Abstract
Members of the RNase III family of double-stranded RNA (dsRNA) endonucleases are important enzymes of RNA metabolism in eukaryotic cells. Rnt1p is the only known member of the RNase III family of endonucleases in Saccharomyces cerevisiae. Previous studies have shown that Rnt1p cleaves dsRNA capped by a conserved AGNN tetraloop motif, which is a major determinant for Rnt1p binding and cleavage. The solution structure of the dsRNA-binding domain (dsRBD) of Rnt1p bound to a cognate RNA substrate revealed the structural basis for binding of the conserved tetraloop motif by α-helix 1 of the dsRBD. In this study, we have analyzed extensively the effects of mutations of helix 1 residues that contact the RNA. We show, using microarray analysis, that mutations of these amino acids induce substrate-specific processing defects in vivo. Cleavage kinetics and binding studies show that these mutations affect RNA cleavage and binding in vitro to different extents and suggest a function for some specific amino acids of the dsRBD in the catalytic positioning of the enzyme. Moreover, we show that 2′-hydroxyl groups of nucleotides of the tetraloop or adjacent base pairs predicted to interact with residues of α-helix 1 are important for Rnt1p cleavage in vitro. This study underscores the importance of a few amino acid contacts for positioning of a dsRBD onto its RNA target, and implicates the specific orientation of helix 1 on the RNA for proper positioning of the catalytic domain.
Keywords: ribonuclease, endonuclease, snoRNA, tetraloop, S. cerevisiae
INTRODUCTION
Double-stranded RNA (dsRNA) endonucleases from the RNase III family are key components of RNA metabolism in eukaryotic cells. They are involved in the maturation of the precursor of rRNAs (Abou Elela et al. 1996; Kufel et al. 1999; Wu et al. 2000) and in the RNA interference and microRNA processing pathways (Bernstein et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Knight and Bass 2001; Lee et al. 2003b). Eukaryotic RNases III also play important roles in the processing of other stable small RNAs, such as spliceosomal snRNAs (Chanfreau et al. 1997; Abou Elela and Ares Jr. 1998; Allmang et al. 1999; Seipelt et al. 1999) and small nucleolar RNAs (snoRNAs) (Chanfreau et al. 1998a; Qu et al. 1999; Lee et al. 2003a; Ghazal et al. 2005). Rnt1p is the only Saccharomyces cerevisiae representative of the RNase III family (Abou Elela et al. 1996). In addition to its functions in the maturation of rRNA, snRNA, and snoRNA precursors, Rnt1p is also involved in degradative pathways for pre-mRNAs and mRNAs (Danin-Kreiselman et al. 2003; Ge et al. 2005). Because of its involvement in a large number of processing or degradative pathways, much attention has focused on studying the substrate specificity of this ribonuclease, which serves as a paradigm for the study of RNA recognition by a eukaryotic RNase III enzyme.
Rnt1p, as well as many fungal RNases III, cleaves in the stem of RNA hairpins containing tetraloops with the consensus sequence AGNN (Chanfreau et al. 2000; Chanfreau 2003). While most bacterial and eukaryotic RNases III cleave dsRNA in a non-sequence-specific manner, the presence of these AGNN terminal tetraloops is a strong determinant for yeast RNase III binding and cleavage (Chanfreau et al. 2000; Nagel and Ares Jr. 2000; Lamontagne et al. 2003; Lamontagne and Abou Elela 2004; Leulliot et al. 2004). The AGNN tetraloop dictates the position of the cleavage site, as the enzyme cleaves the RNA 14–16 bp away from the tetraloop (Chanfreau et al. 2000). This ruler-like mechanism is conserved in at least one other eukaryotic RNase III, Drosha, which also measures the distance from terminal loops to select the cleavage site (Zeng et al. 2005). While the two enzymes use a ruler-type mechanism, it is not known whether similar structural elements in the two proteins are used to determine the site of cleavage within the dsRNA.
Rnt1p contains one RNase III catalytic domain and one double-stranded RNA binding domain (dsRBD) (Lamontagne et al. 2000; Nagel and Ares Jr. 2000). The dsRBD of Rnt1p adopts the α1-β1-β2-β3-α2 fold characteristic of dsRBDs (Bycroft et al. 1995; Kharrat et al. 1995), but contains an additional C-terminal α-helix that has been proposed to indirectly contribute to substrate recognition by stabilizing helix α1 (Leulliot et al. 2004; Wu et al. 2004). Truncation analyses have shown that the Rnt1p dsRBD is at least in part responsible for the specificity of Rnt1p for these terminal tetraloops (Nagel and Ares Jr. 2000; Leulliot et al. 2004). The specificity of the Rnt1p dsRBD for AGNN-capped dsRNA raised the question of how this single dsRBD can achieve such a specific recognition, especially since most of the structural contacts described between dsRBDs and dsRNA involve the sugar–phosphate backbone and are therefore non-sequence-specific (Ryter and Schultz 1998; Ramos et al. 2000; Blaszczyk et al. 2004).
Our recently reported solution structure of the dsRBD of Rnt1p bound to a cognate substrate derived from the snR47 snoRNA precursor revealed that the dsRBD interacts with the minor groove side of the terminal tetraloop and the top of the dsRNA, inducing a minor bend in the RNA substrate at the base of the tetraloop (Fig. 1; Wu et al. 2004). The orientation of helix 1 is different from that observed in other dsRBD structures (Wu et al. 2004). This different orientation, as well as the extended length of this helix compared to other dsRBDs, allows this helix to fit snugly into the minor groove of the tetraloop and adjacent base pairs (Wu et al. 2004). Surprisingly, there are no contacts with the conserved A and G bases that point into the major groove. Rather, the structure shows that the specificity of the interaction between the Rnt1p dsRBD and the model substrate RNA relies on the recognition of the conserved fold of the AGNN tetraloop and the two adjacent base pairs by helix α1 of the dsRBD. This type of interaction between a dsRBD and a terminal loop capping a dsRNA might be a more general feature than previously thought. For example, the dsRBD of PKR has been shown to bind the terminal loop capping a dsRNA region of the EBER1 RNA from the Epstein–Barr virus (Vuyisich et al. 2002). In addition, the RNase III Drosha selects its cleavage site in primary precursors of microRNAs by measuring the distance from terminal loops (Zeng et al. 2005), implying that Drosha also binds directly these terminal loops. However, it is unclear whether the dsRBD of Drosha is directly involved in this process. Finally, the dsRNA editing enzyme ADAR2 also recognizes a terminal loop structure, and the dsRBD of ADAR2 seems involved in that process (Stefl and Allain 2005). Therefore, the study of the recognition of terminal AGNN loops by the Rnt1p dsRBD might provide clues to understand general principles of recognition of specific dsRNAs by a subclass of dsRBD-containing proteins.
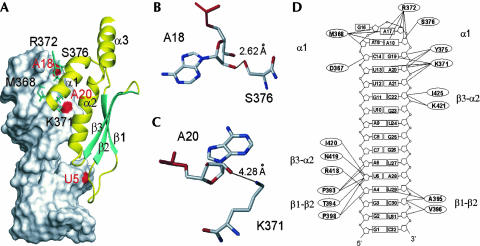
FIGURE 1.
Structure of the Rnt1p dsRBD/snR47h complex (Wu et al. 2004). The corresponding PDB accession code is 1T4L. (A) Overview of the structure of the complex with the RNA shown in surface and the protein, in ribbons. The red spots on the surface of the RNA correspond to the 2′-hydroxyl groups of U5, A18, and A20. The side chains of M368, K371, R372, and S376 are shown in sticks. (B) A close view of the interaction between A18 and S376. The average distance between A18 O2′and S376 OG is indicated. Oxygen and phosphorous atoms are colored in red and nitrogen atoms, in blue. (C) A close view of the interaction between A20 and K371. The average distance between A20 O2′ and K371 NZ is indicated. Colors are as in B. (D) A schematic summary of the interactions between Rnt1p and snR47h observed in the solution structures of the complex. Shown are the secondary structure of snR47h and amino acids of Rnt1p involved in interactions (hydrogen bonding, electrostatic, van der Waals, and hydrophobic interactions) with the RNA.
Among the Rnt1p dsRBD residues that contact the RNA are several that are conserved specifically in fungal RNases III that recognize dsRNA capped by AGNN tetraloops (Wu et al. 2004). These include residues M368, R372, and S376. In contrast, K371, which also contacts the RNA, is conserved among all RNase III dsRBDs, whether they recognize AGNN tetra-loops or not (Wu et al. 2004). Previously, two of these residues (K371 and S376) were shown to be important for processing of a few selected Rnt1p substrates in vivo (Wu et al. 2004). In this study, we have extensively analyzed the roles of both classes of residues, both in vivo, using microarray analyses and in vitro, using cleavage kinetics and binding assays uncoupled from cleavage. In addition, we have investigated the role of specific 2′-hydroxyl groups of the RNA substrate for Rnt1p binding and cleavage in vitro. We show that efficient substrate recognition by eukaryotic RNase III is achieved by a complex set of interactions between specific amino acids of the dsRBD and 2′-hydroxyl groups of the RNA substrate. These results provide a functional framework that illustrates how a protein can use a dsRBD to recognize its physiological targets.
RESULTS
Substitution of methionine 368 of the Rnt1p dsRBD affects processing of a subset of Rnt1p substrates in vivo
The structure of the Rnt1p dsRBD bound to a cognate substrate obtained by NMR (Wu et al. 2004) showed that two of the three residues of helix α1 that interact with the tetraloop, arginine 372 (R372) and serine 376 (S376), contact the sugar–phosphate backbone of the last two nonconserved nucleotides of the loop (Fig. 1A,B,D). The guanidium group of the R372 side chain and the hydroxyl group of the S376 side chain are within hydrogen bond distance of the 2′-hydroxyl groups of adenosine 17 and adenosine 18 (A17 and A18), respectively (Fig. 1B,D). These two amino acids are conserved in fungal RNases III that recognize AGNN-capped dsRNA (Wu et al. 2004). In addition to interacting with the tetraloop, helix α1 also contacts nucleotides of the adjacent dsRNA region of the substrate. In particular, the side chain of lysine 371 (K371) is within van der Waals distance to adenosine 20 (A20), and its amino group is predicted to hydrogen bond to the 2′-hydroxyl group of A20 (Fig. 1C,D) and the O4′ of A21 (Wu et al. 2004). Strikingly, this amino acid is conserved among all RNases III, whether or not they recognize AGNN tetraloops (Wu et al. 2004). The correlation of these structural and phylogenetic observations suggests different roles for these classes of residues in substrate recognition.
M368 is the only amino acid of helix α1 that is positioned in close vicinity to the conserved AG dinucleotide of the tetra-loop (Wu et al. 2004; Fig. 1D). An amino acid with a nonpolar side chain (M or V) is found at this position in all but one of the RNases III that recognize AGNN tetraloops (Wu et al. 2004). The only exception is Debaryomyces hansenii RNase III, which has a lysine at this position, but the dependence of this enzyme on AGNN tetraloops is questionable (Chanfreau 2003). Other RNases III that do not recognize tetraloops often have a lysine residue at this position as well (Wu et al. 2004). In the solution structure of the Rnt1p dsRBD-RNA complex, the nonpolar side chain of M368 stacks on the ribose of the first nucleotide of the tetraloop, A15. A valine would be predicted to stack equally well, corroborating the conservation of methionine or valine residues at this position in RNases III that recognize AGNN tetraloops. These phylogenetic and structural considerations suggest a role for M368 in tetraloop recognition and Rnt1p function.
To further elucidate the role of this conserved residue, we created yeast strains expressing Rnt1p with a substitution of M368 by alanine (M368A) or glutamic acid (M368E) using the “delitto perfetto” method (Storici et al. 2001). Mutant Rnt1p proteins introduced in yeast using this technique are expressed at the endogenous levels from the natural RNT1 promoter in the context of the endogenous chromosomal locus. The M368A substitution results in a shortening of the nonpolar side chain compared to methionine, whereas the M368E mutation converts the methionine to a residue containing a negatively charged side chain. To study the effect of these mutations on Rnt1p function, we analyzed by Northern blot the accumulation of several noncoding RNAs whose expression is known to require Rnt1p (Fig. 2A). The levels of these RNAs were compared to those observed in several control strains grown in the same conditions: a wild-type strain, a strain lacking Rnt1p (rnt1Δ), and a strain expressing the previously published Rnt1pK371A mutant (Wu et al. 2004). Defects in the function of Rnt1p result in a decrease in the accumulation of the mature form of a given species and an accumulation of the corresponding unprocessed precursors (Chanfreau et al. 1998a; Lee et al. 2003a).
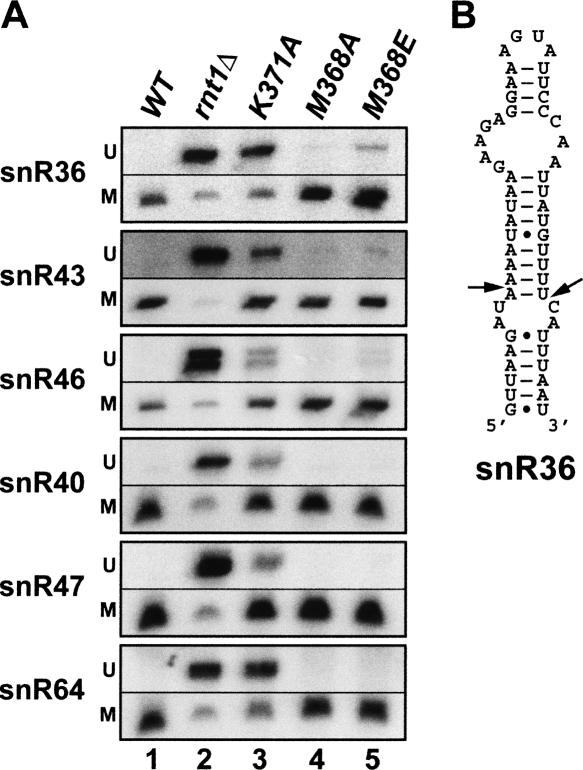
FIGURE 2.
Methionine 368 of the Rnt1p dsRBD is mildly required for processing of a subset of Rnt1p substrates in vivo. (A) Northern blot analysis of the processing of known Rnt1p substrates in a wild-type strain (lane 1), a strain lacking Rnt1p (lane 2), and strains expressing the K371A, M368A, and M368E dsRBD mutant versions of Rnt1p (lanes 3, 4, and 5, respectively). M, mature snoRNAs; U, unprocessed precursors. (B) Predicted secondary structure of the stem-loop recognized by Rnt1p on the precursor of snR36 snoRNA. Arrowheads indicate the cleavage site (Chanfreau et al. 1998a).
As shown in Figure 2A (lane 4), the M368A mutation does not significantly impair the processing of any substrate, as shown by the absence of accumulation of unprocessed precursors. This was not unexpected, as an alanine side chain might be able to fulfill the hydrophobic interactions normally performed by a methionine or a valine side chain, which are found in most fungal RNases III. In contrast, the M368E substitution was found to affect the processing of the precursor of snR36 and to induce a minor defect in the maturation of snR43 and snR46 snoRNAs (Fig. 2A, lane 5). The phenotypes observed with the M368E mutant were less pronounced than that observed with the K371A mutant (Fig. 2A, lane 3). Interestingly, other substrates of Rnt1p (e.g., the precursors of snR40, snR47, or snR64) did not appear to be affected by this mutation. Therefore, the M368E mutation seems to affect specifically the processing of the precursor of snR36 and, to a lesser extent, snR43 and snR46. The same phenotypes were observed when these mutant strains were grown at 37°C (data not shown).
To try to understand why the M368E mutation has stronger effects on the processing of snR36 than on other RNA substrates, we inspected the stem-loop structure recognized by Rnt1p on the snR36 snoRNA precursor to identify specific features that might distinguish this substrate from others. SnR36 contains a large internal loop in the upper part of the stem, 4 bp below the AGNN tetraloop (Fig. 2B). This large loop is predicted to be located where helix α2 of the dsRBD normally interacts on the major groove side of the dsRNA (Wu et al. 2004). In particular, the fifth base pair from the tetraloop, which is normally contacted by K421 of helix α2 (Wu et al. 2004) is an A C mismatch in snR36 (Fig. 2B) that is unlikely to form a protonated pair at physiological pH. The presence of this mismatch and/or of a large internal loop might result in a destabilization of the interaction between α-helix 2 of the dsRBD and the major groove of the dsRNA. This partial loss of interaction with the major groove might result in a weaker binding of the enzyme and render this substrate more sensitive to the M368E mutation in particular, or to any additional perturbations in the dsRBD–dsRNA interactions in general. Indeed, inspection of the processing defects resulting from the other substitutions analyzed previously (K371A, R372A/P, and S376E) (Wu et al. 2004) revealed a more drastic effect of these modifications on the maturation of snR36 compared to snR47, whose precursor has a more canonical stem structure (Chanfreau et al. 1998a). The observed substrate-dependent processing defects are therefore not specific to the M368E substitution, but seem to apply to other Rnt1p dsRBD mutations. In summary, our results indicate that the M368 amino acid of Rnt1p dsRBD is required for efficient processing of a few Rnt1p substrates in vivo. The severity of the processing defects seems to vary in a substrate-dependent manner. The exacerbated sensitivity of the snR36 precursor might be a consequence of the presence of the large bulge in the stem-loop structure that might render this substrate more sensitive to subtle binding defects induced by perturbations of the dsRBD–dsRNA interactions.
Point mutations within α-helix 1 of the Rnt1p dsRBD induce substrate-specific differential processing defects in vivo
The results obtained with the M368 mutants (Fig. 2A), as well as previous results (Wu et al. 2004), suggest that amino acid substitutions in helix 1 of the Rnt1p dsRBD may affect the function of the enzyme in a substrate-specific manner. To test this hypothesis and obtain a more comprehensive view of the in vivo processing defects resulting from these substitutions, we used a genomic approach to compare the accumulation of numerous Rnt1p precursor substrates in a wild-type strain and in strains lacking Rnt1p or expressing mutant versions of the enzyme. Total RNAs were extracted from a wild-type strain and strains lacking Rnt1p or expressing the M368A, M368E, K371A, R372A, R372T, and S376E mutant versions of the enzyme from the endogenous RNT1 gene promoter. As shown previously by Western blot (Wu et al. 2004), the substitutions introduced in the Rnt1p dsRBD do not affect the stability of the mutant proteins expressed in vivo. The purified RNAs were directly labeled as previously described (Hiley et al. 2005). These labeled RNAs were hybridized to custom microarrays containing oligonucleotide probes complementary to the mature and flanking regions of all known yeast noncoding RNAs. These microarrays include, in particular, probes covering a significant number of precursors of snoRNAs whose 5′-end maturation is dependent on Rnt1p (Hiley et al. 2005). Using these arrays, we were able to analyze simultaneously the processing of most Rnt1p substrates in strains lacking Rnt1p or in strains expressing mutant versions of the enzyme. Two strains lacking Rnt1p were used: a strain carrying a knockout of the RNT1 gene (rnt1Δ) (Chanfreau et al. 1998b) and a strain expressing Rnt1p from a repressible promoter grown in repressive conditions (TetO7::RNT1) (Mnaimneh et al. 2004). In these strains, we observed an accumulation of signal corresponding to probes covering the region located upstream from the mature sequence of the snoRNA (Fig. 3, rows 1,2). Although most mature snoRNAs are significantly underaccumulated in the absence of Rnt1p, the overall hybridization to probes corresponding to mature sequences does not vary significantly in RNAs extracted from strains lacking Rnt1p activity (Fig. 3). This is due to the quantitative accumulation of the unprocessed precursors that also include mature sequences (Chanfreau et al. 1998a; Lee et al. 2003a; Peng et al. 2003). Likewise, we did not observe significant variations downstream from the mature sequences, because these regions are efficiently 3′-end processed in the absence of Rnt1p (Chanfreau et al. 1998a). These observations show that the microarray analysis is well adapted to the specific detection of in vivo processing defects resulting from an alteration in Rnt1p function, although defects in the processing of some known snoRNA substrates of Rnt1p could not be detected (e.g., snR52, snR58, snR66). This may be due to problems of sensitivity or alternatively to the fact that an alternative exonucleolytic 5′-end processing pathway compensates for the 5′-processing defect induced by the absence of Rnt1p for some snoRNA substrates (Chanfreau et al. 1998a; Lee et al. 2003a).
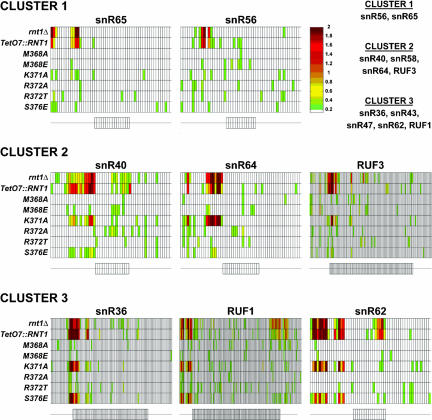
FIGURE 3.
Amino acid substitutions in Rnt1p dsRBD induce substrate-specific processing defects in vivo. Microarray analysis of the accumulation of known substrates of Rnt1p in strains lacking Rnt1p (rnt1Δ and TetO7::RNT1) and strains expressing the M368A, M368E, K371A, R372A, R372T, and S376E dsRBD mutant versions of Rnt1p. The small rectangles aligned in one row correspond to the relative fluorescence of RNA bound to oligonucleotide probes complementary to RNA sequences in the mature or flanking sequences of the snoRNA mentioned above. A schematic representation of the snoRNAs and their flanking sequences are shown below each set of data. On these schematics, rectangles correspond to probes complementary to the mature sequences of the snoRNAs, and thin lines correspond to probes complementary to the 5′ and 3′ flanking regions. The color of each rectangle is indicative of the relative accumulation of the corresponding RNA sequence in the mutant strain indicated on the left compared to the wild-type strain. The color scale is shown on the right; the reported values correspond to the log2 of the ratio over wild type of the fluorescent intensities. The snoRNA precursor substrates can be clustered into three main categories depending on their sensitivities to the Rnt1p dsRBD mutations. Cluster 1 corresponds to substrates that are insensitive to all tested mutations. Cluster 2 corresponds to substrates affected only by the K371A substitution, and Cluster 3, to substrates sensitive to both the K371A and S376E substitutions. The full set of substrates for each cluster is indicated on the upper right part of the figure.
Microarray analysis of strains expressing dsRBD mutations revealed interesting differential processing defects specific to particular substrates (Fig. 3). The snoRNA precursor substrates could be clustered into three main categories based on their differential sensitivity to mutations in the Rnt1p dsRBD. The first category (Cluster 1; Fig. 3) includes the precursors of snR56 and snR65, for which no significant accumulation of the 5′ region of the precursors is observed in any of the strains expressing Rnt1p with point mutations in the dsRBD. The maturation of this cluster of substrates does not seem to be affected by any of the tested amino acid substitutions in Rnt1p, at least in the limits of sensitivity of the method. A second cluster (Cluster 2; Fig. 3) contains precursors of snoRNAs whose processing is affected specifically by the K371A substitution, but not by any of the other mutations. This group includes the precursors of snR40, snR58, snR64, and RUF3. Interestingly, K371 is conserved in all RNases III, and the predicted role of this amino acid is to contact the base pairs in the dsRNA below the tetraloop. Mutation of this residue seems to affect a large number of substrates, in agreement with the hypothesis that this mutation might result in weaker binding to the dsRNA region of most substrates in vivo. The third group of substrates (Cluster 3; Fig. 3) contains precursors such as the ones of snR36, snR62, and RUF1, whose processing is highly sensitive to both the K371A and S376E mutations, but not to others. Thus, this class of substrates is sensitive to mutations in both classes of conserved residues, those conserved in all RNases III such as K371 or specific to RNases III that recognize AGNN tetraloops, like S376.
The microarray analysis did not reveal any strong processing defects in the strains expressing the M368A, M368E, R372A, and R372T mutant versions of Rnt1p, in agreement with previous small-scale Northern blot analyses (Fig. 2A; Wu et al. 2004). This lack of in vivo phenotype is discussed below in the light of the weak in vitro cleavage defects observed for these mutants. The mild processing defects detected by Northern blot for the M368E mutation (Fig. 2A) was not detectable by microarray analysis, possibly because of a lower sensitivity. Overall, the microarray results show that the snoRNA precursor substrates can be clustered into three main groups according to their sensitivity to the K371A and S376E substitutions. We could not correlate these clusters to any particular tetraloop sequence or to a sequence in the base pairs adjacent to the tetraloop (data not shown). Nevertheless, the fact that such clusters exist suggests that substrate recognition by Rnt1p does not obey a single rigid rule but may allow some degree of flexibility depending on substrates. In particular, the molecular contacts that we have identified between the Rnt1p dsRBD and the RNA were derived from the solution structure of the dsRBD bound to one particular RNA substrate (Wu et al. 2004). Although probably globally similar, these contacts may be slightly different from one substrate to another, which could not be revealed by the solution structure of only one substrate complex. These considerations may provide a rationale for the substrate-dependent processing defects resulting from amino acid substitutions in the Rnt1p dsRBD.
Amino acid substitutions in the Rnt1p dsRBD affect Rnt1p cleavage and binding in vitro
The in vivo processing defects observed in the strains expressing the dsRBD mutants could result from the perturbation of many aspects of Rnt1p function in the context of the cellular environment. To demonstrate that these substitutions directly affect the ability of the enzyme to interact with and/or cleave its substrates, we purified recombinant M368A, M368E, K371A, R372A, and S376E mutant versions of Rnt1p and analyzed their in vitro cleavage and binding activities. Cleavage kinetics were performed by incubating the wild-type or mutant enzymes with a radiolabeled synthetic RNA substrate derived from the snR47 snoRNA precursor in single turnover conditions at two different protein concentrations (70 nM and 120 nM). The apparent constants are presented in Table 1, and the cleavage kinetics obtained at 70 nM are shown in Figure 4A. All mutants displayed activities significantly reduced compared to the wild-type enzyme (Table 1). The M368A and R372A mutants are moderately affected, with their apparent constants approximately four and six times slower than the wild type, respectively. However, the M368E, K371A, and S376E enzymes display much more severe cleavage defects in vitro. Their apparent constants at 70 nM are 19, 16, and 18 times slower than wild-type Rnt1p, respectively (Fig. 4A; Table 1). These results show that residues M368, K371, R372, and S376 of Rnt1p dsRBD are required for optimal function of the full-length enzyme in vitro. Moreover, they strongly suggest that the in vivo processing defects observed upon expression of some of the mutant versions of Rnt1p bearing amino acid substitutions in the dsRBD are a direct consequence of the reduced activity of the mutant enzymes. Strikingly, the extent of the in vitro cleavage defects of the mutated proteins correlates well with the extent of the processing defects observed in vivo in the corresponding mutant strains. The most severe phenotypes were observed in vivo with the K371A and S376E mutants, which also exhibit the strongest in vitro cleavage defects (Fig. 4A). No significant defect could be detected in vivo with the M368A and R372A mutants (Figs. 2, 3), which are only moderately affected in vitro (Fig. 4A). This result suggests that the reduction in the catalytic activity of these mutants is not sufficient to affect steady-state levels of processing in vivo, possibly because other steps in the expression pathway of these RNAs that are independent of Rnt1p function (e.g., transcription) are rate limiting in vivo. The M368E substitution induces intermediate phenotypes in vivo but seems more affected in vitro.
TABLE 1.
Apparent kinetic constants kf and ks and dissociation constants (Kd) for the corresponding Rnt1p proteins and RNA substrates
| Rnt1p/RNA | kf ; ks@70 nM (min−1) | F∞@70nM | kf; ks@120 nM (min−1) | F∞@120nM | Kd (nM) |
| WT/WT | 8.40 ±1.08; 0.109 ±0.011 | 0.63 | 7.47 ±1.09; 0.046 ±0.007 | 0.72 | 130 ±13 |
| WT/dU5 | 3.49 ±0.25; 0.071 ±0.006 | 0.60 | 4.48 ±0.25; 0.084 ±0.010 | 0.53 | 70 ±5 |
| WT/dA18 | 0.56 ±0.02; 0.036 ±0.002 | 0.50 | 0.51 ±0.02; 0.033 ±0.003 | 0.54 | 146 ±27 |
| WT/dA20 | 0.52 ±0.05; 0.081 ±0.008 | 0.43 | 0.40 ±0.03; 0.40 ±0.010 | 0.42 | 150 ±6 |
| WT/GAAA | N.D. | N.D. | N.D. | N.D. | 273 ±19 |
| K371A/WT | 0.51 ±0.04; 0.039 ±0.003 | 0.54 | 0.79 ±0.07; 0.085 ±0.006 | 0.60 | 276 ±23 |
| K371A/dA18 | 0.020 ±0.001 | 0.19 | 0.020 ±0.001 | 0.33 | N.D. |
| K371A/dA20 | 0.029 ±0.001 | 0.25 | 0.029 ±0.001 | 0.36 | 412 ±49 |
| K371A/GAAA | N.D. | N.D. | N.D. | N.D. | 525 ±38 |
| S376E/WT | 0.46 ±0.02; 0.027 ±0.001 | 0.49 | 1.04 ±0.14; 0.09 ±0.01 | 0.57 | 172 ±22 |
| S376E/dA18 | 0.022 ±0.002 | 0.08 | 0.022 ±0.001 | 0.23 | 199 ±19 |
| S376E/dA20 | 0.026 ±0.002 | 0.10 | 0.026 ±0.001 | 0.20 | N.D. |
| S376E/GAAA | N.D. | N.D. | N.D. | N.D. | 272 ±42 |
| M368A/WT | 2.20 ±0.20; 0.028 ±0.003 | 0.50 | 5.13 ±0.38; 0.058 ±0.006 | 0.59 | 184 ±60 |
| M368E/WT | 0.44 ±0.03; 0.026 ±0.004 | 0.43 | 2.06 ±0.15; 0.065 ±0.007 | 0.59 | 133 ±8 |
| R372A/WT | 1.44 ±0.05; 0.060 ±0.005 | 0.54 | 4.20 ±0.76; 0.120 ±0.014 | 0.64 | 124 ±25 |
(WT) Wild-type protein or RNA substrate. Apparent kinetic constants are indicated at two concentrations of Rnt1p and are the average of two independent experiments, except for the WT–WT, M368A–WT, M368E–WT, K371A–WT, R372A–WT, and S376E–WT combinations at 120 nM, which are averages from four independent experiments. Most reaction kinetics were fitted to double exponential kinetics: F(t) = F∞ + A1 exp(−kft) + A2 exp(−kst), where F(t) is the fraction reacted at time t, F∞ is the fraction reacted at the endpoint of the kinetics (120 min), A1 and A2 are the amplitudes of the two phases, and kf and ks are the apparent rate constants of the fast and the slow phases, respectively. The K371A/dA18, K371A/dA20, S376E/dA18, and S376E/dA20 kinetics were better fitted to single exponential equations; therefore only one constant is indicated. The Kd values are the average of two independent experiments except for the combinations WT–WT (n = 7), WT–dU5 (n = 4), K371A–WT (n = 3), and S376E–WT (n = 4). (N.D.) Not determined. Errors are the standard deviations obtained from independent experiments.
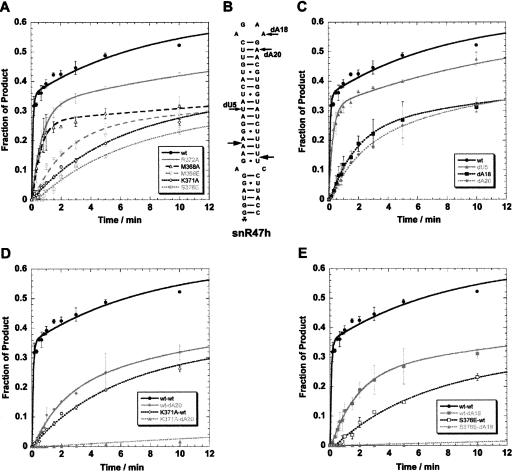
FIGURE 4.
In vitro cleavage of wild-type and 2′-deoxy substituted snR47h synthetic RNA substrates derived from the precursor of snR47 by wild-type and mutant Rnt1p. (A) Single turnover cleavage kinetics of wild-type snR47h substrate by recombinant wild-type and M368A, M368E, K371A, R372A, and S376E dsRBD mutant versions of Rnt1p at a concentration of 70 nM. Error bars represent variations obtained in two independent experiments. (B) Secondary structure of the model snR47h substrate. The location of the 2′-deoxy modifications studied is indicated. Note that the numbering of the nucleotides is according to the model RNA used in the NMR study (Wu et al. 2004) and does not correspond to the numbering of the nucleotides of the substrate used in this study. (C) Single turnover cleavage kinetics of wild-type and dU5, dA18, and dA20 2′-deoxy versions of the snR47h substrate by wild-type Rnt1p at 70 nM. Error bars represent variations obtained in two independent experiments. (D) Single turnover cleavage kinetics of the wild-type or dA20 2′-deoxy modified snR47h substrates by the wild-type or K371A versions of Rnt1p at 70 nM. Error bars represent variations obtained in two independent experiments. (E) Single turnover cleavage kinetics of the wild-type or dA18 2′-deoxy modified snR47h substrates by the wild-type or S376E versions of Rnt1p at 70 nM.
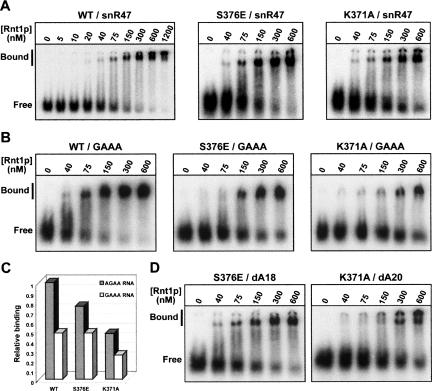
To determine whether the defective cleavage activities of these mutants correlate with a reduced ability to bind the RNA substrate, we measured the approximate dissociation constants (Kd) for the interaction between these enzymes and a wild-type RNA substrate by electrophoretic mobility shift assays (EMSAs) (Fig. 5A; Table 1). Magnesium ions are required for the endonucleolytic activity of Rnt1p (Lamontagne and Abou Elela 2001). In the absence of magnesium, Rnt1p retains its ability to interact with the substrates but loses its catalytic activity, and the binding of the enzymes can be assessed uncoupled from cleavage (Chanfreau et al. 2000; Lamontagne and Abou Elela 2001). In these conditions, the wild-type protein binds to the snR47 precursor stem-loop structure with a Kd of ~130 ±13 nM (Table 1). The M368A, M368E, and R372A mutants do not seem to show any significant binding defect (Table 1). These results show that these Rnt1p mutants retain a robust binding activity, despite the substitutions in the dsRBD. The S376E mutant displays a Kd of ~172 ±22 nM (Fig. 5A; Table 1), indicative of a reduced binding activity compared to the wild-type protein. It is unlikely that this slightly weaker binding is responsible for the drastic cleavage inhibition observed with this mutant. This observation suggests that the S376E mutation might affect the catalytic positioning of the enzyme onto the RNA substrate. The K371A mutant is more severely affected, since it retains only 50% of the wild-type binding efficiency (Fig. 5A; Table 1). These results show that mutation of this residue that is conserved in all RNases III significantly reduces binding of the full-length enzyme.
FIGURE 5.
Binding of wild-type, 2′-deoxy substituted, and tetraloop mutant snR47h RNA substrates by wild-type and mutated versions of Rnt1p. (A) Binding of wild-type Rnt1p and the K371A and S376E mutant versions of the enzyme to a wild-type snR47h substrate. Shown are EMSAs on native gels obtained with the indicated proteins at the indicated concentrations. Free, position of the free substrates; Bound, position of the bound substrates. The presence of multiple bands is consistent with dimeric or multimeric forms of the proteins bound to the substrates (Lamontagne et al. 2000). (B) Binding of the wild-type and K371A and S376E mutants of Rnt1p to a snR47h RNA mutant substrate with a GAAA tetraloop. Legends as in A. (C) Relative binding efficiencies of wild-type Rnt1p and S376E and K371A mutants for a wild-type RNA substrate (AGAA) (gray columns) and a GAAA tetraloop mutant (white columns). The wild-type binding value is set arbitrarily to 1. (D) Binding of the K371A mutant to the dA20 2′-deoxy version of the substrate and of the S376E mutant to the dA18 2′-deoxy substrate. Legends as in A.
Nonspecific binding of a tetraloop mutant RNA by helix 1 dsRBD mutant proteins
To further investigate the recognition of RNA hairpin by Rnt1p, we studied the binding by helix 1 mutants of an RNA substrate with a GAAA tetraloop replacing the canonical AGAA loop sequence of the snR47 substrate. Previous studies have shown that this mutation changes the fold of the tetra-loop from an AGNN-type to a GNRA-type, completely inhibiting cleavage and reducing the binding of Rnt1p or of the dsRBD (Chanfreau et al. 2000; Nagel and Ares Jr. 2000; Lamontagne et al. 2003; Lamontagne and Abou Elela 2004; Leulliot et al. 2004). The RNA substrate used here is long enough so that the dsRBD can bind the RNA without interacting with the GAAA loop. Therefore, any residual binding observed with this mutant RNA is likely to reflect nonspecific binding to dsRNA. Because of the complete inhibition of cleavage observed with the wild-type protein, we did not study the cleavage of this substrate but we restricted our analysis to binding. In our EMSA conditions, the GAAA mutant substrate was bound with a twofold lower efficiency compared to the wild-type substrate by the wild-type protein (Fig. 5B,C; Table 1). Strikingly, this mutant substrate was bound with the same lower efficiency by the wild-type protein and the S376Emutant(Table 1). This result suggests that changing the tetraloop structure results in a disruption of the helix 1 interaction site with the tetraloop region, and that additional mutation of an amino acid that is normally involved in this interaction, such as S376, has no further effect on the over all dsRNA binding efficiency. Therefore, nonspecific binding of dsRNA is equally efficient with the wild-type protein or with the S376E mutant. In contrast, combining the K371A mutation and the GAAA tetraloop mutation resulted in a stronger loss of binding (Fig. 5B,C; Table 1), showing that mutating the tetraloop and K371, which normally binds the dsRNA below the tetraloop, have additive effects. Thus, the K371 residue has an important role in nonspecific binding to the dsRNA but does not contribute significantly to binding of the AGNN tetraloop, in agreement with the dsRBD–RNA structure.
The 2′-hydroxyl groups of residues A18 and A20 of the model substrate are required for efficient cleavage by Rnt1p in vitro
In the solution structure of the Rnt1p dsRBD–snR47 hairpin complex, adenosine 18 (A18), adenosine 20 (A20), and uridine 5 (U5) of the dsRNA interact with the Rnt1p dsRBD (Fig. 1; Wu et al. 2004). The 2′-hydroxyl group of U5 is within H-bond distance of the peptide bond linking proline 393 (P393) to threonine 394 (T394). The 2′-hydroxyl groups of A18 and A20 are within H-bond distance to the terminal groups of the S376 and K371 side chains, respectively (Fig. 1B,C). While these interactions were observed in the solution structure of the dsRBD–hairpin RNA complex, their functional relevance in the context of a catalytically active enzyme–RNA complex remained to be established. To test the importance of these 2′-hydroxyl groups for Rnt1p function, we assessed the cleavage and binding of synthetic substrates containing 2′-deoxy modifications at residues U5, A18, or A20 (dU5, dA18, and dA20, respectively) (Fig. 4B). The absence of the 2′-hydroxyl group of U5 reduces cleavage by Rnt1p by ~60% compared to the wild-type RNA (Fig. 4C; Table 1). Surprisingly, the dU5 mutant substrate is reproducibly bound by Rnt1p with an almost twofold increased affinity compared to the unmodified RNA (Table 1). The in vitro cleavage and binding data relative to this mutant RNA may appear somehow contradictory. However, this apparent increase in binding might reflect a change in the way the enzyme binds the RNA in the absence of the U5 2′-hydroxyl. To retain binding with this substrate, the enzyme possibly binds the RNA in a slightly different conformation, which, albeit stronger, results in a less productive conformation and in a cleavage defect. Substrates lacking the 2′-hydroxyl groups of either A18 or A20 are cleaved by Rnt1p with slower kinetics compared to the wild-type RNA substrate (Fig. 4C). The calculated apparent constants are ~15 times slower for snR47dA18 and snR47dA20 than for the unmodified substrate (Table 1). The binding of these mutant RNAs by Rnt1p does not seem to be significantly less efficient than the binding of the wild-type RNA, in the limits of sensitivity of the EMSA (Table 1). These results indicate that the 2′-hydroxyl groups of residues A18 and A20 are required for efficient cleavage by Rnt1p, but they do not contribute quantitatively to binding. This result is not unexpected, as the contribution of single 2′-hydroxyl groups to binding energy is relatively minor. However, these results suggest that these 2′-hydroxyl groups have important functions in the recognition by Rnt1p, possibly by contributing to the catalytic positioning of the enzyme.
Synergistic effects of α-helix 1 dsRBD mutations and of RNA substrate 2′-deoxy modifications
To further probe the importance of the interaction between helix α1 residues and 2′-hydroxyl groups of the RNA substrate, we studied the cleavage of the snR47dA18 and dA20 substrates by the Rnt1pS376E and Rnt1pK371A mutants. As shown in Figure 4D and in Table 1, very little cleavage product was detected when Rnt1pK371A was incubated with snR47dA20, even after 2 h of incubation (Fig. 4D; Table 1). This cleavage defect is much more dramatic than the cleavage defects observed with the K371A mutation or the dA20 modification alone (Fig. 4D; Table 1). Rnt1pK371A is therefore unable to cleave a model RNA substrate lacking the 2′-hydroxyl group of adenosine 20. This result shows that the K371A mutation and the dA20 modification induce a synergistic defect on Rnt1p cleavage. This effect is not specific to the dA20 modification, as combining the K371A mutant and the dA18 modification also results in a dramatic loss of cleavage activity (Table 1). Thus, we conclude that K371 and the 2′-hydroxyl groups of A18 and A20 contribute to efficient cleavage, and that removing two of these groups result in a synergistic loss of activity, consistent with a loss of multiple interactions between the enzyme and the RNA substrate. The same observation was made when the S376E mutant was combined with the dA18 or dA20 substrates (Fig. 4E; Table 1).
The binding of snR47dA20 by Rnt1pK371A was significantly affected (Fig. 5D; Table 1). The corresponding Kd was almost double that of the Kd of the association between Rnt1pK371A and wild-type snR47 (Fig. 5A; Table 1) and threefold that of the Kd of the interaction between the wild-type enzyme and the unmodified RNA (Fig. 5A; Table 1). The dissociation constant observed for the interaction between Rnt1pS376E and snR47dA18 is only slightly higher than the Kd observed for the binding of Rnt1pS376E to the wild-type substrate (Table 1). Thus, the binding defect induced by the combination of this mutation and of the substrate 2′-deoxy modification is not as dramatic as the defect observed in the cleavage kinetics. It is likely that the residual binding observed with this mutant mostly reflects a nonproductive binding complex, as the combination of this mutant protein and of the RNA substrate is almost completely inactive (Fig. 4E). These observations confirm our hypothesis that K371 contributes quantitatively to binding, while the role of S376 might be limited to positioning the substrate in a conformation required for productive catalysis, without a strong quantitative contribution to binding efficiency.
DISCUSSION
In this study, we have performed an extensive genomic and biochemical analysis of the role of Rnt1p dsRBD amino acids and of the RNA substrate 2′-hydroxyl groups. The amino acids analyzed in this study can be classified in two groups: amino acids conserved in all RNases III, like K371, and residues that are conserved only in RNases III from fungi that recognize dsRNA capped by AGNN tetraloops like M368, R372, and S376. Interestingly, we observe a good correlation between the effects of mutations at these positions on RNA processing in vivo and on in vitro cleavage activity of the recombinant proteins, validating both approaches and strengthening the conclusion that these residues are required for efficient Rnt1p activity. The K371A was the strongest mutation identified, affecting almost all substrates in vivo and showing the most profound effects on binding and cleavage in vitro. This mutation results in a weaker binding of the enzyme to its substrate, probably because essential interactions with the minor groove at the top of the dsRNA are perturbed. The strong effect of this mutation on binding to the dsRNA was expected, because this side chain forms multiple interactions with the RNA via hydrogen bonding and van der Waals and electrostatic interactions (Fig. 1). The loss of these multiple interactions is likely responsible for the fact that this mutation affects the largest number of substrates in vivo. Mutations of amino acids conserved only in RNases specific for AGNN tetraloops show milder effects in vivo, and seem to affect only a subset of substrates. These mutations perturb cleavage in vitro to different extents, but most mutants still display a robust binding to the RNA substrate. These observations suggest that these residues are important for positioning the RNA substrate correctly with respect to the catalytic site, and/or to switch the enzyme–substrate complex into a catalytically active conformation. The EMSA technique used to monitor binding of the enzyme to the wild-type substrate does not discriminate productive binding, leading to the cleavage reaction, from nonproductive binding resulting in a complex in which the enzyme is not properly positioned onto the substrate to carry out the cleavage reaction. Such nonproductive binding has been reported in the crystal structure of Aquifex RNase III mutant in complex with dsRNA (Blaszczyk et al. 2004). Therefore, the calculated affinities of some of the interactions involving mutant proteins and/or modified substrates might not reflect productive binding events.
The effect of dsRBD mutations was analyzed in vivo using classical Northern analysis and using an original genomic method using custom microarrays encompassing the sequences of all the genomic loci supporting the expression of the known noncoding RNAs in yeast (Peng et al. 2003; Hiley et al. 2005). The advantage of this genomic approach relies on the fact that one single experiment enables us to assess the effect of specific point mutations in the dsRBD of Rnt1p on the in vivo processing of the vast majority of the known substrates of the enzyme. One caveat about this approach is that it is not sensitive enough to detect the accumulation of the 5′ region of a significant number of snoRNA precursors known to be Rnt1p substrates in the strains lacking Rnt1p activity (e.g., snR46, snR52, snR60, snR63, snR66, snR68, snR69, and snR71; data not shown). This lack of detection may be due to the low sensitivity of the oligonucleotide probes covering the 5′ extension of these particular substrates combined with the fact that for some of these substrates, 5′-extended precursors accumulate only at very low levels (Lee et al. 2003a). Surprisingly, we found, using this microarray approach, that not all Rnt1p substrates display the same sensitivity to the various mutations in the dsRBD. Therefore, these mutations affect in vivo processing by yeast RNase III in a substrate-specific manner. Several hypotheses could account for the substrate-specific effects observed. In strains lacking Rnt1p, an alternative 5′ →3′ exonucleolytic processing pathway results in the production of mature snoRNAs (Lee et al. 2003a). The relative importance of the Rnt1p-dependent and of the Rnt1p-independent processing pathways is variable depending on the precursors, and this Rnt1p-independent pathway could account for the fact that some of the Rnt1p substrates are insensitive to point mutations in RNT1. However, the snoRNAs that are insensitive to point mutations in the Rnt1p dsRBD (snR56 and snR65) do not correspond to those that are processed predominantly by a Rnt1p-independent pathway, such as snR52, snR58, and snR66 (Lee et al. 2003a). This absence of correlation makes the aforementioned hypothesis rather unlikely. A second hypothesis is that these snoRNA genes are transcribed at different levels. For the snoRNA genes transcribed at low levels, it is possible that the low rate of transcription might be rate-limiting for snoRNA biogenesis in vivo, and that the effects of Rnt1p mutations are masked when only steady-state levels of RNAs are monitored in vivo. A third hypothesis is that substrate recognition by Rnt1p allows some flexibility, and that the contacts established between the dsRBD and the substrates are slightly different from one class of substrate to another. The sequence of the tetraloop and the adjacent base pairs is variable from one substrate to another, and, although the overall structure of the tetraloop region is conserved, some flexibility may occur. This hypothesis is supported by the fact that local dynamics have been observed by NMR in the nonconserved region of a model substrate containing an AGUU tetraloop (Wu et al. 2001). Although helix α1 of the Rnt1p dsRBD is predicted to contact mostly the sugar–phosphate backbone of the substrate, the precise orientation of at least the nonconserved side of the RNA substrate is probably slightly different from one substrate to another, as are the contacts between this backbone and the dsRBD. This variability may account for the substrate-specific processing defects observed in strains expressing the dsRBD mutations. The only substrate for which we can propose a rationale is the precursor of snR36, which contains a rather unusual stem-loop structure that includes a large internal loop in the stem between the tetraloop and the cleavage site. We propose that the presence of this large loop, when combined with mutations in the Rnt1p dsRBD, induces a synergistic defect that strongly hinders recognition of the snR36 precursor by Rnt1p and results in a severe processing defect. Other substrates do not show such extensive internal loops, and none of their structural features can explain their differential sensitivities to dsRBD mutations.
Finally, we show in this study that 2′-hydroxyl groups present in the tetraloop region and in the dsRNA are important determinants for productive cleavage by Rnt1p. The role of these 2′-hydroxyl groups could not be addressed in vivo for obvious reasons. While single 2′-deoxy modifications have moderate effects on cleavage efficiency, the essential nature of some of the interactions with the 2′-hydroxyl groups of the RNA substrate is revealed by combining mutations in residues of the dsRBD and 2′-deoxy modifications of nucleotides of the substrate. The results presented here validate in vivo and in vitro the contacts observed in the NMR structure of the dsRBD of Rnt1p in complex with a model RNA substrate (Wu et al. 2004). This structure was obtained with an RNA substrate that lacked the cleavage site and the dsRBD was not in the context of additional important domains of Rnt1p, such as the catalytic domain, suggesting that the structure is likely to represent the ground state. It is therefore important to emphasize that the contacts seen in the context of the dsRBD–dsRNA ground state complex are shown here to be important for catalytic function of the full-length protein in vivo and in vitro. Our study shows that most of the dsRBD residues and RNA groups at the interface between the two molecules in the context of the solution structure of the dsRBD–dsRNA are major determinants for substrate recognition by the full-length enzyme. This study demonstrates the importance of a few amino acid contacts with the RNA for proper positioning of the dsRBD on the substrate. The specific orientation of helix α1 on the substrate is not only important for substrate binding but it is also likely to be important for the proper positioning of the catalytic domain on the RNA substrate. The precise mechanism by which this positioning of the catalytic domain occurs remains to be established.
MATERIALS AND METHODS
Mutagenesis and in vivo RNA analysis
Yeast strains expressing the M368A or M368E dsRBD mutant versions of Rnt1p were obtained by replacing codon 368 of the RNT1 open reading frame (ATG) by a GCG or a GAG codon, respectively. These mutations were introduced at the chromosomal RNT1 gene locus of the S. cerevisiae strain BMA64 (Chanfreau et al. 1998b) using the “delitto perfetto” two-step homologous recombination method (Storici et al. 2001). The mutations were confirmed by sequencing of an 800-bp PCR fragment spanning the site of mutation. RNA extractions and Northern blot analyses were performed as described previously (Chanfreau et al. 1998b).
Microarray analysis
RNase-free DNase I was used to remove traces of DNA from the RNA preparations. Ten micrograms of DNase I-treated total RNA were labeled with Alexa Fluor 546 or 647 according to the manufacturer’s instructions (Molecular Probes Ulysis kit), ethanol-precipitated, and hybridized to the array as previously described (Peng et al. 2003). Formamide was added to a final concentration of 33%, and hybridizations were carried out in a rotating incubator at 42°C for 16–20 h and washed as described (Hughes et al. 2001). Arrays were scanned on an Axon 4000B instrument. Scanned images were quantitated with GenePix (Axon Instruments). Individual channels were spatially detrended (i.e., over all correlations between spot intensity and position on the slide removed) and dye bias corrected in each slide as previously described (Hiley et al. 2005). After these steps, the normalized intensities were converted in log2 ratios of mutant versus wild type.
Cloning
Mutant RNT1 open reading frames (ORFs) were obtained by PCR from genomic DNAs extracted from the mutant strains bearing point mutations in the endogenous RNT1 chromosomal locus constructed during the course of this study or from the strains published previously (Wu et al. 2004). These PCR reactions were performed using high fidelity Pfu Turbo DNA polymerase (Stratagene) and oligonucleotides introducing NdeI and BamHI restriction sites at the 5′ and 3′ ends, respectively, of the resulting PCR fragments. The PCR products corresponding to the various mutant ORFs were digested by NdeI and BamHI and cloned into the pET16b vector (Novagen) previously digested by the same enzymes. The sequence and the frame of the cloned ORFs were checked by sequencing of the recombinant plasmids.
Protein expression and purification
Recombinant pET16b vectors expressing wild-type or dsRBD mutant Rnt1p proteins fused to a 6-histidine tag at the N terminus were transformed into Escherichia coli strain BL21(DE3) (Novagen). Each strain was grown at 30°C to OD600 0.5 in 4 L of LB medium containing 100 μg/mL of ampicillin. Expression of the recombinant proteins was induced by adding IPTG to the cultures to a final concentration of 0.5 mM. After 3 h of induction, rifampicin was added to a final concentration of 50 μg/mL and incubation was prolonged for another 2 h. Cells were harvested and then resuspended with 20 mL of ice-cold buffer A (20 mM NaPO4 [pH 8.0], 10 mM imidazole, 500 mM NaCl) with one tablet of Complete EDTA-free protease inhibitor cocktail (Roche Diagnostics). Cell suspensions were sonicated on ice for five cycles with a 550 Sonic Dismembrator from Fisher Scientific (one cycle consisting of a succession of 0.2 sec with pulsar on and 0.8 sec with pulsar off for a total of 20 sec, setting 4, followed by 1 min of incubation on ice). Cell extracts were centrifuged at 13,500 rpm for 30 min at 4°C. Supernatants were filtered through 0.45 μm MCE filters (Fisherbrand), then loaded onto 1-mL Hi-Trap Chelating HP columns (Amersham Biosciences) previously loaded with a 100 mM solution of NiSO4 connected to an AKTA prime FPLC (Amersham Pharmacia Biotech). Columns were washed with ~15 mL of buffer containing 20 mM NaPO4 (pH 8.0), 20 mM imidazole, and 500 mM NaCl. Bound proteins were eluted by applying a buffer containing 20 mM NaPO4 (pH 8.0), 500 mM NaCl, with a 20 mM to 500 mM imidazole gradient. Fractions containing the recombinant Rnt1p proteins were pooled and dialyzed in 3500 MWCO Slide-A-Lyser dialysis cassettes (Pierce) overnight at 4°C against 2 L of dialysis buffer containing 50 mM Tris-HCl (pH 7.4), 200 mM KCl, 50% glycerol, and 0.5 mM DTT, and dialyzed for an additional 6 h at 4°C against 2 L of fresh dialysis buffer. The concentration of the purified wild-type and dsRBD mutant protein samples was determined by loading a diluted aliquot of each protein sample on an SDS protein gel along with BSA standards of known concentration.
In vitro cleavage experiments
Chemically synthesized RNA substrates were purchased from Dharmacon. In vitro cleavage of 5′-end-labeled substrates using purified recombinant wild-type or dsRBD mutant versions of Rnt1p was performed as described previously (Chanfreau et al. 2000). RNA was heated to 90°C for 2 min and chilled on ice, and the reactions were initiated by addition of the enzyme. Cleavage reactions were performed in duplicate under single-turnover conditions with 0.3 nM of RNA substrate and 70 nM or 120 nM of enzyme at 23°C. Cleavage was monitored by taking aliquot volumes of the reactions at different time points after the onset of the reaction. Reaction products were fractionated on 10% acrylamide:bis-acrylamide (19:1 ratio, respectively), 0.5× TBE, 8 M urea denaturing gels. The amount of cleaved product relative to full-length substrate was measured using ImageQuant 5.2 software. Data were plotted to produce time course traces, which were then fitted to double or single exponential equations (see Table 1). The unreacted fraction observed at late incubation times is probably due to a fraction of substrates in a head–tail duplex conformation (Wu et al. 2001).
Electrophoretic mobility shift assays
Binding was performed by incubating ~30 fmol of 5′-end-labeled RNA substrates with increasing concentrations of wild-type or mutant versions of Rnt1p for 20 min on ice in a binding buffer containing 50 mM Tris-HCl (pH 7.4), 200 mM KCl, 2.5 mM EDTA (pH 8.0), 5 mM spermidine, 0.5 mM DTT, 20% glycerol, and 100 ng/μL BSA. Binding mixtures were loaded on 4% acrylamide:bis-acrylamide (80:1 ratio, respectively), 50 mM Tris-glycine native gels pre-run for 30 min at 4°C in a 50 mM Tris-glycine (pH 8.5) running buffer. Electrophoresis was carried out for 5–6 h at 8 mA at 4°C. Gels were dried, exposed on Phosphor screens (Molecular Dynamics), and scanned using a PhosphorImager 445 SI (Molecular Dynamics). ImageQuant 5.2 software was used to quantitate the intensity of the bands corresponding to free and bound substrates. The ratio of bound substrate over total (expressed as a percentage) was plotted for each enzyme concentration, and the dissociation constants (Kd) were deduced from the plots.
Acknowledgments
We thank A. Lee and C.Y. Lee for critical reading of the manuscript. A.K.H. was supported by a Human Frontier Science Program Organization postdoctoral fellowship; M.S., by an NSF GK-12 Fellowship; and S.L.H., by a CIHR postdoctoral fellowship. Microarray experiments were funded by CIHR and CFI grants to T.R.H. The work was supported by National Science Foundation grant MCB0111060 (J.F.) and National Institutes of Health grants GM37254 (J.F.) and GM61518 (G.C.).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2760705.
REFERENCES
- Abou Elela, S. and Ares Jr., M. 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17: 3738–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Elela, S., Igel, H., and Ares Jr., M. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85: 115–124. [DOI] [PubMed] [Google Scholar]
- Allmang, C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E., and Tollervey, D. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18: 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Blaszczyk, J., Gan, J., Tropea, J.E., Court, D.L., Waugh, D.S., and Ji, X. 2004. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure 12: 457–466. [DOI] [PubMed] [Google Scholar]
- Bycroft, M., Grunert, S., Murzin, A.G., Proctor, M., and St. Johnston, D. 1995. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 14: 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G. 2003. Conservation of RNase III processing pathways and specificity in hemiascomycetes. Eukaryot. Cell 2: 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Abou Elela, S., Ares Jr., M., and Guthrie, C. 1997. Alternative 3′-end processing of U5 snRNA by RNase III. Genes & Dev. 11: 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Legrain, P., and Jacquier, A. 1998a. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284: 975–988. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., Rotondo, G., Legrain, P., and Jacquier, A. 1998b. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17: 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Buckle, M., and Jacquier, A. 2000. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl. Acad. Sci. 97: 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman, M., Lee, C.Y., and Chanfreau, G. 2003. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11: 1279–1289. [DOI] [PubMed] [Google Scholar]
- Ge, D., Lamontagne, B., and Abou Elela, S. 2005. RNase III-mediated silencing of a glucose-dependent repressor in yeast. Curr. Biol. 15: 140–145. [DOI] [PubMed] [Google Scholar]
- Ghazal, G., Ge, D., Gervais-Bird, J., Gagnon, J., and Abou Elela, S. 2005. Genome-wide prediction and analysis of yeast RNase III-dependent snoRNA processing signals. Mol. Cell. Biol. 25: 2981–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiley, S.L., Jackman, J., Babak, T., Trochesset, M., Morris, Q.D., Phizicky, E., and Hughes, T.R. 2005. Detection and discovery of RNA modifications using microarrays. Nucleic Acids Res. 33: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T.R., Mao, M., Jones, A.R., Burchard, J., Marton, M.J., Shannon, K.W., Lefkowitz, S.M., Ziman, M., Schelter, J.M., Meyer, M.R., et al. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19: 342–347. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharrat, A., Macias, M.J., Gibson, T.J., Nilges, M., and Pastore, A. 1995. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 14: 3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, S.W. and Bass, B.L. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J., Dichtl, B., and Tollervey, D. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamontagne, B. and Abou Elela, S. 2001. Purification and characterization of Saccharomyces cerevisiae Rnt1p nuclease. Methods Enzymol. 342: 159–167. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 279: 2231–2241. [DOI] [PubMed] [Google Scholar]
- Lamontagne, B., Tremblay, A., and Abou Elela, S. 2000. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 20: 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamontagne, B., Ghazal, G., Lebars, I., Yoshizawa, S., Fourmy, D., and Abou Elela, S. 2003. Sequence dependence of substrate recognition and cleavage by yeast RNase III. J. Mol. Biol. 327: 985–1000. [DOI] [PubMed] [Google Scholar]
- Lee, C.Y., Lee, A., and Chanfreau, G. 2003a. The roles of endonucleolytic cleavage and exonucleolytic digestion in the 5′-end processing of S. cerevisiae box C/D snoRNAs. RNA 9: 1362–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003b. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Leulliot, N., Quevillon-Cheruel, S., Graille, M., Van Tilbeurgh, H., Leeper, T.C., Godin, K.S., Edwards, T.E., Sigurdsson, S.T., Rozenkrants, N., Nagel, R.J., et al. 2004. A new a-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III. EMBO J. 23: 2468–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh, S., Davierwala, A.P., Haynes, J., Moffat, J., Peng, W.T., Zhang, W., Yang, X., Pootoolal, J., Chua, G., Lopez, A., et al. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44. [DOI] [PubMed] [Google Scholar]
- Nagel, R. and Ares Jr., M. 2000. Substrate recognition by a eukaryotic RNase III: The double-stranded RNA-binding domain of Rnt1p selectively binds RNA containing a 5′-AGNN-3′ tetraloop. RNA 6: 1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, W.T., Robinson, M.D., Mnaimneh, S., Krogan, N.J., Cagney, G., Morris, Q., Davierwala, A.P., Grigull, J., Yang, X., Zhang, W., et al. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113: 919–933. [DOI] [PubMed] [Google Scholar]
- Qu, L.H., Henras, A., Lu, Y.J., Zhou, H., Zhou, W.X., Zhu, Y.Q., Zhao, J., Henry, Y., Caizergues-Ferrer, M., and Bachellerie, J.P. 1999. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 19: 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, A., Grunert, S., Adams, J., Micklem, D.R., Proctor, M.R., Freund, S., Bycroft, M., St. Johnston, D., and Varani, G. 2000. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 19: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter, J.M. and Schultz, S.C. 1998. Molecular basis of double-stranded RNA–protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17: 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipelt, R.L., Zheng, B., Asuru, A., and Rymond, B.C. 1999. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 27: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl, R. and Allain, F.H. 2005. A novel RNA pentaloop fold involved in targeting ADAR2. RNA 11: 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici, F., Lewis, L.K., and Resnick, M.A. 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19: 773–776. [DOI] [PubMed] [Google Scholar]
- Vuyisich, M., Spanggord, R.J., and Beal, P.A. 2002. The binding site of the RNA-dependent protein kinase (PKR) on EBER1 RNA from Epstein-Barr virus. EMBO Rep. 3: 622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Xu, H., Miraglia, L.J., and Crooke, S.T. 2000. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 275: 36957–36965. [DOI] [PubMed] [Google Scholar]
- Wu, H., Yang, P.K., Butcher, S.E., Kang, S., Chanfreau, G., and Feigon, J. 2001. A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J. 20: 7240–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Henras, A., Chanfreau, G., and Feigon, J. 2004. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. 101: 8307–8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y., Yi, R., and Cullen, B.R. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24: 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]