Abstract
I elements in Drosophila melanogaster are non-long terminal repeat (LTR) retrotransposons of particular interest because high levels of transposition can be induced by appropriate crosses. They use a full-length RNA transposition intermediate as a template for reverse transcription. Detailed molecular characterization of this intermediate is rendered difficult because of the many transcripts produced by defective elements. The use of an active I element marked with a sequence encoding the HA epitope solves this problem. We used an RNA circularization procedure followed by RT–PCR to analyze the transcripts produced by actively transposing tagged I elements. Most start at the 5′ end at the second nucleotide of the I element and all are polyadenylated at a site located in genomic sequences downstream of the 3′ end. One of the tagged I elements, inserted in locus 88A, produces chimeric transcripts that carry sequences from both 5′- and 3′-flanking genomic DNA. We show that synthesis of these chimeric transcripts is controlled by the I element itself. Analysis of full-length transposed copies of this element shows that the extra sequences at the 5′ and 3′ ends are not integrated during retrotransposition. This suggests that initiation and arrest of reverse transcription during retrotransposition are precise processes.
INTRODUCTION
Non-long terminal repeat (LTR) retrotranspons are a very large class of transposable elements present in many eukaryotes. They are thought to transpose according to a mechanism called target primed reverse transcription (TPRT) based on results obtained with R2 elements from the silkmoth, Bombyx mori (1). In this model, the reverse transcriptase uses the 3′ hydroxyl at one end of a nick generated by endonucleolytic cleavage of genomic DNA to prime synthesis of the first cDNA strand, using the RNA transposition intermediate as a template (1–3). R2 elements have a single open reading frame (ORF) and insert at a specific site in the genes coding for the 28S RNA. Very little is known about the reverse transcription and insertion mechanisms of other non-LTR retrotransposons, in particular those having two ORFs such as mammalian L1 and Drosophila I elements. One of the main interests of the I elements is that their mobilization can be induced in vivo by appropriate crosses (4).
I elements control the I-R system of hybrid dysgenesis. Drosophila melanogaster strains can be classified into two categories, inducer strains that contain functional I elements, and reactive strains devoid of functional elements (5). I elements are stable when maintained in inducer strains but transpose at high frequency in the germline of females, called SF females, obtained by crosses between females of reactive strains and males of inducer strains. As a result of I element transposition, SF females are partially sterile, development of their embryos being blocked at early stages (6). The intensity of the sterility of these females, measured as the hatching percentage of the eggs that they lay, correlates to the frequency of transposition of I elements (7). The I element has two ORFs (8,9). The protein encoded by ORF1 is thought to be involved in the formation of ribonucleoprotein complexes (10,11). The protein putatively encoded by ORF2 shows three enzymatic domains: endonuclease, reverse transcriptase and ribonuclease H (8,9,12,13). The 3′ end of the I element is made of tandem TAA repeats. Retro transposition of the I element starts with the synthesis of a full-length RNA transposition intermediate in an amount correlating with the frequency of transposition (14). It is synthesized under the control of an internal RNA polymerase II promoter located in the 5′ untranslated region (UTR) of the element (15). The transcripts produced by I elements at the time of transposition are polyadenylated downstream of the tandem UAA repeats, and reverse transcription is initiated at the level of these repeats (16). How synthesis of the first cDNA strand is arrested at the 5′ end is not known. Previous results indicate that the RNA transposition intermediate of the I element contains the 5′ end of the element and that transcription starts at or near the first nucleotide (14–16). We report here an analysis of the transcripts produced by a population of marked I elements in the ovaries of SF females. We show that one of these I elements located in region 88A produces transcripts that start upstream of the element 5′ end. We provide evidence that the sequences flanking the element that are present in the transcripts are not inserted in the genome after retrotransposition.
MATERIALS AND METHODS
Drosophila melanogaster strains and transgenic lines
All strains were M in the PM system of hybrid dysgenesis. The standard reactive strain used in this work was JA (y, w). The M6.3 transgenic line contains the marked I element I-HA-O1-Δnuc which lacks the nuclease domain and does not transpose autonomously. It was derived from the complete and functional element I-HA-O1 containing a sequence encoding the HA epitope at the beginning of ORF1 (11) (see Fig. 1A). The inducer transgenic line HT1 is described in Seleme et al. (11). It contains several copies of I-HA-O1. Cy/Pm;Dcxf/H is a reactive strain carrying second and third balancer chromosomes marked with Cy and Dcxf, respectively.
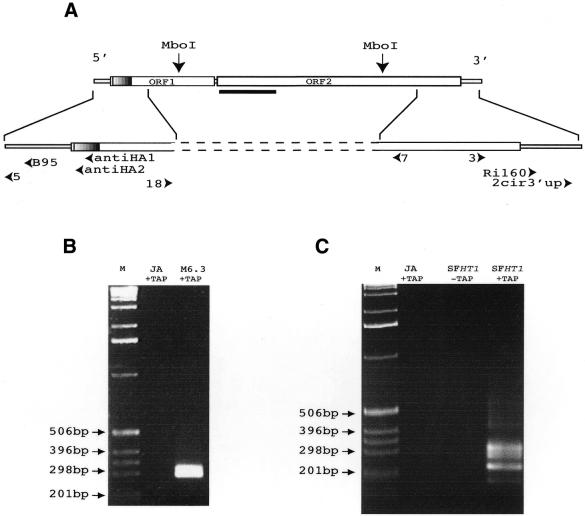
Figure 1.
Analyses of the transcripts produced by the I-HA-O1-Δnuc and I-HA-O1 elements. (A) Structure of the I-HA-O1 and I-HA-O1Δnuc elements. The two large open boxes represent the two ORFs of the I-HA-O1 element, and the small open boxes represent the 5′ and 3′ UTRs. The small hatched region at the beginning of ORF1 indicates the position of the HA epitope which is inserted in frame with the rest of ORF1 (11). The most 5′ and 3′ MboI restriction sites that were used for inverse PCR are indicated. The bold line below the beginning of ORF2 indicates the position of the deletion in I-HA-O1-Δnuc. Arrowheads indicate the positions and orientations of the primers used in RT–PCR and sequencing experiments (see Materials and Methods). (B and C) Analyses on a 1.5% agarose gel of the final products of the RNA circularization/RT–PCR experiments. (B) RNAs were extracted from ovaries of JA females and M6.3 females, transgenic for I-HA-O1-Δnuc. (C) RNAs were extracted from ovaries of JA females and SFHT1 females, issued from crosses between JA females and inducer males of the HT1 line containing five I-HA-O1 elements. RNAs were treated (+TAP) or not (–TAP) with TAP prior to intra-molecular ligation. Reverse transcription was primed using primer antiHA1, and the resulting cDNAs were submitted to two successive PCRs: the first, using primer pair antiHA2/RI160, and the second, using primer pair B95/2cir3′up. M, molecular markers (1 kb ladder from GibcoBRL™).
RNA extraction
Twenty units of Recombinant RNasin Ribonuclease Inhibitor (Promega) were systematically added to all reactions involving RNA. Total RNA was extracted from 50 pairs of ovaries dissected from 1- to 3-day-old females with the RNeasy® Midi kit (Qiagen). Fifty micrograms of RNA was treated with 20 U Rnase-free DNase I (Promega) in 100 µl for 1 h at 37°C. After phenol/chloroform extractions the RNA was ethanol precipitated in 2.5 M ammonium acetate and dissolved in 20 µl of H2O.
RNA circularization, reverse transcription and PCR
Twenty micrograms of RNA was incubated with 2.5 U tobacco acid pyrophosphatase (TAP; Epicentre Technologies) in 20 µl for 1 h at 37°C. TAP treatment removes the Cap at the 5′ end of the transcripts. After ethanol precipitation in 2.5 M ammonium acetate, 5 µg of RNA was incubated with 20 U T4 RNA Ligase (Epicentre Technologies) in 400 µl of optimal buffer (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 20 mM dithiothreitol, 100 µM ATP, 100 µg/ml acetylated BSA) for 16 h at 16°C. After phenol/chloroform extraction the ligated RNA was ethanol precipitated in 2.5 M ammonium acetate. 400 ng of RNA was reverse transcribed using the ThermoScript™ RT–PCR System (Invitrogen) and 10 pmol of primer antiHA1 (5′-CGTAATCTGGGACGTCGTAT GGG-3′) for 1 h at 59°C. The sample was then treated for 20 min at 37°C with 2 U RNase H. Nested PCR amplifications were performed on 2 µl of the RT–PCR, first with primers antiHA2 (5′-TCGTATGGGTAATTTGTCAT-3′) and Ri160 (5′-GTACATAACAAGCCAGCAATTAG-3′) (30 cycles at 94°C for 30 s, 52°C for 30 s, 72°C for 45 s) and second with B95 (5′-GATTTTGCTGATAAGAG-3′) and 2cir3′up (5′-CCCCGTAGCTAATGCTATACTATC-3′) (30 cycles at 94°C for 30 s, 51°C for 30 s, 72°C for 35 s). The PCR products were analyzed on 1.5% agarose gels, cloned using the TOPO TA Cloning® Kit (Invitrogen), and sequenced by Genome Express. The 5′ and 3′ ends of the two I elements studied in this work are the same and the positions of the oligonucleotides on the sequence of I-HA-O1 are shown in Figure 1A.
RT–PCR to detect transcripts from the 88A locus
Five micrograms of total RNA from ovaries was used for reverse transcription with 150 ng of random hexamers for 1 h at 50°C in 20 µl using the ThermoScript™ RT–PCR System (Invitrogen). PCRs were performed during 30 cycles (15 s at 94°C, 15 s at annealing temperature, 15 s at 72°C) using primers 8900 (5′-GTTTTACATTTACAGCATTTA-3′; coordinates 151617–151596 in GenBank accession no. AE003701) and C (5′-AAACTCAAACTCAATCTGGG-3′; coordinates 151404–151424 in GenBank accession no. AE003701) (annealing temperature 54°C), or using primers 8900 and 5 (5′-GAGAGGCACGACTTATCTCTTCGG-3′) (annealing temperature 50°C), or using primers yema-up (5′-CAACGGCGAGCTGAAGTGTGT-3′; coordinates 2677– 2697 in GenBank accession no. X63503), and yema-do (5′-TGTGGTGGGGAGCAGAGTGTC-3′; coordinates 2870– 2890 in GenBank accession no. X63503) (annealing temperature 65°C). For PCR controls, ∼0.5 ng of genomic DNA extracted from JA and HT1 adult flies were also amplified under the same conditions. PCR products were analyzed on 1.5% agarose gels. The positions of the oligonucleotides 8900 and C in the locus 88A and oligonucleotide 5 in the sequence of I-HA-O1 are shown in Figure 3A.
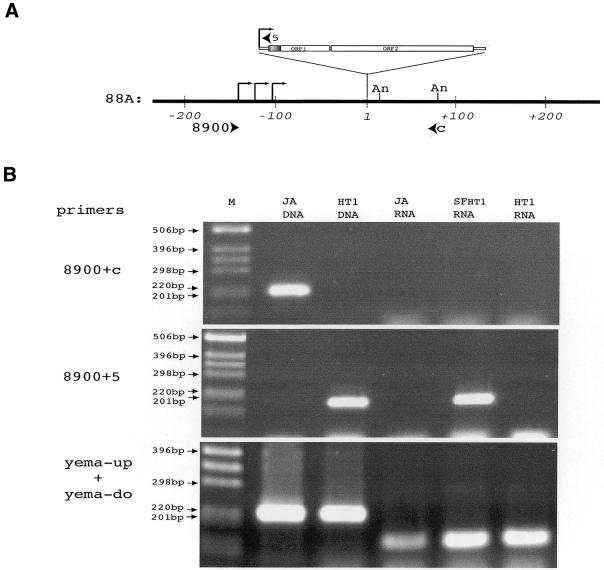
Figure 3.
Transcription of the I-HA-O1(88A) element. (A) Map of the 88A locus around the I-HA-O1(88A) insertion site. Coordinates on genomic DNA are indicated in base pairs, the first nucleotide of the target site duplication generated by the I-HA-O1 insertion being arbitrarily chosen as the origin. The I-HA-O1 element insertion is not shown to scale. The positions of transcription start sites of the transcripts of I-HA-O1(88A) shown in Figure 2B are indicated with broken arrows, and the positions of the polyadenylation sites are indicated with An. Arrowheads indicate the positions and orientations of the primer pairs 8900/5 and 8900/C used for RT–PCR. (B) RT–PCR analysis of the transcripts of the 88A region. RT–PCRs were performed on RNAs extracted from ovaries of JA, SFHT1 and HT1 females as described in Materials and Methods, using random hexamers to prime reverse transcription and primer pairs 8900/C (specific for the empty 88A locus), 8900/5 (specific for the 88A locus containing the I-HA-O1 insertion) and yema-up/yema-do (specific for the gene encoding yemanuclein-α) for PCR amplifications. Genomic DNA extracted from JA and HT1 adult flies were used for PCR controls. PCR products were analyzed on 1.5% agarose gels. M, molecular markers (1 kb ladder from GibcoBRL™).
In situ hybridization to polytene chromosomes of salivary glands of larvae
Salivary gland polytene chromosomes were treated according to a modified version of the protocol of Zink and Paro (17) using as probe the I407 element (5) labeled by nick translation with biotin-14-dATP (BioNick DNA Labelling System; Gibco BRL). Hybridizations were done overnight at 37°C. The chromosomes were mounted in mowiol and analyzed by fluorescent microscopy.
Determination of sequences at the ends of transposed copies
Genomic DNA was extracted from pools of 25 white-eyed males using standard procedures (18). Inverse PCR experiments were performed as previously described (19) except that the restriction enzyme MboI was used to digest genomic DNA, and oligonucleotides antiHA2 and 18 (5′-TTGCCACC AACAGATTTGGG-3′) were used to amplify the 5′ end, and oligonucleotides 7 (5′-TCGCAAGGTCGGCTTTAAGG-3′) and 3 (5′-ACCCTCTAGACCTTCTTAGC-3′) were used to amplify the 3′ end (Fig. 1A). Once an insertion site was defined, the sequences at the other extremity of the element were determined by PCR using a primer specific for the I element sequence and the other specific for the flanking DNA sequence. PCR products were cloned using the Stratagene PCRScriptAmpSK(+) cloning kit, and sequenced.
RESULTS
Capped transcripts of HA-tagged I elements are revealed by an RNA circularization/RT–PCR method
The genome of all strains of D.melanogaster contains defective I elements located in pericentromeric heterochromatic regions (5,20,21). They are ∼93% identical to active I elements. Some of them are constitutively transcribed (14), making analyses of RNAs synthesized by active I elements difficult to perform. In order to overcome these problems we decided to use a marked I element carrying the HA epitope tag at the beginning of ORF1.
We first analyzed the transcripts produced by the I-HA-O1-Δnuc element that is deleted of the nuclease domain. I-HA-O1-Δnuc is unable to transpose autonomously but produces high levels of transcripts in the germ lines of transgenic females derived from the JA reactive strain (4). In order to characterize these transcripts, we used the RNA circularization/RT–PCR method (16,22,23). Total RNAs were extracted from ovaries of M6.3 transgenic females as well as of JA females as a negative control. Intra-molecular ligations after TAP treatment were performed. Reverse transcription was then primed using an oligonucleotide complementary to the DNA sequence encoding the HA tag, and was followed by two sequential PCRs using a set of nested primers (see Materials and Methods and Fig. 1A). The PCR products were analyzed on an agarose gel (Fig. 1B). They should be 166 bp if they derive from circularized transcripts starting at the first nucleotide of the I factor and ending at the last TAA repeat. Two major PCR products of ∼260–340 bp were obtained after circularization/RT–PCR of RNA extracted from M6.3 ovaries. As expected, no PCR product was obtained after circularization/RT–PCR of RNA extracted from the JA strain. This indicates that the experimental procedure allows specific detection of transcripts from the I-HA-O1-Δnuc element in line M6.3, and that these transcripts seem at least 100 nt longer than expected if they correspond only to I-HA-O1-Δnuc sequences.
We then used the same experimental design to study the transcripts produced by active I elements marked with the HA-tag at the beginning of ORF1. HT1 is a typical inducer line derived from the JA reactive strain after propagation of the autonomous I-HA-O1 element in the genome (11). I-HA-O1 elements in the HT1 line were mapped by in situ hybridization to polytene chromosomes of salivary glands of larvae (data not shown); four elements map on chromosome 2 in 23D, 32F, 36A-B and 47F, and one maps on chromosome 3 in 88A. The elements located in 36A-B and 47F correspond to original transgenes. We analyzed the transcripts produced by this population of I-HA-O1 elements in the ovaries of SF females. RNAs were prepared from ovaries of females (named SFHT1) resulting from dysgenic crosses between HT1 inducer males and JA reactive females.
They were circularized and the transcripts of the I-HA-O1 elements were analyzed by RT–PCR as described above. The results are shown in Figure 1C. Several PCR products of discrete sizes were observed with RNAs extracted from SFHT1 females treated with TAP but not when the TAP treatment was omitted. The main products are between 200 and 340 bp, but some minor bands are observed below and above these sizes. This suggests that the I-HA-O1 elements of the HT1 line produce different species of transcripts in the ovaries, generally longer than expected (166 bp; see above) if they contain only I-HA-O1 sequences. The lack of amplification products when TAP treatment is omitted indicates that I-HA-O1 element RNAs carry a Cap structure at the 5′ end like conventional messenger RNAs. No signal was observed with RNA from JA females.
Transcripts of the I-HA-O1 element located in the 88A locus in HT1 contain 5′- and 3′-flanking sequences
In order to obtain information on the sequences at the 3′ and 5′ ends of the transcripts produced by the I element, the RT–PCR products obtained above were cloned and sequenced. The sequences of the RNA species are shown in Figure 2.
Figure 2.
Sequences of the transcripts produced by the I-HA-O1-Δnuc and I-HA-O1 elements. Sequences from the I elements are in bold. Some sequences were omitted for clarity (the numbers of nucleotides are indicated in parentheses). (A) Top, sequence of the I-HA-O1-Δnuc element contained in the transgene used to make line M6.3. Bottom, sequences of the transcripts produced by this transgene. (B) (1–3) Sequences of the three types of transcripts detected in SFHT1 (see text). Sequences of the 5′ and 3′ ends of the I-HA-O1(88A) element and of its flanking sequences are shown on top of the sequences of the transcripts of this element (3). The target site duplication surrounding this element is underlined.
The structures of the transcripts produced by I-HA-O1-Δnuc are consistent with the sizes of the PCR products observed in Figure 1B. They all start at the second nucleotide of the I element, and terminate in the flanking sequences with a poly(A) tail (Fig. 2A). The sites of polyadenylation are 57, 62 and 66 nt downstream of the 3′ end of the I element, and the number of A residues varies between 20 and 50.
Transcripts produced by the I-HA-O1 elements in the ovaries of SFHT1 females belong to one of the three following categories (Fig. 2B), and their structures are also consistent with the sizes of the PCR products observed in Figure 1C. (i) A first category includes transcripts whose ends are similar to the ends of transcripts produced by I-HA-O1-Δnuc. Since flanking sequences of both I-HA-O1-Δnuc and I-HA-O1 are identical in the transgenes, these transcripts are derived from I elements present in the original transgenes used to make HT1, located at 36A-B and 47F. (ii) A second category represents I-HA-O1 transcripts that start at the second nucleotide of the I element and are polyadenylated only a few nucleotides downstream of the 3′ end. They could originate from any I-HA-O1 element. (iii) The third category represents I-HA-O1 transcripts with extra sequences at both the 5′ and 3′ ends. Blast analysis on the sequenced D.melanogaster genome (24) indicates that these flanking sequences match a unique sequence contained in scaffold AE003701. This scaffold overlaps region 88A on chromosome 3 where an I-HA-O1 element was shown to be inserted by in situ hybridization experiments (see above). The element at the origin of these transcripts is therefore the I-HA-O1 element located in 88A. We sequenced the 88A locus in the HT1 line and confirmed that it contains an insertion of an I-HA-O1 element [I-HA-O1(88A)] flanked by a target site duplication of 12 nt (Fig. 2B). Five different transcripts of the I-HA-O1(88A) element containing flanking sequences of variable length were detected (Fig. 2B). They are polyadenylated at two distinct sites in the 3′-flanking sequences. At the 5′ ends, the sequences upstream of the I elements extend up to 144 nt in the flanking DNA. The presence of genomic sequences flanking I element transcripts at the 5′ end has never been reported before.
The 88A transcription unit results from the presence of an I element in this region
The results reported above indicate that RNAs from the I-HA-O1 element in 88A are read-through transcripts initiated in 5′-flanking sequences. In order to study the transcripts of this region in the presence and absence of the I-HA-O1 element, we extracted total RNAs from the ovaries of HT1, JA and SFHT1 females produced by crossing JA females with HT1 males. These RNAs were used for reverse transcription with random hexamers followed by PCR with specific primers pairs (Fig. 3A). The first primer pair was 8900 and C, both specific to sequences of the 88A locus (Fig. 3A). They amplify a genomic DNA fragment of 212 nt, corresponding to the empty 88A locus in the JA strain, but no RT–PCR product was observed with RNAs extracted from ovaries of JA females (Fig. 3B). This indicates that the empty 88A locus is very poorly transcribed, if at all, in ovaries. Similarly, no RT–PCR product was observed using primers designed to amplify a fragment extending 1200 nt upstream of the I-HA-O1 element insertion site in cDNAs originating from JA, HT1 and SFHT1 females (data not shown). The second primer pair was 8900, specific for sequences of the 88A locus, and 5, specific for the I element (Fig. 3A). They amplify a genomic fragment of 183 nt in the 88A locus containing the I-HA-O1 element of the HT1 line (Fig. 3B). In these experiments, a discrete band of the expected size was observed with RNA extracted from SFHT1 ovaries (Fig. 3B). This band is not observed with RNA extracted from HT1 ovaries. As a control, PCRs were also performed using the primer pair yema-up and yema-do. These primers are located in two exonic regions separated by a short intron in the sequence of the yemanuclein-α gene (25). They should amplify a product of 214 bp on the genomic DNA and a product of 154 bp on the cDNA from the yemanuclein-α gene. PCR products of the expected sizes were obtained in all cases (Fig. 3B). Therefore, these experiments confirm that some transcripts containing sequences from I-HA-O1 also contain sequences flanking the element at its 5′ end. Furthermore, they indicate that synthesis of these transcripts occurs in ovaries of dysgenic females, where I elements are active, and is repressed in the ovaries of the inducer HT1 line, where I elements are silenced. Therefore, the I element promoter may initiate transcription at cryptic start sites located upstream in the genomic DNA.
Sequences flanking the I-HA-O1(88A) element are not integrated after retrotransposition
We studied the ability of the I-HA-O1(88A) element to transpose. Since the frequency of I element transposition correlates with the intensity of the sterility of SF females (7), we measured the hatching percentage of the eggs produced by SF females containing only the I-HA-O1(88A) element (SF88A) issued from the crosses represented in Figure 4A. It was 47% which is similar to the hatching percentage of the eggs laid by SF females produced by dysgenic crosses between males of the inducer HT1 line and JA females (11). This suggests that the I-HA-O1(88A) element transposes efficiently.
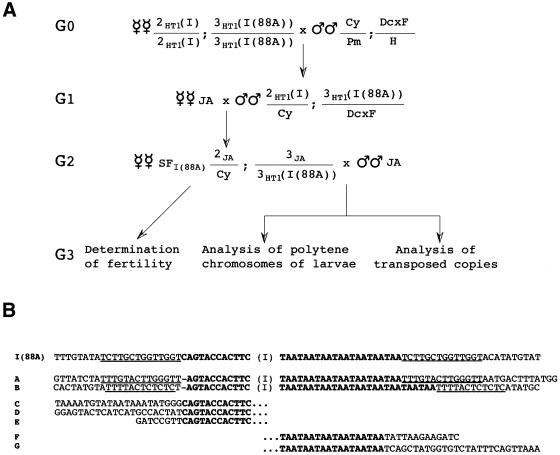
Figure 4.
Transposition of I-HA-O1(88A). (A) Scheme of the crosses performed to produce SF females containing only the I-HA-O1 element located in 88A. 2HT1(I) and 3HT1(I(88A)) indicate chromosomes 2 and 3 of the HT1 stock, and 2JA and 3JA chromosomes 2 and 3 from the JA reactive stock. Cy/Pm;Dcxf/H is a reactive strain that carries balancer chromosomes marked with the Cy and Dcxf mutations. (B) Sequences of transposed copies of I-HA-O1(88A). I element sequences are in bold. The sequences of both ends (A and B), of the 5′ end (C, D and E), or of the 3′ end (F and G) of transposed copies produced by the I-HA-O1(88A) element in the crosses drawn in (A) are shown. Target site duplications are underlined.
Moreover, we studied the presence of transposed copies of I-HA-O1(88A) in the progeny of SF88A females, by in situ hybridization experiments to salivary gland chromosomes of larvae using an I factor probe. We examined 20 larvae issued from SF88A females. Seven of them had new insertions (varying from one to 12) on their chromosomes (data not shown). Therefore, the I-HA-O1(88A) element can transpose autonomously.
We analyzed the sequences at the ends of some transposed copies. For this, we extracted DNA from flies obtained in the progeny of SF88A females (Fig. 4A) and performed inverse PCR experiments. The products were sequenced and the results are shown in Figure 4B. In two cases, we obtained sequences of both the 5′ and 3′ ends of new insertions, and observed the presence of target site duplications as expected. In other cases, we obtained sequences of either the 5′ or the 3′ ends of new insertions. We observed the usual variation of the TAA repeats at the 3′ end, resulting from either a reduction (six repeats) or an increase (nine repeats) of the number of tandem repeats which was seven in the progenitor I-HA-O1(88A) element. In all cases, none of the 88A sequences adjacent to I-HA-O1(88A) in the chimeric transcripts were inserted into chromosomal DNA in the transposed copies.
DISCUSSION
We used a functional I element marked with a unique sequence to detect specifically transcripts and transposed copies of this element. An intra-molecular RNA ligation technique allowed us to determine the 3′ and the 5′ ends of the same RNA molecules produced by the marked elements. We have analyzed the transcripts produced by the population of active I elements in the ovaries of dysgenic females issued from the inducer line HT1. At least three elements out of five are transcribed and are therefore susceptible to transpose. These transcripts contain poly(A) tails added at various sites in the adjacent 3′ sequences as previously observed (16). Our results also provide information about the structure of the transcripts at the 5′ end. Most transcripts initiate at the 5′ end of the I element at the second nucleotide as previously described and discussed (16). Surprisingly, the I-HA-O1 element located in 88A [I-HA-O1(88A)] produces two types of transcripts. One type starts at the 5′ end of the element as usual, while the other type contains flanking sequences adjacent to the element at the 5′ end. These additional sequences extend up to 140 nt. We identified in the Genome Annotation Database of Drosophila (GadFly) only one putative transcription unit (CG14365) encompassing the insertion site of the element, but it is not in the right orientation to produce the transcripts that we have isolated. Furthermore, we did not detect any transcript in this region in the absence of the I element in RT–PCR experiments on RNA extracted from ovaries. This suggests that the I-HA-O1 element in 88A is not inserted within a pre-existing ovarian transcription unit, or that it is inserted into a transcription unit that is poorly active in ovaries. We have also shown that, in ovaries, transcripts from this region are observed only in SF females, in conditions where I elements are actively transcribed, and not in the inducer HT1 line, in conditions where they are repressed. This indicates that synthesis of these transcripts is controlled by regulatory sequences provided by the element itself. The presence of these regulatory sequences might reveal cryptic transcription start sites located upstream of the I element. This is an illustration of the way in which transposable elements can take control of the expression of host sequences.
Although a proportion of the transcripts produced by I-HA-O1(88A) are initiated upstream of the element, we did not detect any chimeric transposed copy of this element. This indicates that either the extra sequences at the 5′ end of the transcripts are not inserted during the retrotransposition process, or that they insert very rarely. This might result from the fact that transcripts containing additional sequences cannot be used as retrotransposition intermediates. However, we cannot exclude that sequences upstream of the I element 5′ end in the RNA transposition intermediate are eliminated by an unknown mechanism during the reverse transcription/insertion process. Very little is known about the arrest of reverse transcription and initiation of second strand cDNA synthesis in the TPRT mechanism according to which I elements are thought to transpose. In the case of R2 elements, it has been proposed that the 5′-end junction results from recombination involving 5′-flanking sequences in the first cDNA strand and the target site (26). R2 is co-transcribed with the 28S RNA gene and the 5′ end of its RNA is homologous to 28S DNA sequences where the element integrates. The I element has no specific target site and therefore cannot use a similar mechanism.
Two I elements that integrated in the genome together with unrelated sequences located between one copy of the target site duplication and the 5′ end of the element have been described (27). Chimeric transcripts similar to those reported here might be at the origin of these hybrid elements. Although such events seem to occur rarely, they might contribute to a new type of exon shuffling different from that observed with L1 elements and sequences located downstream of these elements (28–30).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to François Rassendren and Edouard Bertand for helpful advice during this work. This work was supported by grants from the Association pour la Recherche sur le Cancer (ARC) to A.B. S.C. was the recipient of fellowships from the Fondation pour la Recherche Médicale (FRM) and the Ministère de la Recherche et de la Technologie. S.R. was the recipient of fellowships from la Recherche sur le Cancer (ARC) and the Ministère de la Recherche et de la Technologie.
REFERENCES
- 1.Luan D.D., Korman,M.H., Jakubczak,J.L. and Eickbush,T.H. (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell, 72, 595–605. [DOI] [PubMed] [Google Scholar]
- 2.Luan D.D. and Eickbush,T.H. (1995) RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol., 15, 3882–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J., Malik,H.S. and Eickbush,T.H. (1999) Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl Acad. Sci. USA, 96, 7847–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucheton A., Busseau,I. and Teninges,D. (2002) I elements in Drosophila. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. ASM Press, Washington DC, pp. 796–812.
- 5.Bucheton A., Paro,R., Sang,H.M., Pelisson,A. and Finnegan,D.J. (1984) The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning and properties of the I factor. Cell, 38, 153–163. [DOI] [PubMed] [Google Scholar]
- 6.Lavige J.-M. (1986) I-R system of hybrid dysgenesis in Drosophila melanogaster: further data on the arrest of development of the embryos from SF females. Biol. Cell, 56, 207–216. [Google Scholar]
- 7.Picard G. (1978) Non mendelian female sterility in Drosophila melanogaster: further data on chromosomal contamination. Mol. Gen. Genet., 164, 235–247. [Google Scholar]
- 8.Abad P., Vaury,C., Pelisson,A., Chaboissier,M.C., Busseau,I. and Bucheton,A. (1989) A long interspersed repetitive element—the I factor of Drosophila teissieri—is able to transpose in different Drosophila species. Proc. Natl Acad. Sci. USA, 86, 8887–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fawcett D.H., Lister,C.K., Kellett,E. and Finnegan,D.J. (1986) Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell, 47, 1007–1015. [DOI] [PubMed] [Google Scholar]
- 10.Dawson A., Hartswood,E., Paterson,T. and Finnegan,D.J. (1997) A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J., 16, 4448–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seleme M., Busseau,I., Malinsky,S., Bucheton,A. and Teninges,D. (1999) High-frequency retrotransposition of a marked I factor in Drosophila melanogaster correlates with a dynamic expression pattern of the ORF1 protein in the cytoplasm of oocytes. Genetics, 151, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Q., Moran,J.V., Kazazian,H.H.,Jr and Boeke,J.D. (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell, 87, 905–916. [DOI] [PubMed] [Google Scholar]
- 13.Martin F., Maranon,C., Olivares,M., Alonso,C. and Lopez,M.C. (1995) Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J. Mol. Biol., 247, 49–59. [DOI] [PubMed] [Google Scholar]
- 14.Chaboissier M.C., Busseau,I., Prosser,J., Finnegan,D.J. and Bucheton,A. (1990) Identification of a potential RNA intermediate for transposition of the LINE-like element I factor in Drosophila melanogaster. EMBO J., 9, 3557–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLean C., Bucheton,A. and Finnegan,D.J. (1993) The 5′ untranslated region of the I factor, a long interspersed nuclear element-like retrotransposon of Drosophila melanogaster, contains an internal promoter and sequences that regulate expression. Mol. Cell. Biol., 13, 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chambeyron S., Bucheton,A. and Busseau,I. (2002) Tandem UAA repeats at the 3′-end of the transcript are essential for the precise initiation of reverse transcription of the I factor in Drosophila melanogaster. J. Biol. Chem., 277, 17877–17882. [DOI] [PubMed] [Google Scholar]
- 17.Zink D. and Paro,R. (1995) Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J., 14, 5660–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gloor G.B., Preston,C.R., Johnson-Schlitz,D.M., Nassif,N.A., Phillis,R.W., Benz,W.K., Robertson,H.M. and Engels,W.R. (1993) Type I repressors of P element mobility. Genetics, 135, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaboissier M.C., Finnegan,D. and Bucheton,A. (2000) Retrotransposition of the I factor, a non-long terminal repeat retrotransposon of Drosophila, generates tandem repeats at the 3′ end. Nucleic Acids Res., 28, 2467–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crozatier M., Vaury,C., Busseau,I., Pelisson,A. and Bucheton,A. (1988) Structure and genomic organization of I elements involved in I-R hybrid dysgenesis in Drosophila melanogaster. Nucleic Acids Res., 16, 9199–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaury C., Abad,P., Pelisson,A., Lenoir,A. and Bucheton,A. (1990) Molecular characteristics of the heterochromatic I elements from a reactive strain of Drosophila melanogaster. J. Mol. Evol., 31, 424–431. [DOI] [PubMed] [Google Scholar]
- 22.Couttet P., Fromont-Racine,M., Steel,D., Pictet,R. and Grange,T. (1997) Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA, 94, 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fromont-Racine M., Bertrand,E., Pictet,R. and Grange,T. (1993) A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res., 21, 1683–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 25.Ait-Ahmed O., Bellon,B., Capri,M., Joblet,C. and Thomas-Delaage,M. (1992) The yemanuclein-alpha: a new Drosophila DNA binding protein specific for the oocyte nucleus. Mech. Dev., 37, 69–80. [DOI] [PubMed] [Google Scholar]
- 26.Eickbush D.G., Luan,D.D. and Eickbush,T.H. (2000) Integration of Bombyx mori R2 sequences into the 28S ribosomal RNA genes of Drosophila melanogaster. Mol. Cell. Biol., 20, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busseau I., Pelisson,A. and Bucheton,A. (1989) I elements of Drosophila melanogaster generate specific chromosomal rearrangements during transposition. Mol. Gen. Genet., 218, 222–228. [DOI] [PubMed] [Google Scholar]
- 28.Boeke J.D. and Pickeral,O.K. (1999) Retroshuffling the genomic deck. Nature, 398, 108–109, 111. [DOI] [PubMed] [Google Scholar]
- 29.Eickbush T. (1999) Exon shuffling in retrospect. Science, 283, 1465, 1467. [DOI] [PubMed] [Google Scholar]
- 30.Kazazian H.H. Jr (2000) L1 retrotransposons shape the mammalian genome. Science, 289, 1152–1153. [DOI] [PubMed] [Google Scholar]