Abstract
Ribavirin is a guanosine ribonucleoside analog that displays broad-spectrum anti-viral activity and is currently used for the treatment of some viral infections. Ribavirin has recently been proposed to also be a mimic of the 7-methyl guanosine cap found at the 5′ end of mRNAs. To obtain supporting functional data for this hypothesis, we assessed the ability of ribavirin triphosphate to interfere with the interaction between eIF4E and 7-methyl guanosine capped mRNA. In chemical cross-linking assays, cap-affinity chromatography, and cap-dependent translation assays, ribavirin was unable to function as a cap analog.
Keywords: ribavirin, cap analog, 7-methyl guanosine, translation, RTP
INTRODUCTION
Translation initiation is a highly regulated process, with ribosome recruitment being rate limiting in most instances (for review, see Gingras et al. 1999). The eIF4F complex is involved in this step and is comprised of three subunits: (1) eIF4E, the cap-binding protein responsible for binding of the complex to the mRNA 7-methyl guanosine structure in an ATP-independent fashion; (2) eIF4A, an RNA helicase required for unwinding secondary structure in preparation for 43S ribosomal subunit binding; and (3) eIF4G, a modular scaffold that mediates mRNA binding of the 43S preinitiation complex through interactions with eIF3. Transformation assays in cell culture (Lazaris-Karatzas et al. 1990) and studies with transgenic mice (Ruggero et al. 2004; Wendel et al. 2004) have demonstrated that eIF4E is an oncogene that contributes to cancer progression (Wendel et al. 2004). This is likely the consequence of increased protein production from mRNA species for which ribosome recruitment is highly dependent on eIF4F activity and that are involved in cellular growth and cell cycle progression (Gingras et al. 2001). These findings underscore the importance of better understanding the eIF4E/7-methyl guanosine cap interaction and of targeting eIF4E for possible therapeutic intervention.
Detailed understanding of the interaction between eIF4E and the 7-methyl guanosine cap structure has been obtained from structure/activity studies utilizing cap analogs and from structural studies of eIF4E bound to cap analogs (Marcotrigiano et al. 1997; Matsuo et al. 1997). Cap analogs have been extensively utilized to study the ribosome recruitment step of translation initiation in complete translation assays (Hickey et al. 1977; Adams et al. 1978; Darzynkiewicz et al. 1987, 1989; Cai et al. 1999), in partial reactions with purified initiation factors (Sonenberg et al. 1978; Darzynkiewicz et al. 1981, 1985; Grifo et al. 1983; Tahara et al. 1983), and in direct binding assays with eIF4E (Niedzwiecka et al. 2004). There is a direct correlation between inhibition of overall translation and inhibition of eIF4F activity. There are several structure–activity relationships that have emerged from studies with cap analogs: (1) N7 alkyl and alicyclic substituents larger than ethyl decrease inhibitory activity (Adams et al. 1978; Furuichi et al. 1979; Darzynkiewicz et al. 1989); (2) aryl substitution at N7 improves the efficacy of inhibition (Cai et al. 1999); (3) replacement of the ribose by (hydroxyethoxy)methyl ether (but not by other pentoses) (Darzynkiewicz et al. 1985) or ring opening (Cai et al. 1999) decreases inhibitory activity; and (4) derivatization of the α-phosphate moiety by O-methylation decreases the potency of m7GMP (Darzynkiewicz et al. 1981). In addition, there is preference for the anti-conformation in ligand binding (Darzynkiewicz et al. 1989), there is improved inhibition with increasing phosphate residues (Cai et al. 1999), and the second nucleotide residue in analogs of the form m7GpppN affects inhibitory activity in the order G > C > U > A (Cai et al. 1999). The three-dimensional structure of eIF4E reveals a protein resembling a cupped hand with eight anti-parallel β-sheets and a narrow hydrophobic cap-binding slot on the concave surface and three α-helices on the convex surface (Marcotrigiano et al. 1997; Matsuo et al. 1997). 7-Methyl guanine recognition is mediated by stacking between two conserved tryptophans, as well as hydrogen bonding and van der Waals contacts between the N7-methyl group and a third conserved tryptophan (Marcotrigiano et al. 1997; Matsuo et al. 1997). The stacking interaction is significantly strengthened because of charge transfer between the electron-rich indole groups and the electron-deficient 7-methyl guanine (which carries a delocalized positive charge secondary to methylation) (Ishida et al. 1988).
Ribavirin (1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide) is a guanosine ribonucleoside analog that displays broad spectrum anti-viral activity (Sidwell et al. 1972; De Clercq 1993). In the clinic, it is used for the treatment of several viral infections, including Lassa fever virus, respiratory syncytial virus, hepatitis C virus, and severe acute respiratory syndrome coronavirus (Tam et al. 2001), with low toxicity observed in animals and humans (Sidwell et al. 1972, 1979). In vivo, ribavirin is phosphorylated by cellular kinases, with ribavirin tri-phosphate (RTP) being the major intracellular metabolite (Miller et al. 1977; Page and Connor 1990). A number of mechanisms have been proposed to account for its anti-viral activity. Ribavirin monophosphate inhibits cellular inosine monophosphate dehydrogenase required for de novo synthesis of GTP (Streeter et al. 1973; Muller et al. 1977), and thus depletion of intracellular GTP pools would be detrimental to viral replication. Moreover, ribavirin is a substrate for viral RNA-dependent RNA polymerase, resulting in incorporation opposite cytidines and uridines into the viral genome template and leading to catastrophic lethal mutagenesis (Crotty et al. 2000, 2001; Maag et al. 2001). Ribavirin is also a substrate for viral guanylyl transferases (Goswami et al. 1979; Bougie and Bisaillon 2004) leading to the generation of viral mRNA transcripts that contain a 5′ RpppN structure (where R is ribavirin and N is any nucleotide) instead of 5′ GpppN (Bougie and Bisaillon 2004). In vitro, RpppN-terminated transcripts are not efficiently translated compared to their m7GpppN-terminated counterparts (Bougie and Bisaillon 2004). This finding is at odds with the recent suggestion that ribavirin is a 7-methyl guanosine cap analog (Kentsis et al. 2004). In this report, we have undertaken functional experiments assessing the ability of RTP to interfere with eIF4E/mRNA interactions or to block translation of cap-dependent translation.
RESULTS
Assessing eIF4E/ribavirin interactions
To directly assess if ribavirin can function as a 7-methyl guanosine cap analog (Fig. 1A), we tested the ability of ribavirin triphosphate to inhibit cross-linking of recombinant eIF4E to cap-labeled oxidized mRNA (Fig. 1B). This classic assay was first used to identify eIF4E (Sonenberg and Shatkin 1977; Sonenberg et al. 1978) and consists of generating cap-labeled mRNA containing an oxidized ribose moiety that can be covalently linked to eIF4E (via Schiff base formation and subsequent reduction). Following degradation of the mRNA template with RNAse, the products are analyzed by SDS/PAGE. Proteins covalently bound to the radiolabeled cap structure are readily visualized by autoradiography (Fig. 1B, lane 1). Specificity in this assay is assessed using m7GDP (or m7GTP) as a specific competitor (Fig. 1, cf. lanes 2 and 1) and GDP as a nonspecific competitor (Fig. 1, cf. lanes 3 and 1). In this assay, RTP does not interfere with the eIF4E/7-methyl guanosine interaction (Fig. 1, cf. lanes 4 and 1).
FIGURE 1.
RTP does not inhibit eIF4E/m7G-mRNA cap interaction in functional assays. (A) Chemical structures of the keto forms of 7-methyl-guanosine, guanosine, and ribavirin. R denotes Ribose. (B) Cross-linking of recombinant His6-eIF4E to oxidized 32P-labeled mRNA. Cross-linkings were performed using 1 μg of eIF4E and 50,000 cpm m7G-labeled mRNA in the presence of 0.5 mM m7GDP (lane 2), 0.5 mM GDP (lane 3), and 0.5 mM RTP (lane 4). Following digestion with 10 μg RNAse A, samples were analyzed by electrophoresis on a 10% SDS/PAGE. The gel was dried and exposed to an X-OMAT film (Kodak) at −70°C for 5 h. The position of migration of recombinant His6-eIF4E is indicated by an arrow. (C) Cross-linking of rabbit reticulocyte initiation factors (IF) to oxidized 32P-labeled mRNA. Cross-linkings were performed using 100 μg of IFs in the absence of exogenously supplied ATP (indicated above the panel), since binding of eIF4E to the cap structure is an ATP-independent event (Sonenberg 1981). 7-methyl GDP, GDP, and RTP were used at a final concentration of 0.5 mM in this experiment. The position of migration of eIF4E is indicated by an arrow. (D) Affinity purification of His6-eIF4E from an m7GDP-agarose matrix. Following binding of His6-eIF4E to m7GDP-agarose (synthesized as previously described by Edery et al. [1988]) and extensive washing with LCB (10 mM HEPES-KOH [pH 8.0], 100 mM KCl, 0.2 mM EDTA [pH 8.0]), the resin was successively incubated with 5 bed volumes (100 μL) of 50 μM GDP, 50 μM RTP, and 50 μM m7GDP. The first three 100 μL of each wash were collected and 20 μL processed and loaded onto a 10% SDS/PAGE. Following transfer to Immobillon P, the blot was probed with an anti-His antibody (Cell Signalling) and proteins visualized by chemiluminescence (PerkinElmer Life Sciences, Inc.).
As the above experiment utilized recombinant eIF4E, we repeated the cross-linking assay utilizing initiation factors prepared from rabbit reticulocyte ribosomes to assess the consequence of RTP on the cap binding properties of native eIF4E (Fig. 1C). A greater number of radiolabeled protein species are cross-linked to the cap structure when utilizing initiation factors, indicative of many proteins in the preparation binding nonspecifically (Fig. 1, lane 1). However, the interaction of one of these, eIF4E, is specifically inhibited by the presence of m7GDP in the cross-linking assay (Fig. 1, cf. lanes 2 and 1), and not by GDP (Fig. 1, cf. lanes 3 and 1). In this experiment, RTP did not impair native eIF4E’s ability to interact with 7-methyl guanosine capped mRNA (Fig. 1, cf. lanes 4 and 1).
These results were confirmed in an independent assay involving elution of recombinant eIF4E from an m7GDP-agarose affinity resin (Fig. 1D). In this experiment, eIF4E was bound to m7GDP-agarose and successively washed with 50 μM GDP (lanes 1–3), 50 μM RTP (lanes 4–6), or 50 μM m7GDP (lanes 7–9). Following fractionation by SDS/PAGE, Western blot analysis was used to identify fractions containing eluted His6-eIF4E (Fig. 1D). The results indicate that, under the conditions used, GDP and RTP do not elute eIF4E from m7GDP-agarose—only m7GDP is effective in this task (Fig. 1, cf. lanes 7–9 and 1–6).
In vitro translation assays
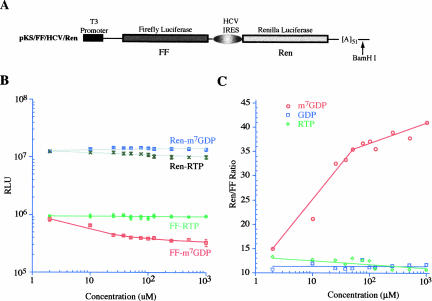
Effectson eIF4E activity by cap analogs can also be scored as inhibition of cap-dependent translation (Darzynkiewicz et al. 1987, 1989; Cai et al. 1999). We utilized translation extracts prepared from Krebs-2 ascites to determine whether RTP could inhibit cap-dependent protein synthesis (Fig. 2). Translation extracts were programmed with the bicistronic FF/HCV/Ren mRNA (Fig. 2A), where production of firefly luciferase is cap dependent and synthesis of Renilla luciferase is cap independent due to internal ribosome recruitment by the HCVIRES. In this experiment, production of firefly luciferase was inhibited by m7GDP in adose-dependent fashion (Fig. 2B). As expected, production of Renilla in the presence of m7GDP was unaffected by concentrations up to 1 mM. In this assay, concentrations of RTP up to 1 mM did not affect the synthesis of either firefly or Renilla luciferases (Fig. 2B). Neither GDP nor RTP affected the ratio of cap-independent to cap-dependent protein synthesis (Fig. 2C). However, increasing concentrations of m7GDP were associated with an increase in this ratio (Fig. 2C), consistent with the observed inhibition of m7GDP on cap-dependent protein synthesis (Fig. 2B).
FIGURE 2.
In vitro translations in the presence of RTP, m7GDP, and GDP. (A) Schematic diagram of the pKS/FF/HCV/Ren bicistronic construct. To generate mRNA for in vitro transcriptions, the plasmid was linearized within the 3′ UTR with BamHI. The firefly luciferase coding region is denoted by a blackened box and the Renilla coding region is denoted by a light gray box. The HCV IRES allows for internal initiation and cap-independent translation of the downstream Renilla cistron. (B) Dose response curve illustrating the effect of RTP and m7GDP on translation of FF/HCV/Ren in Krebs extracts. Translations were performed in the presence of the indicated amounts of RTP or m7GDP and at a final mRNA and K+ concentration of 5 μg/mL and 100 mM, respectively. Control translation reactions contained equivalent amounts of vehicle. The obtained luciferase activities for each different mRNA were normalized to the activity obtained in the control translations of the same mRNA species (which was set at one) and plotted as a function of compound concentration. Each data point represents the average of eight independent translations and the standard error of the mean is shown (too small to be seen for many of the data points). Log scale is used on both axes. (C) The relative ratios of Renilla/firefly luciferase values were taken from B and are plotted for each compound as a function of concentration. Included in this panel are the results from a titration performed with GDP as well.
Assessing RTP activity
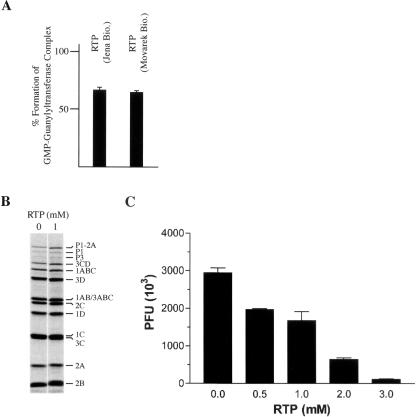
We were concerned by the lack of activity demonstrated by RTP (from Moravek Biochemicals) in these assays, despite having compounds of high purity (see Materials and Methods). Therefore the cross-linking assay (Fig. 1B) and in vitro translation assays (Fig. 2) were performed with a different RTP preparation (Jena Biosciences) and yielded the same results (data not shown). Nonetheless, we also used two independent functional assays to ensure that our RTP preparations were active. First, during the capping process, vaccinia virus guanylyltransferase forms a GMP-enzyme intermediate that can be inhibited by RTP (Bougie and Bisaillon 2004). Using α-32P-GTP, and resolving the enzyme intermediate by SDS/PAGE, our RTP preparations inhibited this reaction by ~35%. This response is dose dependent and similar to previously reported concentrations required to achieve inhibition of this reaction (Bougie and Bisaillon 2004; Fig. 3A). Second, an in vitro replication system for EMC virus has recently been described (Svitkin and Sonenberg 2003) that can be used to score for the mutagenic properties of RTP, since incorporation of RTP into newly replicated EMCV strands is expected to reduce viral fitness (Crotty et al. 2000; Maag et al. 2001). Using this system, EMCV RNA-programmed Krebs extracts were incubated in the presence or absence of RTP. As expected, RTP had no effect on the efficiency or spectrum of viral proteins produced in vitro, indicating that it does not exert nonspecific inhibition on translation of the viral template in this assay (Fig. 3B). However, a dose-dependent reduction in the production of infectious virus was noted when RTP was present in the cell-free synthesis reaction (Fig. 3C). Taken together, these results indicate that our RTP preparations are biologically active.
FIGURE 3.
RTP inhibits Vaccinia virus guanylyltransferase activity and in vitro EMCV production. (A) Inhibition of guanylytransferase-GMP complex formation by RTP. Recombinant guanylytransferase was incubated with α-[32P]GTP and the extent of guanylytransferase–GMP complex monitored as previously described (Bougie and Bisaillon 2004). The extent of nucleotidyl-enzyme formation in the presence of 2 mM RTP, relative to reactions lacking RTP, is shown. (B) RTP does not affect EMCV protein synthesis. Cell-free synthesis of EMCV was performed in the presence of RTP, as previously described (Svitkin and Sonenberg 2003). EMCV RNA (20 μg/mL) was translated in the presence of [35S]methionine. After 1 h, 5-μL aliquots were withdrawn, supplemented with SDS sample buffer, and subjected to SDS-15% PAGE. An autoradiogram of the dried gel is shown, with the assignment of the polypeptides shown. (C) RTP inhibits in vitro cell-free synthesis of EMCV. Krebs-2 S10 extracts were incubated for 20 h at 32°C in the presence of EMCV RNA (20 μg/mL) containing the indicated concentrations of RTP. Samples were treated with a mixture of RNase A and T1, serially diluted, and plaque assays carried out on confluent BHK-21 cells. Indicated are the plaque forming units (PFU) obtained. Plaque assays were performed in duplicate and the error of the mean is shown.
DISCUSSION
In the current study, we used several assays to assess whether the eIF4E/7-methyl guanosine cap interaction could be inhibited by ribavirin triphosphate (Kentsis et al. 2004). Ribavirin triphosphate does not prevent recombinant or native eIF4E from interacting with capped mRNA (Fig. 1B,C) and is incapable of eluting eIF4E from a m7GDP-affinity matrix (Fig. 1D). Whereas m7GDP inhibited cap-dependent protein synthesis, RTP and GDP were not active in this assay (Fig. 2). Consistent with this finding is a report that ribavirin-capped mRNAs (RpppN-terminated) translate much less efficiently than their capped, methylated counterparts (m7GpppN-terminated) (Bougie and Bisaillon 2004).
Our results are different from those reported by Kentsis et al. (2004), although we do not fully understand the reasons. Some of the discrepancies may be a consequence of experimental design or off-target effects of RTP on cellular processes. Additionally, nucleotides may bind to many proteins nonspecifically, as exemplified by the cross-linking assay in Figure 1C. Although several proteins are observed to bind to radiolabeled cap structure in this assay, only the interaction of eIF4E is specific. Hence, experiments assessing nucleotide interaction with eIF4E need to use m7GDP (or m7GTP) as specific competitors to ensure that the observed interactions are mediated through the cap binding site of eIF4E and not due to allosteric effects. As well, nonspecific disruption of protein–RNA interactions by nucleotides have been previously reported (Sonenberg and Shatkin 1978).
Ribavirin has been reported to exert anti-cancer activity in a murine model using FaDu hypopharyngeal squamous cells treated for 20 d at 40 μg/kg/day (Kentsis et al. 2004). This model system is dependent on overexpression of eIF4E for its tumorigenicity, as reduction of eIF4E expression results in suppression of tumor growth. Kentsis et al. (2004) correlated the chemotherapeutic effect of ribavirin to inhibition of eIF4E activity. Our results suggest that the mechanism of action is unlikely to be the consequence of perturbing eIF4E/7-methyl guanosine cap interaction. The efficacy of ribavirin seen on FaDu hypopharyngeal squamous tumors may be related to its effects on nucleo-cytoplasmic shuttling and/or to the transport of cyclin D1 (Kentsis et al. 2004).
We note that the NCI Developmental Therapeutics Program has reported evaluation of ribavirin in several murine cancer models, including B16 melanoma, CX-1 adenocarcinoma, colon carcinoma, mammary adenocarcinoma, ependymoblastoma, L1210 and P388 leukemias, and Lewis and Madison 109 lung carcinoma (http://dtp.nci.nih.gov/dtpstandard/servlet/dwindex?searchtype=namestarts&chemnameboolean=and&outputformat=html&searchlist=ribavirin%0D%0A&Submit=Submit). From over 100 different experiments in mice where the majority of animals survived to the anti-tumor endpoint, and using a variety of treatment schedules with ribavirin doses as high as 900 mg/kg/injection, only two experiments showed anti-tumor activity. Treatment of one cohort of mice with colon carcinoma (implanted subcutaneous) at 50 mg/kg/injection and another cohort with mammary adenocarcinomas CD8F1 (implanted subcutaneous) at 25 mg/kg/injection showed a %T/C (treatment/control survival ratio) of 159 and 288, respectively. However, these data were not reproduced at higher dose escalation or upon repetition of the experiments. Although the NIH program did not test FaDu hypopharyngeal squamous cells, their data suggest that, under the conditions tested and on the tumors assayed, ribavirin is not a general anti-cancer agent.
MATERIALS AND METHODS
Reagents and general methods
Restriction endonucleases and T3 RNA polymerase were purchased from New England Biolabs. 5–3H-cytidine triphosphate (20.5 Ci/mmol), α-[32P]GTP (> 3000 Ci/mmol), and [35S]methionine (> 1000 Ci/mmol) were obtained from Perkin Elmer Life Sciences. Preparation of plasmid DNA, restriction enzyme digestion, and agarose gel electrophoresis of DNA were carried out using standard methods (Sambrook and Russell 2001). m7GDP and GDP were purchased from Sigma. RTP was obtained from two independent sources: One was custom synthesized by Moravek Biochemicals, Inc., and the other was obtained from Jena Biosciences. Compound purity was independently assessed through LC-MS and established to be 96.5% (m7GDP), 88.7% (GDP), and 100% (RTP; Movarek Biochemicals, Inc.). RTP from Jena Biosciences was quality-controlled by the manufacturer and assessed to be > 95% pure by LC-MS.
Purification of recombinant His6-eIF4E
Recombinant His6-eIF4E was expressed from pProEx-4E trans-formed into BL21DE3/pLysS, grown in Terrific Broth/Amp at 37°C until OD600 ~ 0.7, and induced with IPTG (0.25 mM final). Cells were grown for a further 4 h at 37°C prior to harvest. Bacteria cultures were centrifuged 10 min at 5000 rpm in a RC3B centrifuge and the bacterial pellets were resuspended in Buffer A (50 mM HEPES-KOH [pH 7.5], 500 mM KCl, 5mM DTT, 1% Triton X-100, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin, 1 mM PMSF). The suspended cells were sonicated on ice (8 × 30 sec), followed by treatment with DNase I (20 mg/L) and RNase A (5 mg/L) at 30°C for 20 min. Inclusion bodies were sedimented by centrifugation at ~32,000g (18,000 rpm in a RC5B/SS34 rotor) for 45 min. The pellets were washed twice in TBS/Tx (25 mM Tris-HCl[pH 7.2], 137 mM NaCl, 3 mM KCl, 1% Triton X-100) with centrifugation steps of 32,000g between washes.
The final pellets were solubilized in 50 mM HEPES-KOH (pH 7.5), 100 mM KCl, 25 mM DTT, and 6 M guanidine-HCl at 4°C. Insoluble material was removed by ultracentrifugation at 100,000g for 30 min and followed by determination of protein concentration. The protein was diluted in the above buffer to ~1–2 mg/mL and stepwise dialysis performed over a period of 2 d to renature His6-eIF4E (in the above-mentioned HEPES buffer containing 6 M, 4 M, 2 M, and 1 M guanidine-HCl). His6-eIF4E protein was then diluted to > 0.5mg/mL and dialysis continued against guanidine-HCl concentrations of 500 mM, 250 mM, and 125 mM. A final dialysis was performed against LCB (20 mM HEPES-KOH [pH 7.5], 100 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA) and insoluble material separated by centrifugation and filtration through a 0.45 μm filter (Millipore). The filtrate was then purified by m7GDP-agarose affinity chromatography, as previously described (Edery et al. 1995).
Cross-linking assays to oxidized mRNA cap structures
In vitro transcribed mRNA was capped and methylated with α-32P-GTP and SAM (S-adenosyl methionine) using vaccinia guanylyl-transferase, as previously described (Pelletier and Sonenberg 1985). The 5′ and 3′ end ribose moieties were oxidized to dialdehydes using sodium periodate (Sonenberg and Shatkin 1977). Cross-linking reactions with recombinant His6-eIF4E and initiation factors were performed and analyzed as described (Sonenberg and Shatkin 1977).
In vitro translation assays
In vitro transcriptions were performed using BamHI-linearized pKS/FF/HCV/Ren DNA template (Fig. 2A) in Krebs extracts at a final mRNA concentration of 5 μg/mL (Novac et al. 2004; Svitkin and Sonenberg 2004). Firefly and Renilla luciferase activity (RLU) were measured on a Berthold Lumat LB 9507 luminometer (Dyer et al. 2000).
Guanylyltransferase assay
The RNA guanylyltransferase assay was performed with the vaccinia virus D1 protein that had been expressed and purified as described previously (Bougie and Bisaillon 2004). Briefly, 2 μg of the enzyme were incubated with 10 μM [α-32P]GTP in a buffer containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, and 5 mM MgCl2 for 5 min at 37°C. The reactions were stopped by the addition of EDTA to 10 mM and SDS to 1%. Reactions were analyzed by electrophoresis through a 12.5% polyacrylamide gel containing 0.1% SDS and the radiolabeled proteins were visualized by autoradiography of the gel. The extent of covalent complex formation was quantitated by scanning the gel with a PhosphorImager (Molecular Dynamics).
Acknowledgments
We are grateful to Jany Lapointe for experimental expertise during the course of this work. This work was supported by an NCIC grant (no. 014313) to J.P. M.B. is a New Investigator Scholar from the Canadian Institute of Health Research (CIHR). J.P. is a Canadian Institute of Health Research (CIHR) Senior Investigator.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2930805.
REFERENCES
- Adams, B.L., Morgan, M., Muthukrishnan, S., Hecht, S.M., and Shatkin, A.J. 1978. The effect of “cap” analogs on reovirus mRNA binding to wheat germ ribosomes. Evidence for enhancement of ribosomal binding via a preferred cap conformation. J. Biol. Chem. 253: 2589–2595. [PubMed] [Google Scholar]
- Bougie, I. and Bisaillon, M. 2004. The broad spectrum antiviral nucleoside ribavirin as a substrate for a viral RNA capping enzyme. J. Biol. Chem. 279: 22124–22130. [DOI] [PubMed] [Google Scholar]
- Cai, A., Jankowska-Anyszka, M., Centers, A., Chlebicka, L., Stepinski, J., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 1999. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry 38: 8538–8547. [DOI] [PubMed] [Google Scholar]
- Crotty, S., Maag, D., Arnold, J.J., Zhong, W., Lau, J.Y., Hong, Z., Andino, R., and Cameron, C.E. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6: 1375–1379. [DOI] [PubMed] [Google Scholar]
- Crotty, S., Cameron, C.E., and Andino, R. 2001. RNA virus error catastrophe: Direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. 98: 6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Antosiewicz, J., Ekiel, I., Morgan, M.A., Tahara, S.M., and Shatkin, A.J. 1981. Methyl esterification of m7G5′p reversibly blocks its activity as an analog of eukaryotic mRNA 5′-caps. J. Mol. Biol. 153: 451–458. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Ekiel, I., Tahara, S.M., Seliger, L.S., and Shatkin, A.J. 1985. Chemical synthesis and characterization of 7-methylguanosine cap analogues. Biochemistry 24: 1701–1707. [Google Scholar]
- Darzynkiewicz, E., Ekiel, I., Lassota, P., and Tahara, S.M. 1987. Inhibition of eukaryotic translation by analogues of messenger RNA 5′-cap: Chemical and biological consequences of 5′-phosphate modifications of 7-methylguanosine 5′-monophosphate. Biochemistry 26: 4372–4380. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Stepinski, J., Ekiel, I., Goyer, C., Sonenberg, N., Temeriusz, A., Jin, Y., Sijuwade, T., Haber, D., and Tahara, S.M. 1989. Inhibition of eukaryotic translation by nucleoside 5′-mono-phosphate analogues of mRNA 5′-cap: changes in N7 substituent affect analogue activity. Biochemistry 28: 4771–4778. [DOI] [PubMed] [Google Scholar]
- De Clercq, E. 1993. Antiviral agents: Characteristic activity spectrum depending on the molecular target with which they interact. Adv. Virus Res. 42: 1–55. [DOI] [PubMed] [Google Scholar]
- Dyer, B.W., Ferrer, F.A., Klinedinst, D.K., and Rodriguez, R. 2000. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 282: 158–161. [DOI] [PubMed] [Google Scholar]
- Edery, I., Altmann, M., and Sonenberg, N. 1988. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene 74: 517–525. [DOI] [PubMed] [Google Scholar]
- Edery, I., Chu, L.L., Sonenberg, N., and Pelletier, J. 1995. An efficient strategy to isolate full-length cDNAs based on an mRNA cap retention procedure (CAPture). Mol. Cell. Biol. 15: 3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi, Y., Morgan, M.A., and Shatkin, A.J. 1979. Synthesis and translation of mRNA containing 5′-terminal 7-ethylguanosine cap. J. Biol. Chem. 254: 6732–6738. [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 15: 807–826. [DOI] [PubMed] [Google Scholar]
- Goswami, B.B., Borek, E., Sharma, O.K., Fujitaki, J., and Smith, R.A. 1979. The broad spectrum antiviral agent ribavirin inhibits capping of mRNA. Biochem. Biophys. Res. Commun. 89: 830–836. [DOI] [PubMed] [Google Scholar]
- Grifo, J.A., Tahara, S.M., Morgan, M.A., Shatkin, A.J., and Merrick, W.C. 1983. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 258: 5804–5810. [PubMed] [Google Scholar]
- Hickey, E.D., Weber, L.A., Baglioni, C., Kim, C.H., and Sarma, R.H. 1977. A relation between inhibition of protein synthesis and conformation of 5′-phosphorylated 7-methylguanosine derivatives. J. Mol. Biol. 109: 173–183. [DOI] [PubMed] [Google Scholar]
- Ishida, T., Doi, M., Ueda, H., Inoue, M., and Sheldrick, G.M. 1988. Specific ring stacking interaction on the tryptophan-7-methylguanine system: Comparative crystallographic studies of indole derivatives-7-methylguanine base, nucleoside, nucleotide complexes. J. Am. Chem. Soc. 110: 2286–2294. [Google Scholar]
- Kentsis, A., Topisirovic, I., Culjkovic, B., Shao, L., and Borden, K.L. 2004. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc. Natl. Acad. Sci. 101: 18105–18110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas, A., Montine, K.S., and Sonenberg, N. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345: 544–547. [DOI] [PubMed] [Google Scholar]
- Maag, D., Castro, C., Hong, Z., and Cameron, C.E. 2001. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 276: 46094–46098. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.C., Sonenberg, N., and Burley, S.K. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961. [DOI] [PubMed] [Google Scholar]
- Matsuo, H., Li, H., McGuire, A.M., Fletcher, C.M., Gingras, A.C., Sonenberg, N., and Wagner, G. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4: 717–724. [DOI] [PubMed] [Google Scholar]
- Miller, J.P., Kigwana, L.J., Streeter, D.G., Robins, R.K., Simon, L.N., and Roboz, J. 1977. The relationship between the metabolism of ribavirin and its proposed mechanism of action. Ann. NY Acad. Sci. 284: 211–229. [DOI] [PubMed] [Google Scholar]
- Muller, W.E., Maidhof, A., Taschner, H., and Zahn, R.K. 1977. Virazole (1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide; a cytostatic agent. Biochem. Pharmacol. 26: 1071–1075. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka, A., Darzynkiewicz, E., and Stolarski, R. 2004. Thermodynamics of mRNA 5′ cap binding by eukaryotic translation initiation factor eIF4E. Biochemistry 43: 13305–13317. [DOI] [PubMed] [Google Scholar]
- Novac, O., Guenier, A.S., and Pelletier, J. 2004. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 32: 902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, T. and Connor, J.D. 1990. The metabolism of ribavirin in erythrocytes and nucleated cells. Int. J. Biochem. 22: 379–383. [DOI] [PubMed] [Google Scholar]
- Pelletier, J. and Sonenberg, N. 1985. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: Effect of mRNA 5′ secondary structure. Mol. Cell. Biol. 5: 3222–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, D., Montanaro, L., Ma, L., Xu, W., Londei, P., Cordon-Cardo, C., and Pandolfi, P.P. 2004. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 10: 484–486. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Sidwell, R.W., Huffman, J.H., Khare, G.P., Allen, L.B., Witkowski, J.T., and Robins, R.K. 1972. Broad-spectrum antiviral activity of virazole: 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177: 705–706. [DOI] [PubMed] [Google Scholar]
- Sidwell, R.W., Robins, R.K., and Hillyard, I.W. 1979. Ribavirin: An antiviral agent. Pharmacol. Ther. 6: 123–146. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N. 1981. ATP/Mg++-dependent cross-linking of cap binding proteins to the 5′ end of eukaryotic mRNA. Nucleic Acids Res. 9: 1643–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg, N. and Shatkin, A.J. 1977. Reovirus mRNA can be covalently crosslinked via the 5′ cap to proteins in initiation complexes. Proc. Natl. Acad. Sci. 74: 4288–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1978. Nonspecific effect of m7GMP on protein-RNA interactions. J. Biol. Chem. 253: 6630–6632. [PubMed] [Google Scholar]
- Sonenberg, N., Morgan, M.A., Merrick, W.C., and Shatkin, A.J. 1978. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl. Acad. Sci. 75: 4843–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter, D.G., Witkowski, J.T., Khare, G.P., Sidwell, R.W., Bauer, R.J., Robins, R.K., and Simon, L.N. 1973. Mechanism of action of 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide (Virazole), a new broad-spectrum antiviral agent. Proc. Natl. Acad. Sci. 70: 1174–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y.V. and Sonenberg, N. 2003. Cell-free synthesis of encephalomyocarditis virus. J. Virol. 77: 6551–6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2004. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 257: 155–170. [DOI] [PubMed] [Google Scholar]
- Tahara, S.M., Morgan, M.A., and Shatkin, A.J. 1983. Binding of inosine-substituted mRNA to reticulocyte ribosomes and eukaryotic initiation factors 4A and 4B requires ATP. J. Biol. Chem. 258: 11350–11353. [PubMed] [Google Scholar]
- Tam, R.C., Lau, J.Y., and Hong, Z. 2001. Mechanisms of action of ribavirin in antiviral therapies. Antivir. Chem. Chemother. 12: 261–272. [DOI] [PubMed] [Google Scholar]
- Wendel, H.-G., de Stanchina, E., Fridman, J.S., Malina, A., Ray, S., Kogan, S., Cordon-Cardo, C., Pelletier, J., and Lowe, S.W. 2004. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428: 332–337. [DOI] [PubMed] [Google Scholar]