Abstract
The kink-turn is a widespread motif in RNA consisting of a three-nucleotide bulge flanked on one side by consecutive A3G mismatches. Important examples are found in the ribosome, U4 RNA, and in snoRNAs involved in RNA modification. The motif is a common protein binding site, and the RNA has been found to adopt a tightly kinked conformation in crystal structures. However, in free solution there is a dynamic exchange between kinked and extended conformations, with the equilibrium driven toward the kinked form by the addition of metal ions. Here we used fluorescence resonance energy transfer (FRET) to show that the L7Ae protein of Archaeoglobus fulgidus binds to RNA containing a kink-turn with nanomolar affinity, and induces folding into the tightly kinked conformation even in the absence of metal ions. Thus this RNA may act as a relatively flexible hinge during RNA folding, until fixed into its ultimate kinked structure by the binding of L7 or related protein.
Keywords: RNA structure, RNA–protein interaction, K-turn
INTRODUCTION
Kink-turns (K-turns) are commonly occurring sequences that can introduce a sharp kink into double-helical RNA (Klein et al. 2001). They were first identified as a novel motif occurring at several locations in crystal structures of ribosomal subunits, but have also been found in mRNA (Mao et al. 1999; Winkler et al. 2001; Allmang et al. 2002), guide RNAs (Kuhn et al. 2002; Watkins et al. 2002; Bortolin et al. 2003; Marmier-Gourrier et al. 2003; Rozhdestvensky et al. 2003), and spliceosomal RNA (Vidovic et al. 2000; Watkins et al. 2000). They are thus important in translation, RNA modification, spliceosome assembly, and the control of gene expression.
The archetypal K-turn comprises a three-nucleotide bulge (of which the 5′ is generally a purine) preceded by two consecutive A·G mismatches (the N-C stem), and followed by a section of regular basepairing (the C stem) (Fig. 1; Klein et al. 2001). This motif has been observed to generate a large kink in the RNA in a number of crystal structures, with an included angle between the axes of the N-C and C stems of ~60°, i.e., a 120° deviation from a linear duplex. This is stabilized by A-minor interactions of the stacked adenosines of the A·G mismatches with the C stem, and by stacking of the 5′ (often purine) and following bases of the bulge on the ends of the C and N-C stems respectively. The 3′ base of the bulge is extruded away from the RNA structure.
FIGURE 1.
The kink-turn motif in RNA. (A) The sequence of an archetypical K-turn. The Kt-7 sequence found in 23S rRNA. (B) The structure of the Kt-7 motif (Klein et al. 2001) taken from the crystal structure of the 50S ribosomal subunit. The conserved G·A mismatches define the noncanonical helix (NC), and the fully base-paired stem is called the canonical (C) helix. The coloring matches that of the sequence in part A.
However, the structure of the K-turn that emerges from X-ray crystallography is only part of the description of these motifs. It has emerged that the free RNA structure is dynamic, and dependent on the binding of metal ions. RNA fragments containing K-turn sequences were found to exhibit strongly retarded electrophoretic mobility provided that divalent metal ions were present, but in the absence of added metal ions the RNA migrated at a speed that is typical of a normal 3-nucleotide bulge, i.e. much less tightly kinked (Goody et al. 2003; Matsumura et al. 2003). This suggested that the strongly kinked structure of the K-turn in free RNA required the binding of metal ions. These observations were extended by steady-state fluorescence resonance energy transfer (FRET) measurements (Goody et al. 2003) showing that K-turn sequences undergo an ion-induced two-state transition to the strongly kinked state, with a half-magnesium concentration of 1 mM. The same degree of kinking could also be obtained using monovalent ions, with a half-sodium concentration of 72 mM. Time-resolved FRET analysis of the system further underlined the dimorphic nature of the K-turn as free RNA, showing the presence of the kinked and extended forms under all conditions. For example, in the presence of 50 mM Mg2+ ions, 70% of the RNA was in the tightly kinked conformation, with calculated interfluorophore distances that were in good agreement with those calculated from K-turn structures observed in the crystal. However, the remaining 30% of the molecules were in the extended form under the same conditions. At more physiologically relevant ionic concentrations this equilibrium would be less biased toward the kinked geometry. Molecular dynamics simulations have also suggested that K-turn structures are flexible (Razga et al. 2004; Cojocaru et al. 2005).
Most natural K-turn sequences serve as binding sites for proteins; five of the six K-turns of the H. marismortui 50S ribosomal subunit are bound by proteins, for example (Klein et al. 2001). The most common K-turn binding protein is the ribosomal L7 and related proteins. These form a family of RNA-binding proteins including the eukaryotic and archaeal proteins L7Ae, L30e, and S12e (Koonin et al. 1994), the yeast Nhp2 and Snu13p proteins, and the human 15.5-kDa protein (Nottrott et al. 1999). All these proteins are known to bind to K-turn motifs in RNA, and some functional substitutions are tolerated (Rozhdestvensky et al. 2003). K-turns are found in the box C/D and H/ACA guide RNAs that direct methylation and pseudouridylation respectively of RNAs in archaea and eukaryotes (Kiss-Laszlo et al. 1996; Ganot et al. 1997). These modifications require the formation of a nucleoprotein complex, but in each case the initiation of the assembly requires the binding of an L7Ae (in archaeal sRNA) or 15.5 kDa (in eukaryotic snoRNA) to the K-turn motif. Telomerase RNA also contains box H/ACA sequences within the stability domain (Mitchell et al. 1999) and bind some proteins in common with the editing snoRNAs (Filipowicz and Pogacic 2002), and thus similar interactions may be important in telomere function. The 15.5-kDa protein also binds a K-turn within the U4 stem-loop within the U4-U6.U5 tri-snRNP (Nottrott et al. 1999). Crystal structures have been obtained for the complexes of Archaeoglobus fulgidus L7Ae and box C/D RNA (Moore et al. 2004), Methanococcus jannaschii L7Ae and box H/ACA RNA (Hamma and Ferré-D’Amaré 2004), and the human 15.5-kDa protein and the U4 snRNA (Vidovic et al. 2000). In each case closely similar protein folds bind to the K-turns in the same manner. The RNA adopts the tightly kinked conformation observed in the ribosome (Klein et al. 2001). The protein is bound to the major groove side of the K-turn making contact with both C and NC stems, with the loop region and the extended central base held in a pocket within the protein.
The structural dimorphism of the K-turn suggests that is unlikely to act by itself as an organizing feature of RNA conformation. It may therefore be that the strongly kinked structure is stabilized by the binding of proteins to generate the functional RNA structure within the ribosome, snoRNP complexes, or elsewhere. We have therefore sought to explore whether the kinked structure of the near-consensus Kt-7 RNA (Klein et al. 2001) is stable when bound by the A. fulgidus L7Ae protein as an archetypal K-turn binding protein. Using steady-state and time-resolved FRET measurements, we found that the tightly kinked structure is indeed induced upon binding of L7Ae protein, even in the absence of added metal ions.
RESULTS
Preparation of the L7Ae protein of Archaeoglobus fulgidus
The L7Ae protein was expressed as an N-terminal hexahistidine fusion in Escherichia coli BL21 cells from a plasmid containing a copy of the ribosomal gene from A. fulgidus (Rozhdestvensky et al. 2003). The protein was purified using Ni-NTA agarose and the oligohistidine tag removed by cleavage with thrombin. The purified L7Ae protein migrated as a single species on electrophoresis in an SDS-containing polyacrylamide gel (Fig. 2A), and had a molecular mass of 13,490 (calculated = 13,484) by MALDI-TOF mass spectrometry.
FIGURE 2.
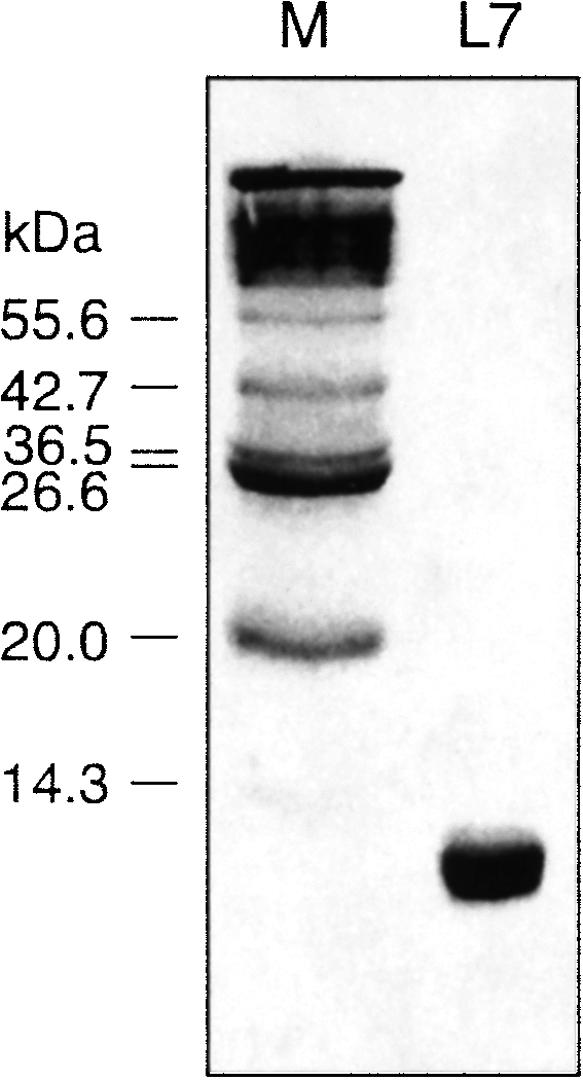
Purified L7Ae protein from Archaeoglobus fulgidus analyzed by PAGE in the presence of SDS. The left track (M) contained a mixture of proteins (2–212 kDa) to act as size markers (molecular masses indicated at left).
Protein-induced Kt-7 kinking revealed by steady-state FRET measurements
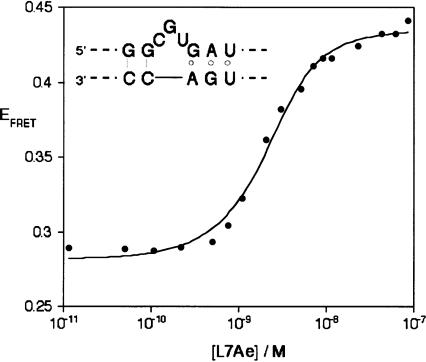
We explored the effect on the conformation of the RNA using steady-state FRET. For this purpose we constructed a duplex containing a Kt-7 sequence flanked by helices of 12 base pairs (C helix) and 13 bp (NC helix, including the A·G mispairs) (Fig. 3A). Initially, we considered the possibility that the binding of the protein might restrict the motion of the fluorescein, and thereby complicate the interpretation of FRET efficiencies in terms of distances. We therefore made a version of the K-turn-containing RNA that was singly labeled with fluorescein at the 5′ terminus of the bulge-containing strand, and we measured the fluorescence anisotropy as a function of L7Ae concentration (Fig. 3A). In the absence of protein the fluorescein had an anisotropy of r = 0.091, consistent with free mobility of the terminally attached fluorophore. The anisotropy remained low (r ≤ 0.10) over the range of concentration 0–80 nM. We conclude that the binding of the L7Ae to the K-turn-containing duplex does not affect the mobility of the fluorescein attached to the 5′ terminus. We could therefore proceed to measurements of FRET confident that restriction of fluorophore mobility would not complicate the interpretation.
FIGURE 3.
Binding of L7Ae protein to RNA containing a K-turn analyzed by FRET in the steady state. The schematic at the top shows the RNA construct used in the fluorescence studies. This contains a central Kt-7 sequence with helical arms of 12 bp (C helix) and 13 bp (N-C helix, including the A·G mispairs). Fluorescein (FRET donor) is attached at the 5′ terminus of the bulge-containing strand, and Cy3 (FRET acceptor) to the 5′ terminus of the nonbulge-containing strand. A version of this species lacking the Cy3 was also prepared for measurement of fluorescein anisotropy. (A) Addition of L7Ae to the RNA in the absence of Mg2+ ions. Plot of fluorescein anisotropy (r, ○) and FRET efficiency (EFRET, •) as a function of added L7Ae concentration. Note the pronounced change in FRET efficiency with addition of nanomolar concentrations of L7Ae, indicating a shortening of the end-to-end distance. Over the same range there is little change in fluorescein anisotropy. The data were fitted to a model assuming stoichiometric binding of L7Ae to the RNA. (B) Addition of L7Ae to the RNA in the presence of 10 mM Mg2+ ions. Plot of FRET efficiency (EFRET) as a function of added L7Ae concentration. Under these conditions the RNA is significantly folded in the absence of protein, but addition of L7Ae induces an increase in EFRET from 0.4 to 0.5, indicating a further stabilization of the kinked conformation by the protein. The data were fitted to the same stoichiometric binding model.
For FRET studies we constructed an RNA of the same sequence as that above, differing only in having Cy3 attached to the 5′ terminus of the nonbulged strand. Using this species we could estimate the axial distortion of the RNA at the K-turn by the efficiency of energy transfer between the donor (fluorescein) and acceptor (Cy3) fluorophores. The FRET efficiency (EFRET) depends on the distance between the fluorophores (R) according to (Förster 1948):
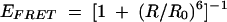 |
(1) |
where R0 is the Förster length, which is 56 Å for this donor–acceptor pair (Norman et al. 2000). The interfluorophore distance will be reduced by significant kinking of the helical axis at the K-turn motif. We showed previously that FRET efficiency in this construct increases from 0.2 to 0.4 on addition of 10 mM Mg2+ ions, due to stabilization of the tightly kinked conformation of the K-turn (Goody et al. 2003).
We studied the energy transfer in the absence of divalent metal ions. Without added protein, a FRET efficiency of 0.2 was obtained. However, on addition of L7Ae protein the efficiency increased markedly, reaching a plateau value of 0.5 (Fig. 3A). The change in FRET reveals the binding of the L7Ae protein to the RNA, and the change in efficiency could be well fitted by a model involving the stoichiometric binding of the protein. Repeated titrations with different RNA and protein preparations have given an apparent dissociation constant of Kd = 0.7 ± 0.2 nM. The constant low value of fluorescein anisotropy over the concentration range where the FRET efficiency changes permits us to interpret the change in terms of the interfluorophore distance. Significantly, this indicates that the binding of the L7Ae protein induces the kinked conformation of the K-turn under these conditions, i.e., in the absence of divalent metal ions.
The titration of FRET efficiency against L7Ae concentration was also performed in the presence of 10mM Mg2+ ions (Fig. 3B). In the absence of protein the FRET efficiency was 0.4, as observed with steady-state FRET measurements on the free RNA previously (Goody et al. 2003). Addition of L7Ae protein resulted in a small increase in EFRET, to reach a value of 0.55. The free RNA is known to exist in an equilibrium between kinked and less-kinked conformations in the presence of 10 Mg2+ ions (Goody et al. 2003), so it is likely that the binding of the L7Ae protein fixes the kinked conformation, thereby driving the equilibrium toward that form. The data were fitted to the same stoichiometric binding model, giving a similar Kd = 0.3 nM.
We conclude that the L7Ae protein binds to the K-turn-containing RNA, inducing a marked change in conformation consistent with the strongly kinked conformation (see Discussion). This does not require the presence of magnesium ions, and binding occurs with a similar affinity in the presence or absence of divalent metal ions.
Protein-induced Kt-7 kinking analyzed by time-resolved FRET measurements
Time-resolved FRET measurements provide additional data on the distributions of different species in solution. We therefore carried out frequency-domain fluorescence lifetime measurements on the donor–acceptor-labeled K-turn RNA in the presence and absence of a saturating concentration of L7Ae protein.
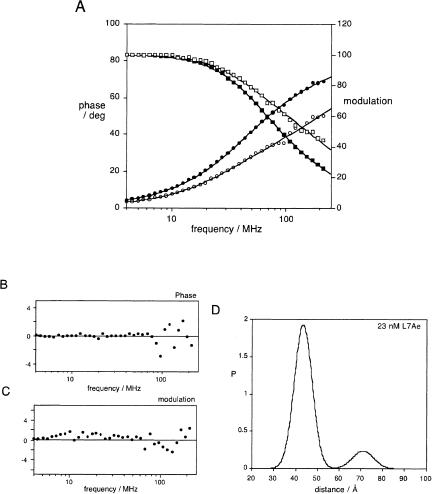
We measured the phase shift and demodulation of fluorescein emission as a function of the frequency of modulation of the excitation intensity in the presence of 0 or 23 nM L7Ae protein, in the presence of 1 nM Mg2+ ions (Fig. 4A). Simple inspection of the data shows the shift of the data to higher frequencies on addition of L7Ae, indicative of a shortening of the fluorescein lifetime. This is consistent with increased energy transfer due to protein-induced folding of the K-turn RNA.
FIGURE 4.
Time-resolved FRET analysis of the structure of the K-turn-containing RNA bound by L7Ae protein in 1 nM Mg2+. (A) Plots of phase shift and demodulation for fluorescein emission as a function of modulation frequency in the presence (open symbols) or absence (filled symbols) of 23 nM L7Ae. The phase shift is plotted as circles, while the modulation is plotted as squares. The best fits using two Gaussian distributions of donor–acceptor lengths with and without L7Ae are shown by the lines. These fits were performed nonglobally on the individual data for free or bound RNA. (B,C) Plots of phase and modulation residuals respectively for the data in the presence of 23 nM L7Ae. (D) Donor–acceptor length distributions for the K-turn RNA in the presence of 23 nM L7Ae, modeled as two Gaussian distributions of donor–acceptor lengths. Under these conditions 87% of the RNA is in the tightly kinked conformation with a donor–acceptor length of 43.7 Å.
Our previous studies of the free RNA (Goody et al. 2003) indicated that this was well described by a two-state equilibrium of kinked and extended structures, and the current steady-state FRET measurements indicated that a two-state equilibrium also provides a good description of the RNA in the presence of the L7Ae protein. At saturating protein concentrations only one species should be present, so our new data have therefore been analyzed in terms of two Gaussian distributions of fluorescent donor–acceptor lengths, corresponding to short (folded) and long (extended) distances. Although it was clear that the folded population was dominant, it was not possible to determine independently all five variable parameters (i.e. the center and half-width of each distribution and the fractional population), due to the high correlation between parameters. We therefore fitted the data with and without 23 nM protein globally, assuming that both distributions had the same center and half-width in the presence and absence of protein (Fig. 4A). This gave distances of 43 and 71 Å, with full width at half maximum (fwhm) of 9.8 and 11.3 Å, respectively. In the absence of protein, the distribution was biased toward the longer distances by a ratio of 6.0:1, while in the presence of the protein, this was inverted with a bias toward a shorter distance by a ratio of 6.7:1. The results and statistics of all the data fitting are presented in Table 1.
TABLE 1.
Values obtained from the time-resolved FRET data for the RNA-L7Ae complex, and the fitting statistics
| Model | Parametera | Valueb | 95% C.I.c | χ2 |
| 1 Gaussiand | r | 40.02 | 39–41.1 | 11.7 |
| fwhm | 25.2 | 24.1–26.3 | ||
| 2 Gaussian globale | r1 | 43.4 | 42.1–44.1 | 10.9 |
| fwhm1 | 9.8 | 2.9–16.9 | ||
| r2 | 70.6 | 69.5–71.5 | ||
| fwhm2 | 11.6 | Fixed | ||
| p | 0.149 | 0.072–0.205 | ||
| 2 Gaussianf | r1 | 43.4 | 42.0–44.2 | 10.7 |
| fwhm1 | 8.8 | 0.0–16.9 | ||
| r2 | 69.6 | Fixed | ||
| fwhm2 | 14.6 | Fixed | ||
| p | 0.169 | 0.079–0.221 |
ar, r1 and r2 are donor-acceptor (D-A) distances and fwhm are full widths at half maxima. p-values are the amplitudes of the long distance, relative to a short distance amplitude of unity.
bAll distances are given in Å.
c95% confidence interval.
dResult of fitting the data for the RNA in the presence of L7Ae to a single Gaussian function of D-A distances.
eResult of fitting the data for the RNA both in the presence and absence of L7Ae globally to two Gaussian functions of D-A distances. The results are only shown for the RNA in the presence of L7Ae. The halfwidth of the longer distance was fixed for this calculation.
fResult of fitting the data for the RNA in the presence of L7Ae to two Gaussian functions of D-A distances. The longer distance and halfwidth were fixed for this calculation.
The assumption that the folded structure of the RNA is the same in the presence and absence of protein may not be valid; therefore the data recorded in the presence of the L7Ae protein were then subjected to a further round of fitting without inclusion of the protein-free data. In this analysis we fixed the longer distance and half-width to that obtained from the global analysis. The residuals are shown in Figure 4B and 4C, and the resulting distance profile is shown in Figure 4D. Under these conditions, 85% of the RNA is folded into the tightly kinked conformation. Since the concentration of L7Ae is much higher than our determined Kd, the RNA should be bound stoichiometrically, so the 15% of the RNA giving the longer distance is likely to be incompetent for binding for some reason, such as mis-hybridization. The parameters determined from this analysis were almost identical, with a distance of 43.4 Å and a fwhm of 8.8 Å. The distance was well determined, with a 95% confidence range of 41–44 Å. This donor–acceptor separation is in excellent agreement with that expected for the structure of a K-turn bound by L7Ae protein (see below). Fixing the parameters of the long distance distribution to other values within the limits determined from the global fit did not significantly alter the estimates for the parameters of the shorter distance distribution (data not shown).
Box C/D and related K-turns are induced to fold by the binding of L7Ae protein
Although Kt-7 is closest to the consensus K-turn sequence, it is not naturally bound by L7Ae in the ribosome, despite demonstrably being induced to adopt the kinked conformation by the protein in vitro. The three-base bulge of all the natural box C/D K-turns have a U at the 3′ end, whereas this is an A in Kt-7. Using electrophoretic and filter binding assays, Kuhn et al. (2002) found significant changes in L7Ae affinity, with no detectable binding when a G was placed at this position. However, on replacing the corresponding A in Kt-7 by U or G we observed changes in steady-state FRET efficiency indicative of full folding, with affinities similar to those of Kt-7 (data not shown). To extend this analysis we performed the analysis using an RNA containing a natural box C/D sequence, equivalent to that used to crystallize the A. fulgidus complex (Moore et al. 2004). Although the C stem of this construct is extended, no protein contacts are made with the loop in the crystal structure. We found that the box C/D RNA is induced to fold into the fully kinked conformation by the binding of L7Ae, with a closely similar asymptotic FRET efficiency and a similar apparent binding affinity of Kd = 0.9 ± 0.2 nM (Fig. 5). It thus appears that the binding and induced folding of all K-turns studied are closely similar, including the box C/D sequences.
FIGURE 5.
Binding of L7Ae protein to RNA containing a box C/D K-turn analyzed by steady-state FRET. Addition of L7Ae to the RNA in the absence of Mg2+ ions. Plot of FRET efficiency (EFRET) as a function of added L7Ae concentration. The data were fitted to a model assuming stoichiometric binding of L7Ae to the RNA. The change in conformation induced by the binding of the L7Ae protein is closely similar to that of the Kt-7 sequence—compare with the data shown in Figure 3A.
DISCUSSION
Steady-state and time-resolved FRET experiments clearly show that the K-turn motif is induced to fold by the binding of the ribosomal L7Ae protein in solution. In the absence of protein, the RNA exists in a dynamic equilibrium between the tightly kinked and a more extended structure, and the equilibrium is driven toward the kinked conformation in the presence of metal cations (Goody et al. 2003). However, our new data reveal that the kinked structure is stabilized in the complex with the A. fulgidus L7Ae protein, even in the absence of added metal ions. The binding of the protein to K-turn-containing RNA is a clear example of an induced fit in RNA structure.
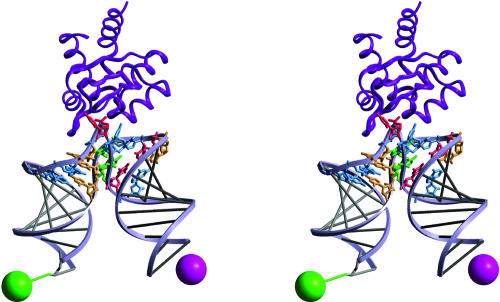
The fluorescein-Cy3 distance calculated from the time-resolved FRET data for the major form of the RNA present in the presence of the L7Ae protein was 43 Å. This can be compared with a value calculated from the model of the extended box C/D K-turn bound by the A. fulgidus L7Ae taken from the structure in the crystal. We took the structure reported by Li and coworkers (Moore et al. 2004) and superimposed A-form RNA helices with the base–paired arms of that structure to generate a model with extended arms of length and sequence equivalent to the species on which we performed the FRET measurements. Fluorophores were then added at their known positions (Norman et al. 2000) and the interfluorophore distance measured (Fig. 6). This was found to be 44 Å, in good agreement with that measured by time-resolved FRET, i.e., 43Å. Although more distances would need to be measured to define the structure better, the agreement between the single distance measured in solution and in the extended crystal structure indicates that the structure of the K-turn in the bound complex in solution is closely similar to that found in the crystal. The width of the distribution for the interfluorophore distance (9 Å) can be well accounted for by the length of the fluorescein tether, suggesting that the structure of the K-turn is well constrained in the complex with the L7Ae protein.
FIGURE 6.
A model of the complex of the RNA species used for fluorescence analysis bound by L7Ae, based on the crystal structure of the box C/D complex (Moore et al. 2004). The green and magenta spheres are placed at the expected average locations of fluorescein and Cy3, respectively. The calculated distance between the fluorophores is 44 Å. The nucleotides are color-coded: guanine, blue; adenine, green; cytosine, yellow; and uracil, red.
The great majority of K-turns found in nature are bound by proteins. These include most of the ribosomal examples (Klein et al. 2001), as well as the box C/D (Moore et al. 2004) and H/ACA (Hamma and Ferré-D’Amaré 2004), snoRNPs and the U4 snRNP (Vidovic et al. 2000). The change in conformation of the K-turn on binding L7Ae protein provides a very sensitive method of measuring the affinity of binding that can be performed at low RNA concentration. Previous estimates of dissociation constants for L7Ae and the 15.5-kDa protein by electrophoretic retardation or filter binding have been in the range of 5–50 nM, but it was noted that these values could overestimate the Kd, and the real affinity could be at least an order of magnitude higher (Kuhn et al. 2002). The subnanomolar affinity measured in our study accord well with the role of these proteins in initiating the formation of snoRNP complexes.
Our results show that the RNA by itself may be a relatively flexible structure that is then induced to fold by the binding of an L7 or related protein. Further changes in RNA structure might then be induced, and/or assembly of an RNA–protein complex. This manner of folding could permit the free RNA to explore conformations that are then fixed by the binding of protein during assembly of complex structures. In the case of the RNAs that guide site-directed modifications in eukaryotic and archaeal RNA, binding of the Nhp2, L7Ae, or 15.5-kDa protein is the first stage of the assembly of an RNA–protein complex (Kuhn et al. 2002; Szewczak et al. 2002; Watkins et al. 2002). As we have shown, this induces the formation of the tightly kinked structure, and this should then be the form that recruits further proteins. For example, in the case of the archaeal box C/D sRNA, having bound the L7Ae protein with high affinity, the complex recruits the Nop5p protein and ultimately the fibrillarin methyl transferase. Thus the tight binding and manipulation of the RNA by the L7Ae protein can be seen as the critical first step in assembling the RNA structure required to catalyze site-specific methylation. Similarly, the binding of the human 15.5-kDa protein to the U4 snRNA K-turn of the U4/U6.U5 complex is an important step in spliceosome assembly (Nottrott et al. 1999), and it is likely that this protein plays a similar role in organizing the geometry of the RNA by means of fixing the kinked structure. A recent molecular dynamics simulation suggested that the K-turn structure could be stabilized by binding the 15.5-kDa protein (Cojocaru et al. 2005).
The abundance of K-turn sequences in natural RNA and the flexible and multifunctional nature of the L7 class of proteins points to the importance of the self-assembling architectural scaffold formed by the protein-induced folding of this element.
MATERIALS AND METHODS
Preparation of A. fulgidus L7Ae protein
A pET28-b+ vector containing the gene for a hexahistidine-L7Ae fusion protein was obtained from Prof. A. Hüttenhoffer. The plasmid was transformed into Escherichia coli BL21-Gold (DE3) pLysS cells (Stratagene), and protein expression was induced by the addition of IPTG to 1 mM. The cells were shaken for 4 h at 37°C and harvested by centrifugation at 4000 rpm for 30 min at 4°C. The cells were immediately lysed in 20 mM phosphate buffer (pH 7.0), 0.5 M NaCl, 2.5 mM imidazole (buffer A) containing 0.1 mg/mL lysozyme (Sigma) and Complete protease cocktail (Roche). The suspension was heated at 85°C to denature endogenous protein, and this was removed by centrifugation at 10,000 rpm for 30 min at 4°C. L7Ae was purified from the cleared supernatant by application to a Ni2+-chelated HiTrap column (Amersham Biosciences) installed on a Bio-Cad Sprint HPLC apparatus (Perspective Biosystems). The bound protein was dissociated from the column by a linear 0–500 mM imidazole gradient in buffer A, with the protein eluting at 200 mM imidazole. Pooled fractions were then dialyzed against 20 mM HEPES-KOH (pH 7.4), 150 mM KCl, 1.5mM MgCl2 (buffer C) containing 5% v/v glycerol before incubation with bovine thrombin (Sigma) for 16 h at 4°C. After inactivation of the protease by heat treatment, L7Ae was concentrated and stored at −20°C in buffer C containing 40% v/v glycerol. Protein concentration was measured using a bicinchoninic acid assay (Smith et al. 1985), using a kit purchased from Perbio with bovine serum albumin as a standard. The purified protein was analyzed by electrophoresis in polyacryl-amide in the presence of SDS, alongside a mixture of proteins (2–212 kDa, New England Biolabs) to act as size standards.
Chemical synthesis of RNA
Kt-7 oligoribonucleotides were synthesized using tBDMS phosphoramidite chemistry (Beaucage and Caruthers 1981), as described in Wilson et al. (2001). Other oligoribonucleotides were synthesized using ACE chemistry (Scaringe 2000) implemented on a modified 394 synthesizer (Applied Biosystems). Phosphoramidites (Dharmacon) were supplied in the form of oils, and were diluted to a concentration of 0.1 M in dry acetonitrile (Glen Research) and filtered through a 0.45-μm filter (Aldrich) before use. In addition, 0.5 M S-ethyltetrazole (ETT) in dry acetonitrile (Link Technologies) was used as activator. Deprotection of 5′ silyl-protecting groups was carried out using a solution of 1.1 M hydrofluoride, 2.9 M triethylamine in dimethylformamide. Oxidation was performed using a solution of 1M tert-butyl hydroperoxide in toluene. Capping reagents were 10% (v/v) N-methylimidazole and 10% (v/v) acetic anhydride each in dry acetonitrile. Fluorescein and Cy3 fluorophores were attached at 5′ termini as appropriate during synthesis.
The Kt-7 sequences (written 5′ to 3′) used in the FRET analyses were:
Bulge strand—Fluorescein-CCAGUCAGUGGCGAAGAACCAUGUCAGG;
Nonbulge strand—Cy3-CCUGACAUGGGGAGCCACUGACUGG.
Additional 5′-fluorescein-labeled bulge strands with G or U replacing the A in the 3′ position of the GAA bulge were also synthesized.
Two strands were synthesized to create an RNA duplex in which the K-turn was that of box C/D:
Bulge strand—Fluorescein-CCUCAGUGGGCGUGAUGCAUGUCUAG;
Nonbulge strand—Cy3-CUAGACAUGCUGACCCACUGAGG.
Steady-state fluorescence
Fluorescence spectra were recorded in 90 mM Tris-borate (pH 8.3) plus added metal salts as required at 4°C using an SLM-Aminco 8100 fluorimeter. Spectra were corrected for lamp fluctuations and instrumental variations, and polarization artifacts were avoided by setting excitation and emission polarizers crossed at 54.74°. Values of EFRET were measured using the acceptor normalization method. Fluorescence anisotropy (r) was determined from:
 |
(2) |
G is the correction factor given by the ratio FHV/FHH, where the subscripts V and H refer to fluorescence with vertical and horizontal polarizers respectively, in the order excitation, emission.
Changes in FRET efficiency with protein concentration were fitted by a stoichiometric binding model:
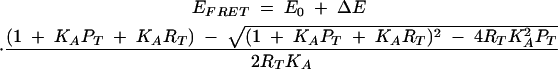 |
(3) |
where E0 is the FRET efficiency in the absence of protein, ΔE is the maximum change in FRET efficiency, PT is the total protein concentration (free + bound), RT is the total RNA concentration (free + bound), and KA is the association constant for the binding of the L7Ae protein to the RNA.
Time-resolved fluorescence
Time-resolved fluorescence lifetime measurements were performed using a K2-Digital Phase Fluorimeter (ISS). Excitation at 488 nm was provided by a 200-mW vertically polarized, argon ion laser (Melles-Griot), intensity modulated at 36 frequencies between 4 and 200 MHz. The cross-correlation frequency was 400 Hz. Donor emission was measured using a 10-nm bandpass filter centered at 520 nm (520DF10, Omega Optical) to exclude scattered incident light and acceptor fluorescence, and a polarizer set at 54.7° to remove instrumental artifacts. Measurements were referenced to fluorescein in 10 mM NaOH, with a lifetime of 4.05 nsec. A minimum of six and a maximum of 12 measurements were performed at each frequency, with data collection terminating when the phase error was below 0.1° and the modulation error was below 0.002. Experiments were performed at 4°C using 25 nM RNA in 90 mM Tris-borate (pH 8.3), 1 nM Mg2+ with or without 23 nM L7Ae protein.
Phase shift and modulation data were analyzed with the parameter estimation program CFS_LS, according to the theory given in Melcher et al. (2003). The data were weighted using the standard error of each datum. The effects of energy transfer on the donor lifetime were fitted by models where the donor–acceptor distances were described by one or two Gaussian distributions, assuming an R0 of 56 Å (Norman et al. 2000). The fractional labeling of the acceptor was determined from the absorbance of the labeled oligonucleotide, using extinction coefficients for Cy3 of 5000 at 260 nm and 150,000 at 547 nm. Donor decay lifetimes were determined independently in parallel experiments using RNA where an unlabeled oligonucleotide was substituted for the Cy3-labeled oligonucleotide. Goodness of fit was evaluated by χ2, the distribution of residuals, and by a runs test. Confidence intervals were estimated by the Bootstrap method (Efron and Tibshirani 1993).
Modeling the structure of the extended K-turn species based on crystallographic data
The model of the L7Ae-complexed RNA was made using InsightII (Accelerys). The structure of the complex of the box C/D RNA and A. fulgidus RNA of Li and coworkers (Moore et al. 2004; PDB coordinate file 1rlg) was the starting point. Canonical A-form RNA helices with the sequences of the arms of the RNA used in our fluorescence measurements were superimposed with the 4 terminal bp of the N-C arm and the 2 bp (ignoring the terminal loop) of the C arm. This was then used to generate a continuous structure containing the core of the crystal structure, and the extended arms of the fluorescence construct. Pseudo-atoms were added at positions corresponding to those determined for fluorescein and Cy3 attached to double-stranded nucleic acid (Norman et al. 2000).
Acknowledgments
We thank Drs. T. Rozhdestvensky and A. Hüttenhofer for provision of a plasmid for the expression of A. fulgidus L7Ae protein, Drs. Alasdair Freeman and Jia Liu for discussion, and the Cancer Research UK for financial support.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2680605.
REFERENCES
- Allmang, C., Carbon, P., and Krol, A. 2002. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: Identification of SBP2 amino acids important to SECIS RNA binding. RNA 8: 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaucage, S.L. and Caruthers, M.H. 1981. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 22: 1859–1862. [Google Scholar]
- Bortolin, M.L., Bachellerie, J.P., and Clouet-d’Orval, B. 2003. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp). Nucleic Acids Res. 31: 6524–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru, V., Nottrott, S., Klement, R., and Jovin, T.M. 2005. The snRNP 15.5K protein folds its cognate K-turn RNA: A combined theoretical and biochemical study. RNA 11: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron, B. and Tibshirani, R.J. 1993. An introduction to the Bootstrap. Chapman Hall, New York.
- Filipowicz, W. and Pogacic, V. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14: 319–327. [DOI] [PubMed] [Google Scholar]
- Förster, T. 1948. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Phys. 2: 55–75. [Google Scholar]
- Ganot, P., Bortolin, M.L., and Kiss, T. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809. [DOI] [PubMed] [Google Scholar]
- Goody, T.A., Melcher, S.E., Norman, D.G., and Lilley, D.M.J. 2003. The kink-turn motif in RNA is dimorphic, and metal ion dependent. RNA 10: 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma, T. and Ferré-D’Amaré, A.R. 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8Å resolution. Structure 12: 893–903. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M., and Kiss, T. 1996. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V., Bork, P., and Sander, C. 1994. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 22: 2166–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, J.F., Tran, E.J., and Maxwell, E.S. 2002. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 30: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, H., White, S.A., and Williamson, J.R. 1999. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat. Struct. Biol. 6: 1139–1147. [DOI] [PubMed] [Google Scholar]
- Marmier-Gourrier, N., Clery, A., Senty-Segault, V., Charpentier, B., Schlotter, F., Leclerc, F., Fournier, R., and Branlant, C. 2003. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA 9: 821–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura, S., Ikawa, Y., and Inoue, T. 2003. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 31: 5544–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher, S.E., Wilson, T.J., and Lilley, D.MJ. 2003. The dynamic nature of the four-way junction of the hepatitis C virus IRES. RNA 9: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.R., Cheng, J., and Collins, K. 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, T., Zhang, Y., Fenley, M.O., and Li, H. 2004. Molecular basis of box C/D RNA-protein Interactions; Cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure 12: 807–818. [DOI] [PubMed] [Google Scholar]
- Norman, D.G., Grainger, R.J., Uhrin, D., and Lilley, D.M.J. 2000. The location of Cyanine-3 on double-stranded DNA; importance for fluorescence resonance energy transfer studies. Biochemistry 39: 6317–6324. [DOI] [PubMed] [Google Scholar]
- Nottrott, S., Hartmuth, K., Fabrizio, P., Urlaub, H., Vidovic, I., Ficner, R., and Luhrmann, R. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBOJ. 18: 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razga, F., Spackova, N., Reblova, K., Koca, J., Leontis, N.B., and Sponer, J. 2004. Ribosomal RNA kink-turn motif—a flexible molecular hinge. J. Biomol. Struct. Dyn. 22: 183–194. [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky, T.S., Tang, T.H., Tchirkova, I.V., Brosius, J., Bachellerie, J.-P., and Hüttenhofer, A. 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 31: 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaringe, S.A. 2000. Advanced 5′-silyl-2′-orthoester approach to RNA oligonucleotide synthesis. Methods Enzymol. 317: 3–18. [DOI] [PubMed] [Google Scholar]
- Smith, P.K., Krohn, R.I., Hermanson, G.T., Mallia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, B.J., and Klenk, D.C. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150: 76–85. [DOI] [PubMed] [Google Scholar]
- Szewczak, L.B., DeGregorio, S.J., Strobel, S.A., and Steitz, J.A. 2002. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9: 1095–1107. [DOI] [PubMed] [Google Scholar]
- Vidovic, I., Nottrott, S., Hartmuth, K., Luhrmann, R., and Ficner, R. 2000. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol. Cell 6: 1331–1342. [DOI] [PubMed] [Google Scholar]
- Watkins, N.J., Segault, V., Charpentier, B., Nottrott, S., Fabrizio, P., Bachi, A., Wilm, M., Rosbash, M., Branlant, C., and Luhrmann, R. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466. [DOI] [PubMed] [Google Scholar]
- Watkins, N.J., Dickmanns, A., and Luhrmann, R. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22: 8342–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T.J., Zhao, Z.-Y., Maxwell, K., Kontogiannis, L., and Lilley, D.M.J. 2001. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 40: 2291–2302. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Grundy, F.J., Murphy, B.A., and Henkin, T.M. 2001. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 7: 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]