD2 (E). (F, G) Three hours of seizure activity induced by pentylenetetrazole (PTZ; 50 mg/kg intraperitoneal), increased ARC expression in dendritic and cellular layers of the dentate gyrus (F). (The dark area over the granule cell layer is overexposed silver grains). In contrast, PTZ treatment did not change the abundance or distribution of mGLD2 mRNA (G).
We next focused on the cerebellum, where mGLD2 mRNA also is abundant (Fig. 7). mGLD2 mRNA was present at the highest levels in the granule (GCL) and Purkinje (PCL) cell layers of cerebellar cortex (Fig. 7A). mGLD2 mRNA was also present at a lower level, and in a diffuse distribution, throughout the molecular cell layer (MCL) (Fig. 9A). mCPEB1 mRNA, in contrast, was expressed almost exclusively within the large Purkinje neurons in the cerebellum (Fig. 9B). Interestingly, the pattern of mGLD2 expression in the cerebellum was more similar to mCPEB2, 3, and 4, all of which were present in the major cerebellar neuronal layers (Fig. 9C–E). These data imply regional specificity of cytoplasmic polyadenylation mechanisms in the brain.
FIGURE 9.
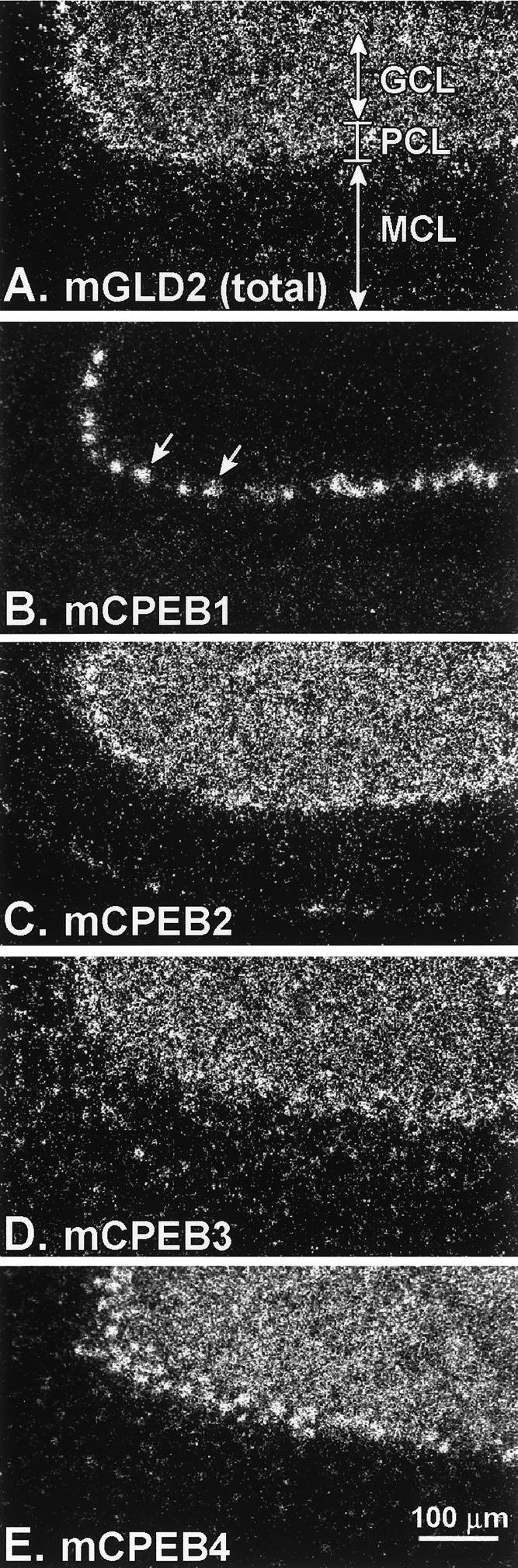
mRNAs encoding CPEB isoforms are differentially expressed in mouse cerebellum. Darkfield microscopy of 35S-labeled RNA probes on emulsion-coated sections. (A–E) Bar, 100 μm. (A) mGLD2 mRNA is found in the major neuronal cell populations in the cerebellum, including the granule cell layer (GCL) and the Purkinje cell layer (PCL), with less prominent expression in the cell-sparse molecular cell layer (MCL). (B) mCPEB1 expression was confined to Purkinje neurons (arrows). (C) mCPEB2 was expressed in a similar pattern to mGLD-2. (D) mCPEB3 was expressed in a similar pattern to mGLD-2 except that more prominent labeling was evident in cells of the MCL. (E) mCPEB4 was expressed in a similar pattern to mGLD-2 except that more prominent labeling was present in cells within the PCL.
DISCUSSION
We have focused in this paper on three vertebrate GLD2 proteins. The Xenopus enzyme, which exists in two closely related forms, polyadenylates RNAs to which it is tethered and enhances their translation. Furthermore, it interacts with cytoplasmic polyadenylation factors, including CPSF and CPEB, and with target mRNAs. These findings confirm and extend a recent report that a GLD2 enzyme is the long-sought PAP responsible for cytoplasmic polyadenylation in oocytes (Barnard et al. 2004). Previous work identified cytoplasmic PAPs closely related to the nuclear enzyme, but their biological roles have not been identified unambiguously (Ballantyne et al. 1995; Gebauer and Richter 1995).
XlGLD-2 protein is both cytoplasmic and nuclear (Fig. 2). The role of the nuclear enzyme is unclear. However, the two GLD2 homologs in S. cerevisiae, TRF4 and TRF5, both are nuclear proteins (Huh et al. 2003), and have been strongly implicated in RNA quality control. They appear to polyadenylate abberant initiator tRNA molecules, triggering their decay by the nuclear exosome (Kadaba et al. 2004). Nuclear GLD2 protein in oocytes may have an analogous role. It is unlikely that the nuclear enzyme is required for cytoplasmic polyadenylation events after nuclear breakdown during oocyte maturation, since those reactions proceed unabated in enucleated oocytes (Fox et al. 1989).
XlGLD-2 interacts with CPEB and CPSF but does not interact with maskin or Pumilio. Maskin, Pumilio, and CPEB are associated with repressed mRNAs in the oocyte (for review, see Mendez and Richter 2001; Nakahata et al. 2001). Our findings imply that at least two complexes exist in resting, stage VI oocytes (Fig. 4F). One contains GLD2, CPEB, and CPSF. We term this the G complex (for GLD2). The other complex contains CPEB, maskin, and Pumilio, and so is termed MP (for maskin and Pumilio). The finding that XlGLD-2 associates with symplekin implies that it too is in the G complex, which may contain other polyadenylation factors as well (Barnard et al. 2004).
Cyclin B1 mRNA is associated with GLD-2, and thus the G complex (the current study). It also may be present in the MP complex, since the cyclin B1 3′UTR can bind Pumilio in extracts of stage VI oocytes (Nakahata et al. 2001). Although cyclin B1 mRNA is polyadenylated and activated during oocyte maturation (Sheets et al. 1994), some cyclin B1 mRNA molecules must already be active in oocytes since oocytes contain cyclin B1 protein (Kobayashi et al. 1991). Active and inactive mRNA molecules may be partitioned differently between G and MP complexes. For example, the G complex may contain actively translated mRNAs. Alternatively, the mRNAs in G and MP complexes may be regulated differently during maturation.
The presence of XlGLD-2 on mRNAs destined to receive poly(A), but not yet doing so, implies that the enzyme is kept inactive in the G complex prior to the onset of meiotic maturation. Activation could involve the removal of inhibitory modifications of GLD2; recombinant, presumably unmodified GLD2, purified from bacteria, is active. Alternatively, the G complex may contain specific repressors of GLD2’s enzyme activity, whose action is relieved during oocyte maturation. In C. elegans, protein partners of GLD-2 have been identified that stimulate its activity and interact with gld-2 genetically (Wang et al. 2002). Whatever the repression mechanism, it is striking that XlGLD-2 produced in oocytes and tethered to an mRNA is active, and so must escape the inhibition.
The widespread distribution of mammalian GLD2 mRNA suggests that regulated polyadenylation occurs in many tissues. In cultured somatic cells, repressed mRNAs are deadenylated and almost certainly can be readenylated and reactivated (Muckenthaler et al. 1997). Likely examples of regulated increases in poly(A) length on cellular and viral mRNAs have been described (for example, Dehlin et al. 1996). It will be of interest to determine whether the GLD2 PAP is required for these events.
We focused on mammalian GLD2 mRNAs in the brain, because the clearest instances of regulated cytoplasmic polyadenylation in the soma occur in the nervous system (see Introduction). Three lines of evidence suggest that the mammalian GLD2 plays an important role in synaptic plasticity. First, human GLD2 protein physically interacts with Xenopus CPEB and is active in vitro (Figs. 3, 4). Further, as tethered proteins, mouse and human GLD2 potentiate translation in the oocyte via polyadenylation and are PAPs in vitro (Fig. 3; Kwak et al. 2004). Second, the spatial distribution of mGLD2 mRNA in the hippocampus strikingly parallels that of mCPEB1 and Pumilio1 (Fig. 8); in the cerebellum, mGLD2 mRNA colocalizes with those of other CPEB isoforms (Fig. 9). Proteins related to Pumilio (PUF proteins) are required for learning in Drosophila (Dubnau et al. 2003), are found in complexes with CPEB in Xenopus (Nakahata et al. 2001), and localize to dendrites in mammals (Ye et al. 2004). Third, mCPEB1, whose mRNA colocalizes with that of mGLD2 in the hippocampus (Fig. 8), is required for the late phase of long-term potentiation (Alarcon et al. 2004).
The colocalization in the brain of GLD2 mRNA with Pumilio and CPEB mRNAs is striking. We suggest that Pumilio-mediated repression is coordinated with cytoplasmic polyadenylation via GLD2 and CPEB. The proteins could co-occupy an mRNA, silence its translation until synaptic stimulation, and then activate it. Alternatively, as appears to be the case in Xenopus oocytes (Figs. 4, 5), GLD2 and “repression” proteins may reside in distinct complexes prior to stimulation, perhaps associated with different mRNA targets. The fact that mCPEB1 colocalizes with mGLD2 in the hippocampus, while mCPEB2, 3, and 4 colocalize in the cerebellum, implies regional differences in GLD2/CPEB complexes. Region-specific interactions of the multiple CPEBs with GLD2 may yield special properties. For example, mCPEB3 mRNA increases dramatically in abundance after drug-induced seizures (Theis et al. 2003) and resembles the Aplysia neuronal CPEB, which possesses prion-like properties (Si et al. 2003b). We suggest that GLD2 is required for regulated polyadenylation and sustained translation of dendritic mRNAs.
MATERIALS AND METHODS
Cloning Xenopus GLD-2
Degenerate primers for Xenopus GLD-2 were designed based on multiple sequence alignment of C. elegans GLD-2 and known homologs (Wang et al. 2002). The template used for PCR was DNA from an oligo(dT)-primed, Xenopus oocyte cDNA library (Romanowski et al. 1996). Seminested PCR was performed using primers DG1 and DG3, and then DG2 and DG3, at 50°C (for primer sequences, see Table 1). A single prominent DNA product was obtained, cloned into pT7Blue (Novagen), and sequenced. Two clones sequenced in their entirety were identical and showed extensive similarity to C. elegans and mammalian GLD-2 proteins (Kwak et al. 2004), but little similarity to conventional PAPs. To identify the 5′ and 3′ ends of the cDNA, PCR was performed with primer pairs that anneal to the 5′ end of the cDNA (XGLD-2-5) and 5′ flanking vector (pVP16-1); or to 3′ end of the cDNA (xgld2-10) and to 3′ flanking vector sequences (pVP16-2). The full-length ORF was then amplified by high-fidelity PCR from the cDNA library, using primers xgld2SmaN and xgld2SmaC, and inserted into the Sma1 site of pGEX6P1, to generate pLW071.
TABLE 1.
Oligonucleotides
| Oligo | Sequence | Use | |
| 1 | DG1 | GCIAARGTICCIATHRTIAARTTY | degenerate PCR |
| 2 | DG2 | CCIYTIGTIYTNGTIRTIAARAARTGG | degenerate PCR |
| 3 | DG3 | RTCRAAIGGITCYTCIAYRCAIAY | degenerate PCR |
| 4 | xgld2-10 | TTGACTGGAGCAAAGACATC | Forward primer, XIGLD-2A (1448–1267) |
| 5 | xgld2-5 | TGGCCCACTTTTTAATGACC | Reverse primer, XIGLD-2A (1170–1189) |
| 6 | pVP16-1 | CTACGGCGCTCTGGATATGG | XIGLD-2 identification (pVP16 MCS) |
| 7 | pVP16-2 | CCTCTACAAATGTGGTATGG | XIGLD-2 identification (pVP16 MCS) |
| 8 | xgld2SmaN | CGCCCGGGGATGTACCCTAACTCCCCGAGCC | Forward primer, XIGLD-2A (178–199) |
| 9 | xgld2SmaC | CGCCCGGGTCATAAAGAGTTCATTTTTTTCACG | Reverse primer, XIGLD-2A (1683–1707) |
| 10 | xD242f | GAATCAGTGATGCAGcTTTGTGCCTGGTTTTAAAAGAG | Forward primer, XIGLD-2A active-site mutagenesis (887–924) |
| 11 | xD242r | CTCTTTTAAAACCAGGCACAAAgCTGCATCACTGATTC | Reverse primer, XIGLD-2A active-site mutagenesis (887–924) |
| 12 | Xlgld-2aNhel | CCGCTAGCGCTAGCATGTACCCTAACTCCCCCAGCC | Forward primer, XIGLD-2A (178–199) |
| 13 | Xlgld-2aXhol | CCCCTCGAGCTCGAGTCATAAAGAGTTCATTTTTTTCACG | Reverse primer, XIGLD-2A (1683–1707) |
| 14 | Xlgld-2bNhel | CCGCTAGCGCTAGCATGTACCCTAACTCCCCCAGCCTGGGCCGC | Forward primer, XIGLD-2B (139–168) |
| 15 | Xlgld-2bXhol | CCCCTCGAGCTCGAGTCATAACGAGTGCATTTTTTTCATGATTCC | Reverse primer, XIGLD-2B (1639–1568) |
| 16 | cycB1-74 | GCAGGTTTGCGCTTGAGAAAATGTCAC | Forward primer, cyclin B1 (accession no. J03166, 74–100) |
| 17 | cycB1-1010anti | GGAGGAAGCAGCTGCTATTTGGGAAGGCG | Reverse primer, cyclin B1 (accession no. J03166, 1010–1038) |
| 18 | cmos-361 | TACAGAGGGGAGACGGTGGCGCTG | Forward primer, cmos (accession no. X13311, 361–384) |
| 19 | cmos-3′anti | CCACTTAAACAGCAATGCAACAC | Reverse primer, cmos (accession no. X13311, 3073–3095) |
| 20 | Actin-1801 | CAACTGGAATAAGGGCAGACTTCC | Forward primer, actin (accession no. M24769, 1801–1824) |
| 21 | Actin-2227anti | GGGCCTCAGTATCAATTTCCAACC | Reverse primer, actin (accession no. M24769, 2227–2251) |
| 22 | L1-1033 | GCTGAACCCATATGCAAAGACCGC | Forward primer, L1 (accession no. BC054956, 1033–1056) |
| 23 | L1-3′anti | ACAGAATTTATTGAGTAAATCC | Reverse primer, L1 (accession no. BC054956, 1279–1300) |
| 24 | GLD-2(L) F | TTATGGCATGACTTTTCAGCA | Forward primer, mGLD-2(L) (1907–1928) |
| 25 | GLD-2(L) R | GGCCAGTGAATTGTAATAGCACTCACTATAGGGAGGCGTCCGTTTTCTGCTGTTTCCT | Reverse primer, mGLD-2(L) probe (2484–2504) |
| 26 | CPEB-1 F | CATCTTGGGACCTTCTTGGA | Forward primer, CPEB1 (808–828) |
| 27 | CPEB-1 R | AAGACCCAAGGGATTACC | Reverse primer, CPEB1 (1246–1266) |
| 28 | CPEB-2 F | GGTCGCTCTTCCCTATTTCC | Forward primer, CPEB2 (773–793) |
| 29 | CPEB-2 R | TCCAAAGGCTGAGAACCATC | Reverse primer, CPEB2 (1198–1218) |
| 30 | CPEB-3 F | CCCTTCTCCAGCAATGTGAT | Forward primer, CPEB3 (958–978) |
| 31 | CPEB-3 R | CTCGTTCCCCATTTTGACAT | Reverse primer, CPEB3 (1412–1432) |
| 32 | CPEB-4 F | CCTCACTGCTTCACTCACCA | Forward primer, CPEB4 (97–117) |
| 33 | CPEB-4 R | AACAAAGCGGGCACTGATAG | Reverse primer, CPEB4 (643–663) |
Numbering of sequences (last column) are based on the following: oligonucleotides 4, 5, and 8–13, GenBank accession BC082438 (IMAGE: 50848776); oligonucleotides 14 and 15, GenBank accession BC076832 (IMAGE:6643643); oligonucleotides 16–23, indicated GenBank accession numbers; and oligonucleotides 24–33, sequences in the Unigene database.
The cDNA XlGLD-2 sequence was used as a reference sequence in BLAST to search databases from National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/BLAST), the National Institute for Basic Biology (NIBB), and the X. laevis EST project (http://xenopus.nibb.ac.jp). These searches yielded multiple ESTs that were members of either Contig026310 (11 EST sequences) or Contig029930 (six EST sequences) from the NIBB database; UniGene cluster Xl.1354 (42 EST sequences) or Xl.15643 (19 EST sequences) from the NCBI database. ESTs of each group were obtained from NIBB, purchased for the IMAGE consortium (Open Biosystems), and fully sequenced. Clones NIBB:XL084p08, NIBB:XL080o08, IMAGE:5084876, and IMAGE:6638921 are representative of NIBB Contig026310 and UniGene cluster Xl.1354, which represent our original clone, and whose gene was called XLGLD-2A. Clones NIBB:XL074e24, IMAGE:5077998, and IMAGE:6643643 are representative of NIBB Contig029930 and UniGene cluster Xl.15643, whose gene was called XlGLD-2B. XlGLD-2B corresponds to the clone described in Barnard et al. (2004). ESTs representative short and long 3′UTRs also were identified. These include XL009c13 and XL058d09 (XlGLD-2(S)), and XL084p08 and IMAGE:35492925 (XlGLD-2(L)). Both XlGLD-2A and XlGLD-2B encode a 509-amino-acid-long protein.
XlGLD-2A and XlGLD-2B ORFs were tested for sequence similarity to Xenopus tropicalis cDNA and ESTs via BLAST searching against the Sanger X. tropicalis EST database (www.sanger.ac.uk/cgi-bin/blast/submitblast/x-tropicalis). Both X. laevis proteins were related to NCBI Unigene XP–342174.1 (i.e., TEgg044c10, TEgg130h16, and TEgg037n22).
Cloning mGLD2 and hGLD2
Clones corresponding to full-length mGLD2 and hGLD2 proteins have been described (Kwak et al. 2004). hGLD2Δ8 and hGLD2Δ11 cDNAs were obtained by reverse transcription and PCR from total RNA, using hGLD2 gene-specific primers. mRNA and EST sequences representative of mGLD2 are represented in UniGene cluster Mm.242865, with polyadenylated representatives for mGLD2(S) (IMAGE:3595512 and NM–133905), and for mGLD2(L) (BB787817). mRNA and EST sequences representative of hGLD2 are represented in UniGene cluster Hs.418198, with polyadenylated representatives for hGLD2(S) (GenBank entry NM–173797), and for hGLD2(L) (IMAGE:4824607 and GenBank entry BC047581.1).
DNA constructs
pLW071
To express GST-XlGLD-2A in bacteria, the entire XlLD-2A ORF was inserted into SmaI-digested pGEX6P-1.
pLW071(D242A)
To generate the active site mutant of GST-XlGLD-2A, site-directed mutagenesis was performed on pLW071 and primers xD242f and xD242r.
pLW073
To express MS2/XlGLD-2A in Xenopus oocytes, the XlGLD-2A ORF was amplified with Xlgld-2aNheI and Xlgld-2aXhoI, and inserted into the NheI and XhoI sites of p3HA-MSP-CeGLD-2 (Kwak et al. 2004). This replaces CeGLD-2 with XlGLD-2A. Transcripts were produced by using a T7 promoter.
pLR073
To express MS2-XlGLD-2B in Xenopus oocytes, the XlGLD-2B ORF was amplified with Xlgld-2bNheI and Xlgld-2bXhoI primers, and inserted into the NheI and XhoI sites of p3HA-MSP-CeGLD2 (Kwak et al. 2004). This replaces CeGLD-2 with XlGLD-2B. The mRNA was transcribed from a T7 promoter.
pLW078
To generate the active site mutant of MS2-XlGLD-2A, site-directed mutagenesis was performed on pLW073 and primers xD242f and xD242r.
pCS2+/HA/XLGLD-2A
To express HA-XlGLD-2A in Xenopus oocytes, the XlGLD-2A ORF preceded by two HA epitope tag sequences, was inserted into EcoRI digested pCS2+, and can be transcribed from a SP6 promoter.
pCS2+/MS2/hGLD2
To express MS2-hGLD2 in Xenopus oocytes, a PCR fragment from p3HA-hGLD2 (Kwak et al. 2004), which contained MS2/hGLD2 ORF, was inserted into BamH1 and Xho1 sites of pCS2+ and can be transcribed from SP6 promoter.
pCSMS2-hGLD2
To detect the expression of MS2-hGLD2 protein in Xenopus oocytes, triple HA tags were inserted into pCS2+/MS2/hGLD2 at the BamH1 site.
pCSMS2-hGLD2Δ8 and pCSMS2-hGLD2Δ11
To express and detect alternative splicing variants of hGLD2 in Xenopus oocytes, hGLD2 ORF in pCSMS2-hGLD2 was replaced with spliced variants hGLD2Δ8 and hGLD2Δ11 ORFs, using Nhe1 and Xho1, and can be transcribed from a SP6 promoter.
Constructs for tethered assay reporters are pLG-MS2, pLGMS2-LucHS, and pJK350, which have been previously described (Kwak et al. 2004).
In vitro transcription
pCS2+ based constructs were linearized with Not1 and transcribed with Megascript SP6 transcription kit (Ambion). pLW073, pLR073, and pLW078 were linearized with EcoR1 and transcribed with Megascript T7 transcription kit (Ambion).
Oocyte manipulations, injections, and tethered assays
Oocyte injection and progesterone treatment were performed as described previously (Ballantyne et al. 1997), as were tethered function assays (Kwak et al. 2004). mRNAs were injected at 0.7 μg/μlL. Enucleations were performed under mineral oil (Lund and Paine 1990; Dickson et al. 2001).
Northern blotting
One microgram of total RNA (unless otherwise stated) was electrophoresed in individual lanes on a 1.2% agarose/formaldehye/1× MOPS gel. The gel was transferred to GeneScreen plus (Perkin Elmer Life Sciences, Inc.) using Stratagene’s PosiBlotter (Strata-gene, Inc.). 32P-labeled cDNA (2 × 106 c.p.m./mL) was hybridized to the membrane in Hybrisol I (Intergen, Inc.). Northern blots were washed and exposed to a phosphorimager screen for 1 d. Screens were scanned on a Storm phosphorimager (Molecular Dynamics Inc.).
XLGLd-2
Northern blotting was performed using 25 μg of total RNA, 25 μg of nonpolyadenylated RNA, and 800 ng of oligo(dT)-purified oocyte RNA. The 32P-dCTP–labeled DNA probe comprised the entire XlGLD-2A ORF.
mGLD2
Northern blotting was performed by using 20μg of total RNA or oligo(dT)-purified RNA from NIH-3T3 cells, 10μg of total RNA from various mouse tissues, and on 1 μg of total RNA from mouse brain, cerebellum, P1, and synaptosomal preps.
hGLD2
Northern blotting was performed by using 2μg of total RNA from various human tissues (Ambion), and a 32P-UTP–labeled probe anti-sense to the entire hGLD2 ORF.
In vitro PAP assay
One hundred nanograms of purified recombinant GST-XlGLD-2A, GST/XlGLD-2A(D242A), or GST/hGLD2 proteins, were incubated with 32P-end–labeled L1 RNA substrate (sequence UUAUCUCAU GUUCAGCACUUUGGAUUUACUCAAUAAAUUCUGUU (Integrated DNA Technologies), 20 U RNAsin (Promega), and 1 mM rNTPs, in PAP buffer (25 mM Tris-HCl at pH 7.0, 40 mM KCl, 0.5 mM MnCl2, 0.05 mM EDTA, 0.5 mM DTT, and 0.2 mg/mL BSA; USB). Reactions were stopped by with 2μ RNA loading dye (Ambion).
α-XGLDLD-2A antibodies
Recombinant GST-XlGLD-2A was purified as described for GST-FBF-1 in Bernstein et al. (2005). Purified protein was injected into guinea pigs (Cocalico Biologicals). Antibody specificity was tested by using the TnT T7-Quick couple transcription/translation system (Promega). This antibody is referred to in the text as αXlGLD-2.
Interactions with GST fusion proteins
All steps were performed at 4°C; 4μg of GST-XlGLD-2A, GST-PUF-8, or GST was bound to glutathione-Sepharose 4B (Amersham Pharmacia Biotech) and equilibrated with MSB. Twenty stage VI oocytes were homogenized in 200μL MSB and centrifuged at 3000 rpm in a Eppendorf 5415C microcentrifuge for 10 min. The soluble fraction was incubated overnight with recombinant protein bound to 20μL of glutathione beads. Samples were then centrifuged at 3000 rpm for 5 min and washed thrice with 500μL of MSB, and the fraction associated with glutathione beads was eluted by boiling in 50μL of SDS-PAGE loading dye for 5 min.
Westerns
The presence of XlGLD-2 protein in stage VI oocytes, eggs, and different stages of embryogenesis was monitored by lysing groups of 10 oocytes, eggs, or embryos in 100μL of MSB; centrifuging 3000 rpm in a eppendorf 5415C microcentrifuge for 10 min at 4°C; and mixing the soluble fraction with 2 × SDS loading dye. Westerns were done for protein representative of one oocyte/egg/embryo. αXlGLD-2 antibody was used at 1:1000 dilution in blotto. HRP-conjugated anti-Guinea pig secondary antibody (Sigma) was used at a 1:10,000 dilution. Western blotting with other antibodies were done following the providers’ protocol.
Immunoprecipitations
For immunoprecipitations, antibodies were purified and bound on Protein A–Sepharose (Sigma). 15μL of serum was incubated with 25μL of Protein A–Sepharose following in 150 μL of PBS, for 4 h at 4°C. Sepharose was then centrifuged at 3000 rpm and washed three times with PBS.
Antibodies directed against the following proteins have been described previously and were gifts of the indicated laboratories: α-CPEB (Dickson et al. 2001), αCPSF100 (Jenny et al. 1994); α-CPSF73 (gift of Dr. D.L. Bentley, University of Colorado HSC), αXenopus Pumilio (Nakahata et al. 2003), α-XlPABP2 (Good et al. 2004), and α-maskin (gift of Dr. C. Wiese).
Protein A–Sepharose bound antibodies were equilibrated with MSB (150 mM NaCl, 0.1% NP-40, 50 mM Tris-Cl at pH 8) and a protease inhibitor cocktail (Boehringer Manneheim) (Dickson et al. 2001) and incubated overnight at 4°C with oocyte extract (0.1 oocyte/μL). Samples were centrifuged at 3000 rpm in a Eppendorf 5415C microcentrifuge for 5 min and washed three times with 500μL modified MSB, and the bound fraction was eluted by boiling in 50μL of SDS loading dye for 5 min.
For RNA coimmunoprecipitations and RNA dependence analysis of the CPEB-XlGLD2 interaction, modified MSB was made with DEPC-treated water, 1 mM DTT, and 1 U/μL rRNAsin, with or without 1 μL RNAseA/T1 mix (Ambion) per 10 μL of lysate. The Protein A–Sepharose bound antibodies were incubated with oocyte lysate (0.04 oocytes/μL) 5 h at 4°C and washed three times with 200μL of DEPC-based MSB. RNA was eluted from beads by extraction with TRI reagent (Sigma) and solubilized in 20 μL of DEPC water.
Where indicated, oocytes were injected with mRNAs coding for tagged forms of GLD2 and incubated at room temperature for 6 h, prior to preparation of extracts.
RT-PCR
Reverse transcriptase reactions were performed using one third of the total coimmunoprecipitated sample or 1 μg of total oocyte RNA. Oligo(dT)-primed reverse transcription reactions were performed by using the GeneRacer Superscript II reverse transcription module (Invitrogen), as described by manufacturer. Three microliters of the Reverse transcription reaction was used for PCR with gene-specific primers for 35 cycles with annealing temperatures of 55°C (c-mos and actin), 65°C (cyclinB1), and 45°C (L1).
In situ hybridizations
Sections on slides were fixed in 4% paraformaldehyde in PBS for 2 h at 4°C. Slides were then washed for 5 min in 2 × SSC three times, and incubated in 0.2 μg/mL Proteinase K (Qiagen) in 0.1 M Tris base and 50 mM EDTA (pH 8.1) for 10 min at 37°C. Slides were washed in 2 × SSC at room temperature for 2 min and incubated in 0.1 M TEA at room temperature with rapid stirring, and acetic anhydride was added to a final concentration of 0.25% (v/v) with rapid stirring for 10 min. Slides were then washed in 2 × SSC for 5 min. Finally, sections were dehydrated in an ascending ethanol series and air-dried for 15 min.
Templates for generating 35S-labeled RNAprobes were generatedby PCR of a mouse brain cDNA library using T7 anchored primer pairs (Table 1). In vitro transcription was carried out in 1 × Transcription OptimizedBuffer,10mMDTT;1U/mLRNasin;0.375mMATP,CTP, and GTP; 1 U/mL T7 RNA polymerase (all Promega); 3.5 mCi/mL [α-35S]UTP (PerkinElmer); and 100 ng template DNA and incubated for 2 h at 37°C. RQ1 RNase free DNase (Promega) was added at a concentration of 0.15 U/mL and incubated for an additional 15 min at 37°C. The labeled probes were purified using ProbeQuant G-50 Micro columns (Amersham Biosciences). Probes were diluted in hybridization solution (3 × SSC, 10% dextran sulfate, 1 × Denhardt’s solution, 0.2 mg/mL tRNA, 50 mM NaPO4 buffer, and freshly added DTT to 50 mM final concentration) to −106 cpm/100 μL). One hundred microliters of the hybridization solution at 55°C with labeled probe was applied to each slide. Slides were then covered with coverlips and incubated at 55°C in a hybridization chamber saturated with 75% formamide for 16 h.
After hybridization, coverslips were removed and slides were washed three times in 2 × SSC with 2 mM DTT at room temperature for 10 min. Slides were incubated in 1.5 U/mL RNase A (Qiagen) in RNase buffer (10 mM Tris-HCl and 0.5 M NaCl at pH 8.0) at 37°C for 1 h followed by washes in 1 × SSC with 1 mM DTT at room temperature for 5 min, 0.5 × SSC with 1 mM DTT at room temperature for 5 min, and 0.1 × SSC with 2 mM DTT at 70°C for 1 h. The sections were then dehydrated in an ascending series of ethanol and then were air-dried. Sections were exposed to a phosphorimager screen and were subsequently scanned on a Storm phosphorimager. Slides were then covered with NTB2 emulsion (Eastman Kodak Co.) and exposed for 28 d for analysis of silver grain distribution. After development, slides were counterstained with Nissl stain and dehydrated through a graded series of ethanol and xylene. A coverslip was then applied. Images were taken with a Leica DC 300F digital camera linked to Image Pro-Plus software on a PC through a Leica DMRX microscope.
Acknowledgments
We thank Judith Kimble for suggestions on the manuscript and the work, and we thank members of the Wickens and Kimble laboratory for advice and assistance. We also appreciate Jerry Yin’s insights and suggestions. We are grateful to Dr. Steve Liebhaber (University of Pennsylvania School of Medicine) for mouse RNAs and blots, and to Drs. W. Keller (Biozentrum, Basel, Switzerland), D. Bentley (University of Colorado Health Sciences Center), M. Yamashita (Hokkaido University, Sapporo, Japan), M. Sheets (University of Wisconsin, Madison), and C. Wiese (University of Wisconsin-Madison) for antibodies. This work was supported by an NIH fellowship to C.S. (DA016503), and by NIH grants to M.W. (GM50942) and C.L. (DA019153).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2630205.
REFERENCES
- Alarcon, J.M., Hodgman, R., Theis, M., Huang, Y.S., Kandel, E.R., and Richter, J.D. 2004. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn. Mem. 11: 318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L. and Koonin, E.V. 1998. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 26: 3746–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne, S., Bilger, A., Astrom, J., Virtanen, A., and Wickens, M. 1995. Poly (A) polymerases in the nucleus and cytoplasm of frog oocytes: Dynamic changes during oocyte maturation and early development. RNA 1: 64–78. [PMC free article] [PubMed] [Google Scholar]
- Ballantyne, S., Daniel Jr., D.L., and Wickens, M. 1997. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol. Biol. Cell 8: 1633–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard, D.C., Ryan, K., Manley, J.L., and Richter, J.D. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651. [DOI] [PubMed] [Google Scholar]
- Bernstein, D., Hook, B., Hajarnavis, A., Opperman, L., and Wickens, M. 2005. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA 11: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilger, A., Fox, C.A., Wahle, E., and Wickens, M. 1994. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes & Dev. 8: 1106–1116. [DOI] [PubMed] [Google Scholar]
- Dehlin, E., von Gabain, A., Alm, G., Dingelmaier, R., and Resnekov, O. 1996. Repression of β interferon gene expression in virus-infected cells is correlated with a poly(A) tail elongation. Mol. Cell Biol. 16: 468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, K.S., Bilger, A., Ballantyne, S., and Wickens, M.P. 1999. The cleavage and polyadenylation specificity factor in Xenopus laevis oocytes is a cytoplasmic factor involved in regulated polyadenylation. Mol. Cell Biol. 19: 5707–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, K.S., Thompson, S.R., Gray, N.K., and Wickens, M. 2001. Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem. 276: 41810–41816. [DOI] [PubMed] [Google Scholar]
- Dubnau, J., Chiang, A.S., Grady, L., Barditch, J., Gossweiler, S., McNeil, J., Smith, P., Buldoc, F., Scott, R., Certa, U., et al. 2003. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 13: 286–296. [DOI] [PubMed] [Google Scholar]
- Ebersole, T.A., Chen, Q., Justice, M.J., and Artzt, K. 1996. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 12: 260–265. [DOI] [PubMed] [Google Scholar]
- Eichenbaum, H. 2004. Hippocampus: Cognitive processes and neural representations that underlie declarative memory. Neuron 44: 109–120. [DOI] [PubMed] [Google Scholar]
- Fox, C.A., Sheets, M.D., and Wickens, M.P. 1989. Poly(A) addition during maturation of frog oocytes: Distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes & Dev. 3: 2151–2162. [DOI] [PubMed] [Google Scholar]
- Frey, U., Krug, M., Reymann, K.G., and Matthies, H. 1988. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res. 452: 57–65. [DOI] [PubMed] [Google Scholar]
- Fukazawa, Y., Saitoh, Y., Ozawa, F., Ohta, Y., Mizuno, K., and Inokuchi, K. 2003. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron 38: 447–460. [DOI] [PubMed] [Google Scholar]
- Gebauer, F. and Richter, J.D. 1995. Cloning and characterization of a Xenopus poly(A) polymerase. Mol. Cell Biol. 15: 1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, P.J., Abler, L., Herring, D., and Sheets, M.D. 2004. Xenopus embryonic poly(A) binding protein 2 (ePABP2) defines a new family of cytoplasmic poly(A) binding proteins expressed during the early stages of vertebrate development. Genesis 38: 166–175. [DOI] [PubMed] [Google Scholar]
- Hake, L.E., Mendez, R., and Richter, J.D. 1998. Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol. Cell Biol. 18: 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M.W. 1995. Translational regulation: Versatile mechanisms for metabolic and developmental control. Curr. Opin. Cell. Biol. 7: 393–398. [DOI] [PubMed] [Google Scholar]
- Huang, Y.S., Jung, M.Y., Sarkissian, M., and Richter, J.D. 2002. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and αCaMKII mRNA polyadenylation at synapses. EMBO J. 21: 2139–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O’Shea, E.K. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Jenny, A., Hauri, H.P., and Keller, W. 1994. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol. Cell Biol. 14: 8183–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.W., Errington, M.L., French, P.J., Fine, A., Bliss, T.V., Garel, S., Charnay, P., Bozon, B., Laroche, S., and Davis, S. 2001. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 4: 289–296. [DOI] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A.M., Hinnebusch, A.G., and Anderson, J. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae Genes & Dev. 18: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, H., Minshull, J., Ford, C., Goldsteyn, R., Poon, R., and Hunt, T. 1991. On the synthesis and destruction of A- and B-type cyclins during oogenesis and meiotic maturation in Xenopus laevis. J. Cell. Biol. 114: 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer, B., Crittenden, S., Gallegos, M., Moulder, G., Barstead, R., Kimble, J., and Wickens, M. 1999. NANOS-3 and FBF proteins physically interact to control the spermoocyte switch in Caenorhabditis elegans. Curr. Biol. 9: 1009–1018. [DOI] [PubMed] [Google Scholar]
- Kuersten, S. and Goodwin, E.B. 2003. The power of the 3′ UTR: Translational control and development. Nat. Rev. Genet. 4: 626–637. [DOI] [PubMed] [Google Scholar]
- Kwak, J.E., Wang, L., Ballantyne, S., Kimble, J., and Wickens, M. 2004. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. 101: 4407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. and Schwartz, J.H. 2003. The cytoplasmic polyadenylation element binding protein and polyadenylation of messenger RNA in Aplysia neurons. Brain Res. 959: 68–76. [DOI] [PubMed] [Google Scholar]
- Lund, E. and Paine, P.L. 1990. Nonaqueous isolation of transcriptionally active nuclei from Xenopus oocytes. Methods Enzymol. 181: 36–43. [DOI] [PubMed] [Google Scholar]
- Malkani, S., Wallace, K.J., Donley, M.P., and Rosen, J.B. 2004. An egr-1 (zif268) antisense oligodeoxynucleotide infused into the amygdala disrupts fear conditioning. Learn. Mem. 11: 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G., Keller, W., and Doublie, S. 2000. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 19: 4193–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G., Moglich, A., Keller, W., and Doublie, S. 2004. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J. Mol. Biol. 341: 911–925. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, M., Honkura, N., Ellis-Davies, G.C., and Kasai, H. 2004. Structural basis of long-term potentiation in single dendritic spines. Nature 429: 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez, R. and Richter, J.D. 2001. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell. Biol. 2: 521–529. [DOI] [PubMed] [Google Scholar]
- Mendez, R., Murthy, K.G., Ryan, K., Manley, J.L., and Richter, J.D. 2000. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell 6: 1253–1259. [DOI] [PubMed] [Google Scholar]
- Miller, S., Yasuda, M., Coats, J.K., Jones, Y., Martone, M.E., and Mayford, M. 2002. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36: 507–519. [DOI] [PubMed] [Google Scholar]
- Muckenthaler, M., Gunkel, N., Stripecke, R., and Hentze, M.W. 1997. Regulated poly(A) tail shortening in somatic cells mediated by cap-proximal translational repressor proteins and ribosome association. RNA 3: 983–995. [PMC free article] [PubMed] [Google Scholar]
- Nakahata, S., Katsu, Y., Mita, K., Inoue, K., Nagahama, Y., and Yamashita, M. 2001. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 276: 20945–20953. [DOI] [PubMed] [Google Scholar]
- Nakahata, S., Kotani, T., Mita, K., Kawasaki, T., Katsu, Y., Nagahama, Y., and Yamashita, M. 2003. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 120: 865–880. [DOI] [PubMed] [Google Scholar]
- Nguyen, P.V., Abel, T., and Kandel, E.R. 1994. Requirement of a critical period of transcription for induction of a late phase of LTP. Science 265: 1104–1107. [DOI] [PubMed] [Google Scholar]
- Otmakhov, N., Tao-Cheng, J.H., Carpenter, S., Asrican, B., Dosemeci, A., Reese, T.S., and Lisman, J. 2004. Persistent accumulation of calcium/calmodulin-dependent protein kinase II in dendritic spines after induction of NMDA receptor-dependent chemical long-term potentiation. J. Neurosci. 24: 9324–9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, P.T., Teng, H.K., Zaitsev, E., Woo, N.T., Sakata, K., Zhen, S., Teng, K.K., Yung, W.H., Hempstead, B.L., and Lu, B. 2004. Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science 306: 487–491. [DOI] [PubMed] [Google Scholar]
- Pawlak, R., Nagai, N., Urano, T., Napiorkowska-Pawlak, D., Ihara, H., Takada, Y., Collen, D., and Takada, A. 2002. Rapid, specific and active site-catalyzed effect of tissue-plasminogen activator on hippocampus-dependent learning in mice. Neuroscience 113: 995–1001. [DOI] [PubMed] [Google Scholar]
- Read, R.L., Martinho, R.G., Wang, S.W., Carr, A.M., and Norbury, C.J. 2002. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. 99: 12079–12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, J.D. 2000. The influence of polyadenylation-induced translation on metazoan development and neuronal synaptic function. In Translational control (eds. J.W.B. Hershey et al.), pp. 785–805. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- ———. 2001. Think globally, translate locally: What mitotic spindles and neuronal synapses have in common. Proc. Natl. Acad. Sci. 98: 7069–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski, P., Madine, M.A., and Laskey, R.A. 1996. XMCM7, a novel member of the Xenopus MCM family, interacts with XMCM3 and colocalizes with it throughout replication. Proc. Natl. Acad. Sci. 93: 10189–10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, S., Chabes, A., McDonald, W.H., Thelander, L., Yates, J.R., and Russell, P. 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109: 563–573. [DOI] [PubMed] [Google Scholar]
- Scoville, W.B. and Milner, B. 1957. Loss of recent memory after bilateral hippocampal lesions. J. Neurochem. 20: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets, M.D., Fox, C.A., Hunt, T., Vande Woude, G., and Wickens, M. 1994. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes & Dev. 8: 926–938. [DOI] [PubMed] [Google Scholar]
- Shin, C.Y., Kundel, M., and Wells, D.G. 2004. Rapid, activity-induced increase in tissue plasminogen activator is mediated by metabotropic glutamate receptor-dependent mRNA translation. J. Neurosci. 24: 9425–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si, K., Giustetto, M., Etkin, A., Hsu, R., Janisiewicz, A.M., Miniaci, M.C., Kim, J.H., Zhu, H., and Kandel, E.R. 2003a. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell 115: 893–904. [DOI] [PubMed] [Google Scholar]
- Si, K., Lindquist, S., and Kandel, E.R. 2003b. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell 115: 879–891. [DOI] [PubMed] [Google Scholar]
- Simon, P., Schott, K., Williams, R.W., and Schaeffel, F. 2004. Posttranscriptional regulation of the immediate-early gene EGR1 by light in the mouse retina. Eur. J. Neurosci. 20: 3371–3377. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N. 1994. mRNA translation: Influence of the 5′ and 3′ untranslated regions. Curr. Opin. Genet. Dev. 4: 310–315. [DOI] [PubMed] [Google Scholar]
- Sonoda, J. and Wharton, R.P. 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes & Dev. 13: 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins-Boaz, B., Cao, Q., de Moor, C.H., Mendez, R., and Richter, J.D. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell 4: 1017–1027. [DOI] [PubMed] [Google Scholar]
- Steward, O. and Worley, P.F. 2001. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron 30: 227–240. [DOI] [PubMed] [Google Scholar]
- Tang, S.J. and Schuman, E.M. 2002. Protein synthesis in the dendrite. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis, M., Si, K., and Kandel, E.R. 2003. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc. Natl. Acad. Sci. 100: 9602–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Eckmann, C.R., Kadyk, L.C., Wickens, M., and Kimble, J. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. [DOI] [PubMed] [Google Scholar]
- Wells, D.G., Richter, J.D., and Fallon, J.R. 2000. Molecular mechanisms for activity-regulated protein synthesis in the synaptodendritic compartment. Curr. Opin. Neurobiol. 10: 132–137. [DOI] [PubMed] [Google Scholar]
- Wickens, M., Bernstein, D.S., Kimble, J., and Parker, R. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18: 150–157. [DOI] [PubMed] [Google Scholar]
- Wu, L., Wells, D., Tay, J., Mendis, D., Abbott, M.A., Barnitt, A., Quinlan, E., Heynen, A., Fallon, J.R., and Richter, J.D. 1998. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron 21: 1129–1139. [DOI] [PubMed] [Google Scholar]
- Ye, B., Petritsch, C., Clark, I.E., Gavis, E.R., Jan, L.Y., and Jan, Y.N. 2004. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 14: 314–321. [DOI] [PubMed] [Google Scholar]