Abstract
The 5′-leader sequence (called Ω) of tobacco mosaic virus (TMV) functions as a translational enhancer in plants. A poly(CAA) region within Ω is responsible for the translation enhancement and serves as a binding site for the heat shock protein, HSP101, which is required for the translational enhancement. Genetic analysis of the HSP101-mediated enhancement of translation from Ω-containing mRNA suggested that two eukaryotic initiation factors (eIFs), i.e. eIF4G and eIF3, were necessary. In this study, the functional interaction between Ω and other RNA elements known to participate in the recruitment of eIF4G, i.e. the 5′-cap and the poly(A) tail, was examined. Ω exhibited functional overlap with the 5′-cap and the poly(A) tail but not with the native TMV 3′-UTR which contains an independent translational enhancer. Consistent with the role of HSP101 in mediating the translational function of Ω, the enhancement afforded by Ω increased following a heat shock, which elevates expression of HSP101. The use of a fractionated translation lysate revealed that of the two eIF4F proteins present in plants, eIF4F was specifically required for the activity of Ω. The data suggest that Ω is functionally similar to a 5′-cap and a poly(A) tail in that it serves to recruit eIF4F in order to enhance translation from an mRNA.
INTRODUCTION
Efficient translation initiation from most cellular mRNAs requires the action of the 5′-cap structure (m7GpppN, where N represents any nucleotide) and the 3′-terminal poly(A) tail which promote binding of 40S ribosomal subunits to an mRNA (1). The 5′-cap structure serves as the binding site for the eukaryotic initiation factor (eIF) 4F which is composed of three subunits: eIF4E, eIF4A and eIF4G. eIF4E functions as the cap-binding subunit, eIF4A possesses RNA helicase activity required to remove secondary structure within the 5′-leader sequence that would otherwise inhibit scanning of the 40S ribosomal subunit, and eIF4G is a large subunit that interacts with eIF4E and eIF4A. eIF4G also recruits other proteins involved in stimulating the 40S ribosomal subunit binding to an mRNA such as eIF3 and the poly(A)-binding protein (PABP). The interaction between eIF4G and PABP is conserved in plants, yeast and animals and serves to stabilize the binding of eIF4F to the 5′-cap (2–4). In plants and animals, PABP also interacts with eIF4B, a factor that assists eIF4A and eIF4F activities (4–6). The 5′-cap and poly(A) tail, therefore, serve to recruit eIF4G through the proteins that bind each element, i.e. eIF4E and PABP, respectively.
Tobacco mosaic virus (TMV) is an RNA virus whose genome is a single-strand, positive sense RNA that serves as the mRNA for the 5′-proximal cistron encoding the replicase. The genomic mRNA is released from the virion particle through a co-translation disassembly process in which 40S ribosomal subunits of the host cell are recruited to the 5′-leader of the viral RNA to which coat protein only loosely binds (7–10). During translation elongation, ribosomes synthesize replicase protein from the 5′-cistron and simultaneously strip the coat protein from the viral RNA. Although the genomic RNA is capped, it is an unusual mRNA in that it does not terminate with a poly(A) tail but instead contains a 204 nt 3′-untranslated region (3′-UTR). Despite the lack of a poly(A) tail, the TMV genomic mRNA is efficiently translated, including the first round of translation which is completed within 2–3 min following entry of the virus into a host cell (11). Two translation enhancers probably responsible for the efficient translation of TMV genomic mRNA have been identified. One is present within the 3′-UTR, which, like a poly(A) tail, is dependent on the 5′-cap in order to enhance translation (12,13). That the 3′-end of the virion particle does not undergo disassembly until after replicase protein is synthesized (11) suggests that the 3′ translational enhancer is prevented from participating in the first round of translation. The second translational enhancer is located within the 68 nt 5′-leader sequence (called Ω) (14–16). Ω is required for the release of RNA from the coat protein by enhancing the translation of RNAs and thus enhancing disassembly of the virion particle (17). Ω also enhances the translation of free RNAs, i.e. not present in virion particles, and its activity requires no other viral sequence or viral protein (14–16). The presence of Ω as the 5′-leader facilitates ribosome recruitment even in the absence of the participation of the 3′-translational enhancer (12–14,16) enabling it to promote translation initiation of encapsidated RNA without assistance from any 3′-terminal regulatory element (17). The translational enhancer within Ω is recognized by the heat shock protein, HSP101, which is sufficient to mediate the translational enhancement associated with Ω (18). Genetic analysis has suggested that the translational activity of HSP101 requires eIF4G and eIF3 (18), two factors that promote the recruitment of 40S ribosomal subunits to an mRNA.
In this study, the functional interaction between Ω and those elements of an mRNA [i.e. a 5′-cap and a poly(A) tail] known to participate in the recruitment of eIF4G has been investigated. Functional overlap was observed between Ω and a cap or between Ω and the poly(A) tail but not between Ω and the TMV 3′-UTR translational enhancer. The translational enhancement afforded by Ω increased following a heat shock, which elevates expression of HSP101. Of the two eIF4F proteins, i.e. eIF4F and eIFiso4F, present in plants, the ability of Ω to enhance translation preferentially required eIF4F. The data suggest that the HSP101/Ω complex is functionally similar to the eIF4E/5′-cap and PABP/poly(A) tail complexes in that it serves to recruit eIF4F to an mRNA in order to enhance translation.
MATERIALS AND METHODS
Plasmid constructs and in vitro RNA synthesis
The pT7-luc and pT7-Ω-luc constructs, in which the firefly luciferase-coding region is under the control of the T7 promoter in a pBluescript-derived vector, have been described previously (16). The 68 nt Ω sequence was introduced into the HindIII and SalI sites within the polylinker and the luc reporter introduced into the SalI and BamHI sites. The poly(A)50 tract was introduced into the SmaI and EcoRI sites downstream of the luc reporter gene. Linearization of DNA with DraI, a site immediately downstream of the poly(A)50 tract, allowed synthesis of polyadenylated transcripts whereas linearization with BamHI allowed the synthesis of poly(A)– mRNA. DNA concentration was quantified spectrophotometrically following linearization and brought to 0.5 mg/ml. In vitro transcription was carried out as described previously (19) using 40 mM Tris–HCl pH 7.5, 6 mM MgCl2, 100 µg/ml BSA, 0.5 mM each of ATP, CTP, UTP, GTP, 10 mM DTT, 0.3 U/µl RNasin (Promega) and 0.5 U/µl T7 RNA polymerase. The constructs terminated in a poly(A)50 tail, the 204 nt TMV 3′-UTR or a control sequence. Capped RNAs were synthesized using 3 µg of template in the same reaction mix as described above except GTP was used at 160 µM and 1 mM of either GpppG or m7GpppG was included. Under these conditions >95% of the mRNA is capped.
mRNA delivery to plant protoplasts
Protoplasts were isolated from carrot (RCWC) cell suspension by digesting with 0.25% CELF cellulase, 1% cytolase, 0.05% pectolyase Y23, 0.5% BSA and 7 mM β-mercaptoethanol in protoplast isolation buffer (12 mM sodium acetate pH 5.8, 50 mM CaCl2, 0.25 M mannitol) for 90–120 min. Protoplasts were washed with protoplast isolation buffer followed by electroporation buffer (10 mM HEPES pH 7.2, 130 mM KCl, 10 mM NaCl, 4 mM CaCl2, 0.2 M mannitol) and resuspended in electroporation buffer to ∼106 cells/ml. Equal amounts of mRNAs (∼2.5 µg) were added to 400 µl of cell suspension immediately before electroporation (250 µF, 300 V, 0.2 mm electrode) using an IBI GeneZapper. The electroporated cells were incubated in protoplast growth media (MS salts pH 5.8, 30 g/l sucrose, 100 mg/l myo-inositol, 0.1 mg/l 2,4 dichlorophenoxyacetic acid, 1.3 mg/l niacin, 0.25 mg/l thiamine, 0.25 mg/l pyridoxine, 0.25 mg/l calcium penthotenate) supplemented with 20% of cultured medium (protoplast growth medium conditioned with carrot cells for 3 days) overnight prior to assaying for reporter gene activity. For each experiment, an mRNA was delivered to triplicate samples of protoplasts and each sample assayed in duplicate. The average value for the constructs of a typical experiment is reported.
Protein purification and in vitro translation assays
Wheat eIF4F, eIFiso4F (20), eIF4B (21), eIF4A (22) and recombinant eIFiso4G and eIFiso4E (23) were purified as described. The purification of eIF4G and eIF4E will be described elsewhere.
The preparation and characterization of the eIF4F/eIFiso4F or PABP-depleted lysates has been described previously (24,25). The eIF4F/eIFiso4F or PABP-depleted lysates were prepared by adding 200 µl wheat germ extract (Promega) to 300 µl of m7GTP-Sepharose (Pharmacia) or to 100 µl poly(A)–agarose (Sigma), respectively, and incubated with rotation at 4°C for 30 min. The lysate was collected by centrifugation (800 g for 1 min) through a spin column (Promega) and used immediately. The extent of depletion of eIF4G, eIF4E, eIFiso4G, eIFiso4E, eIF4A, eIF4B, eIF3, eEF2, PABP or HSP101 was determined by western analysis following resolution of the extract by SDS–PAGE. mRNA constructs were translated using complete or depleted wheat germ lysate as described by the manufacturer except all amino acids were unlabeled. The lysates were supplemented with recombinant initiation factors or factors purified from wheat germ extract as indicated. The reactions were incubated for 3 h and 2 µl aliquots assayed for luciferase activity.
Luciferase assay
Carrot protoplast extract or wheat germ lysate in luciferase assay buffer (25 mM Tricine pH 8, 5 mM MgCl2, 0.1 mM EDTA supplemented with 33.3 mM DTT, 270 µM coenzyme A, 500 µM ATP) was assayed for luciferase activity following injection of 0.5 mM luciferin using a Monolight 2010 Luminometer (Analytical Luminescence Laboratory). Each mRNA construct was translated in triplicate and the average value for each construct is reported.
RESULTS
Ω exhibits functional overlap with the 5′-cap and the poly(A) tail
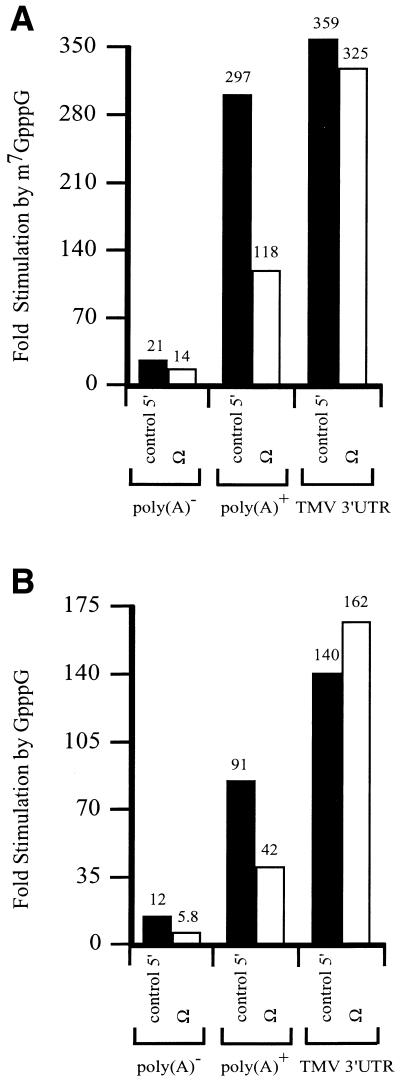
The eIF4E/5′-cap and PABP/poly(A) tail complexes function to recruit eIF4G to an mRNA. Ω serves as the leader sequence of a viral mRNA that is naturally capped but lacks a poly(A) tail. If the translational enhancement associated with Ω involves recruitment of eIF4G, the presence of Ω in an mRNA would be predicted to reduce the degree to which a 5′-cap or poly(A) tail promotes translation. To test this hypothesis, luciferase (luc) mRNA with or without Ω as the 5′-leader was synthesized in vitro with or without a 5′-cap or poly(A) tail. mRNAs terminating in the TMV 3′-UTR were also synthesized. The degree to which each mRNA was translated was examined in carrot protoplasts following RNA delivery by electroporation. Addition of a cap, Ω, poly(A) tail, or the TMV 3′-UTR to luc mRNA increased expression individually or when present in combination (Table 1). As observed previously (12,13,26), a cap and poly(A) tail or a cap and the TMV 3′-UTR functioned synergistically to increased expression. Addition of a methylated cap increased expression from poly(A)– luc mRNA by 21-fold, from poly(A)+ luc mRNA by 297-fold, and from luc mRNA terminating in the TMV 3′-UTR by 359-fold (Fig. 1A). However, the addition of a cap increased expression from poly(A)– and poly(A)+ Ω-luc mRNAs by only 14- and 118-fold, representing a significant decrease in cap-stimulation relative to the corresponding constructs with the control 5′-leader. Similar observations were made when a non-methylated cap was used, although in the absence of methylation, the degree of cap-mediated stimulation for all mRNAs was lower (Fig. 1B). The degree to which the presence of Ω affected the function of a poly(A) tail could also be determined: the addition of a poly(A) tail increased expression from methylated capped luc mRNA with a control leader by 21-fold, but only 11-fold when Ω was present.
Table 1. Functional interaction between the cap, Ω, TMV 3′-UTR and a poly(A) tail during translation.
| mRNA | Luciferase expression (light units/min/mg/protein) | ||
|---|---|---|---|
| Uncapped | GpppG | m7GpppG | |
| luc | 2940 | 35 400 | 63 000 |
| luc-TMV 3′ | 5210 | 727 000 | 1 870 000 |
| luc-A50 | 4480 | 407 000 | 1 330 000 |
| Ω-luc | 90 600 | 524 000 | 1 270 000 |
| Ω-luc-TMV 3′ | 201 000 | 32 600 000 | 65 300 000 |
| Ω-luc-A50 | 122 000 | 5 170 000 | 14 500 000 |
Capped or uncapped luc or Ω-luc mRNAs that were synthesized as poly(A)–, or terminating in a poly(A)50 tail, or the TMV 3′-UTR were delivered to carrot protoplasts. Expression from each mRNA construct was determined by measuring luciferase activity.
Figure 1.
The presence of Ω reduces the function of the 5′-cap. luc mRNA with either a control 5′-leader or Ω was synthesized in vitro as poly(A)–, terminating in a poly(A)50 tail, or terminating in the 204 nt TMV 3′-UTR. The mRNAs were synthesized without a cap, with m7GpppG (A) or with GpppG (B) and delivered to carrot protoplasts by electroporation. The degree to which the m7GpppG or the GpppG structures stimulated expression from the luc mRNAs with either a control 5′-leader or Ω relative to the corresponding uncapped constructs was calculated from the data in Table 1.
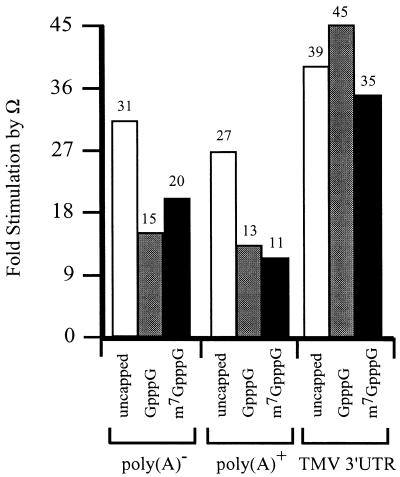
These observations suggest that the presence of Ω reduces the function of the cap and poly(A) tail. The function of Ω was similarly affected by a cap or a poly(A) tail. In the absence of a cap, Ω stimulated expression from poly(A)– and poly(A)+ luc mRNAs to a similar extent (Fig. 2). However, the degree to which Ω-stimulated expression was reduced when the mRNAs were capped with GpppG or m7GpppG. In contrast, the degree to which Ω stimulated expression from luc mRNA terminating in the TMV 3′-UTR was similar for capped and uncapped mRNA. These observations suggest that the function of Ω is reduced for luc mRNA terminating in a poly(A) tail but not with the TMV 3′-UTR. They also suggest that the negative effect of a cap on Ω function is overcome when the mRNA terminates in the TMV 3′-UTR.
Figure 2.
The presence of a 5′-cap reduces the function of Ω. luc mRNA with either a control 5′-leader or Ω was synthesized in vitro as poly(A)–, terminating in a poly(A)50 tail, or terminating in the 204 nt TMV 3′-UTR. The mRNAs were synthesized without a cap, with GpppG, or with m7GpppG and delivered to carrot protoplasts by electroporation. The degree to which Ω stimulated expression from the luc mRNAs without a cap or were capped with either GpppG or the m7GpppG relative to the corresponding constructs containing a control 5′-leader was calculated from the data in Table 1.
To examine whether Ω affects the ability of the cap to stimulate the rate of translation, the translational efficiency and the functional stability of capped and uncapped luc-A50 and Ω-luc-A50 mRNAs could be separately quantified by following the kinetics of their translation after their delivery to carrot cells. The rate of luciferase protein production was used as a measure of translational efficiency and the length of time over which luciferase protein continued to accumulate was used to calculate message stability. Luciferase protein has a half-life of 20 h in plant cells (26). Following delivery of each mRNA construct, aliquots of cells were removed at time intervals and luciferase assays performed. The kinetics of luc mRNA translation were determined by following the appearance of protein as measured by enzyme activity plotted as a function of time (Fig. 3). Translation of the introduced mRNA begins immediately following mRNA delivery, as there is detectable luciferase enzyme activity within 2 min following electroporation. Once the mRNA is recruited onto ribosomes, translation proceeds at a rate (i.e. the slope of each curve) that is dictated by its translational efficiency and for a period of time that is determined by the stability of the mRNA. The eventual degradation of the mRNA results in a decreased rate of protein accumulation. Following degradation of the mRNA, further accumulation of luciferase protein ceases, represented by the plateau of each curve at the later time points in Figure 3. Between the loading of the mRNA onto the polysomes and its eventual degradation, there is a phase of steady-state translation in which the rate of luciferase production (i.e. the maximum slope) is both maximal and constant. This represents the translational efficiency of each mRNA, which is quantified separately from the stability of the mRNA. By comparing the rates for each luc mRNA construct, the effect of Ω on the ability of the cap to stimulate translational efficiency could be determined.
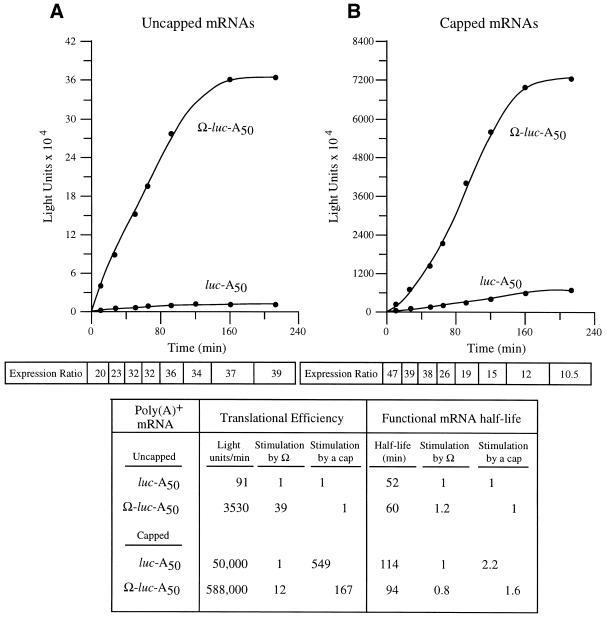
Figure 3.
The presence of Ω reduces the translational stimulatory function of the 5′-cap. Kinetic analysis of the translation of luc-A50 and Ω-luc-A50 mRNAs without (A) or with (B) a cap in carrot cells. Following their delivery using electroporation, aliquots of cells were removed at time intervals and assayed. The resulting luciferase activity was plotted as a function of time. The translational efficiency was determined from the slope of each line during the transient steady-state phase of translation and the values shown in the table. The functional mRNA half-life was determined as the amount of time required to complete a 50% decay in the capacity of the luc mRNA to synthesize luciferase. The fold increase in expression conferred by Ω relative to the control mRNA at each time point analyzed is indicated as the expression ratio below each graph. The degree to which the 5′-cap or Ω stimulated the rate of translation or functional stability is shown in the table.
Following the delivery of capped and uncapped luc-A50 and Ω-luc-A50 mRNAs, aliquots of cells were taken at time intervals and assayed for luciferase activity. The addition of a cap to luc-A50 mRNA stimulated the rate of translation 549-fold but stimulated the rate of translation from Ω-luc-A50 mRNA by only 167-fold (Fig. 3). This difference is due specifically to differences in the translational efficiency of the two mRNAs and does not include substantial differences in mRNA stability (see below), indicating that the presence of Ω reduces the ability of the cap to function to promote translation initiation. When the mRNAs were uncapped, the presence of Ω stimulated the steady-state rate of translation of uncapped mRNA 39-fold (Fig. 3). In contrast, Ω stimulated the rate of translation of capped mRNA 12-fold, data suggesting that the presence of a cap reduced Ω function just as the presence of Ω reduced the function of the cap. Interestingly, examination of the degree of stimulation conferred by Ω during the first rounds of translation revealed that Ω stimulated translation to a greater extent for capped mRNA than uncapped mRNA but that this advantage was quickly loss with subsequent translation (see expression ratio, Fig. 3B and also see Fig. 4A and B). The data suggest that the functional overlap between the cap and Ω occurs following the first rounds of translation of an mRNA.
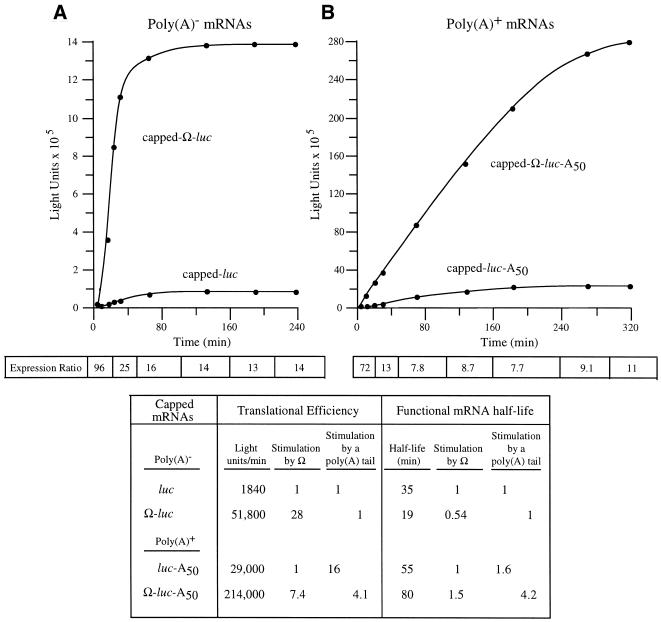
Figure 4.
The presence of a poly(A) tail reduces the translational stimulatory function of Ω. Kinetic analysis of the translation of capped luc and Ω-luc mRNAs without (A) or with (B) a poly(A) tail in carrot cells. The resulting luciferase activity was plotted as a function of time. The translational efficiency was determined from the slope of each line during the transient steady-state phase of translation and the values shown in the table. The functional mRNA half-life was determined as the amount of time required to complete a 50% decay in the capacity of the luc mRNA to synthesize luciferase. The fold increase in expression conferred by Ω relative to the control mRNA at each time point analyzed is indicated as the expression ratio below each graph. The degree to which Ω stimulated the rate of translation or functional stability for poly(A)– or poly(A)+ mRNA is shown in the table.
The functional stability of each construct was also measured. Those forms of an mRNA that are more stable will be translationally active longer, represented in the kinetic analysis by a longer period of time over which the protein will continue to accumulate. From visual inspection of the data in Figure 3, it is clear that the presence of Ω does not substantially alter the length of time over which an mRNA is translationally active. The stability of each mRNA could be quantified by measuring the functional half-life, which is a measure of the integrity of the message as determined by the length of time over which it is translationally active. As the functional half-life measures the stability of only that mRNA which is undergoing active translation, it more accurately describes the stability of message that is polysome-associated than does physical half-life measurements. The functional half-life is defined as the amount of time needed to complete 50% decay in the capacity of an mRNA to synthesize protein (27,28). Although the addition of a 5′-cap does increase the functional half-life of the mRNA as reported previously (13), the presence of Ω did not substantially or consistently alter the half-life of uncapped and capped mRNAs. The data suggest that Ω and a cap structure are partially functionally redundant in promoting translation.
In order to examine the functional interaction between Ω and a poly(A) tail, capped luc and Ω-luc mRNAs were synthesized as poly(A)– and poly(A)+ mRNAs and their translation followed in protoplasts. For capped, poly(A)– mRNAs, the presence of Ω stimulated the rate of translation 28-fold whereas Ω stimulated the rate of translation of poly(A)+ mRNA by only 7.4-fold (Fig. 4). In contrast to the effect of a cap on Ω function during the first rounds of translation observed in Figure 3, the reduction in Ω function in poly(A)+ mRNA was evident from the earliest time points and continued throughout the translation of the mRNA (Fig. 4). The addition of a poly(A) tail to capped luc mRNA increased the rate of translation by 16-fold but stimulated that of Ω-luc mRNA by only 4.1-fold. The addition of a poly(A) tail increased the functional mRNA half-life in good agreement with previous reports (13) and the presence of Ω reduced the functional half-life of poly(A)– mRNA but increased the functional half-life of poly(A)+ mRNA. The data demonstrate that the ability of Ω to promote translation is substantially reduced in a capped mRNA that is polyadenylated and suggest that Ω and a poly(A) tail are also partially functionally redundant in promoting translation.
Ω and a 5′-cap enhance first-round translation from an mRNA
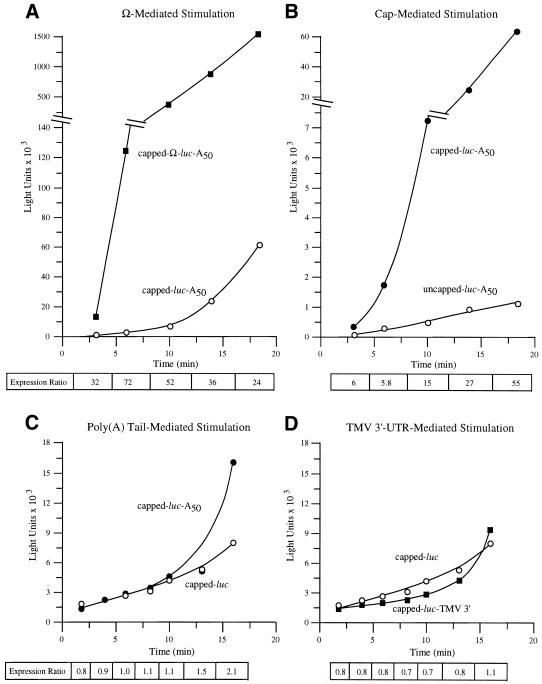
As the binding site for eIF4F, the cap structure promotes ribosome recruitment and would be expected to do so for even the first round of translation initiation. To examine whether Ω is similar to a 5′-cap in this respect, capped Ω-luc-A50 and luc-A50 mRNAs were delivered to protoplasts and their translation followed during the first rounds of translation. With a coding region of 1653 nt, the translation of luc mRNA is estimated to take 2 min, given an elongation rate of 5 codons/s (29). Three minutes following its delivery, the presence of Ω increased expression from luc-A50 mRNA 32-fold, which increased to 72-fold by 6 min and declined during subsequent translation (Fig. 5A). The translation of capped and uncapped luc-A50 mRNAs was also measured following their delivery. Expression from capped luc-A50 mRNA was 6-fold higher 3 min following RNA delivery than from uncapped luc-A50 mRNA, which increased progressively during subsequent translation (Fig. 5B). Consequently, Ω and a 5′-cap each increase translation from an mRNA even in the first rounds of translation. However, they differ in that a cap increases in its stimulatory function as an mRNA undergoes further rounds of translation whereas the stimulatory effect of Ω decreases over the same period. In contrast, the presence of a poly(A) tail (Fig. 5C) or the TMV 3′-UTR (Fig. 5D) did not confer a translational advantage to capped luc mRNA for 10–15 min of translation. Following this initial period, the rate of translation of an mRNA terminating in a poly(A) tail or the TMV 3′-UTR increased substantially whereas the rate of translation of the control, poly(A)– mRNA did not. These observations suggest that the poly(A) tail or the TMV 3′-UTR, unlike the 5′-cap and Ω, do not participate in the first rounds of translation following RNA delivery.
Figure 5.
Ω and a 5′-cap stimulate first-round translation. Translation from capped and uncapped Ω-luc-A50 (A) or luc-A50 (B) mRNAs was examined immediately following their delivery to carrot cells. The translation from capped, poly(A)– or poly(A)+ luc mRNAs (C) or from luc mRNA terminating in the TMV 3′-UTR (D) was also determined. The resulting luciferase activity was plotted as a function of time. The fold increase in expression conferred by Ω (A), 5′-cap (B), poly(A) tail (C) or TMV 3′-UTR (D) relative to the control mRNA at each time point analyzed is indicated as the expression ratio below each graph.
Ω function increases following a heat shock
Heat shock results in the loss of cap-dependent translation (30), a loss of eIF4G from the cap-binding fraction, and a loss in the functional and physical interaction between PABP and eIF4F (5). Ω serves as the binding site for the heat shock protein, HSP101, which is required to mediate the translational enhancement associated with viral leader sequence (18). eIF4G was implicated as necessary for the HSP101-mediated translational regulatory function of Ω (18). Although HSP101 is expressed at a low level under non-heat shock conditions, its expression increases following exposure to the stress (31,32). Because of the role of HSP101 in the Ω-mediated enhancement of translation, it might be predicted that expression from an Ω-containing mRNA would increase following a heat stress, which increases expression of HSP101. To examine this possibility, capped Ω-luc-A50 and luc-A50 mRNAs were delivered to protoplasts which were then subjected to short heat treatments that varied in severity and which had been used previously to illustrate the loss of cap function and loss in the functional interaction between the cap and poly(A) tail (30). The level of expression from each mRNA was measured following a 15 h incubation at room temperature. The presence of Ω stimulated expression from capped, polyadenylated mRNA by 4.5-fold in non-stressed cells (Table 2). Following even short exposures to moderate heat stress, an increase in Ω function was observed and the degree to which Ω stimulated expression increased with the severity of the stress. Under mild to moderate stress conditions, this increase resulted from a preferential increase in the absolute level of expression from capped Ω-luc-A50 mRNA relative to capped luc-A50 mRNA (Table 2). Following a longer exposure to severe temperatures (e.g. 40–60 min at 44°C), expression from the control mRNA was repressed (Table 2) which correlated with the previously observed loss of eIF4G from eIF4F, loss in cap function and loss in the functional interaction between a cap and poly(A) tail under similar conditions (5,30). Expression from Ω-luc-A50 mRNA was not repressed under these same conditions, which resulted in up to a 42-fold increase translational enhancement conferred by Ω. The observation that an increase in HSP101 expression and Ω function following a heat stress is consistent with the notion that HSP101 mediates Ω function and that recruitment of eIF4G to an mRNA becomes increasingly important as the function of the cap is inhibited.
Table 2. Enhancement conferred by the TMV 5′-leader as a function of the severity of a heat shock.
| Temperature |
Luciferase expression (light units/min/mg/protein) |
Fold stimulation by Ω |
|
|---|---|---|---|
| luc-A50 | Ω-luc-A50 | ||
| 24°C | |||
| 60 min | 119 000 | 538 000 | 4.5 |
| 42°C | |||
| 5 min | 118 000 | 853 000 | 7.2 |
| 10 min | 146 000 | 1 600 000 | 11 |
| 20 min | 181 000 | 1 860 000 | 10 |
| 30 min | 241 000 | 3 890 000 | 16 |
| 40 min | 273 000 | 3 110 000 | 11 |
| 50 min | 284 000 | 4 360 000 | 15 |
| 60 min | 251 000 | 5 130 000 | 20 |
| 43°C | |||
| 5 min | 145 000 | 981 000 | 6.8 |
| 10 min | 190 000 | 1 590 000 | 8.4 |
| 20 min | 247 000 | 2 110 000 | 8.6 |
| 30 min | 225 000 | 3 100 000 | 14 |
| 40 min | 236 000 | 4 100 000 | 17 |
| 50 min | 194 000 | 4 310 000 | 22 |
| 60 min | 146 000 | 4 050 000 | 28 |
| 44°C | |||
| 5 min | 177 000 | 868 000 | 4.9 |
| 10 min | 213 000 | 2 160 000 | 10 |
| 20 min | 269 000 | 4 640 000 | 17 |
| 30 min | 188 000 | 4 010 000 | 21 |
| 40 min | 71 300 | 2 330 000 | 33 |
| 50 min | 30 100 | 821 000 | 27 |
| 60 min | 19 000 | 800 000 | 42 |
Capped luc-A50 or Ω-luc-A50 mRNAs were synthesized and delivered to carrot protoplasts. Expression from each mRNA construct was determined by measuring cell extract in a luminometer.
Ω specifically requires eIF4G to enhance translation
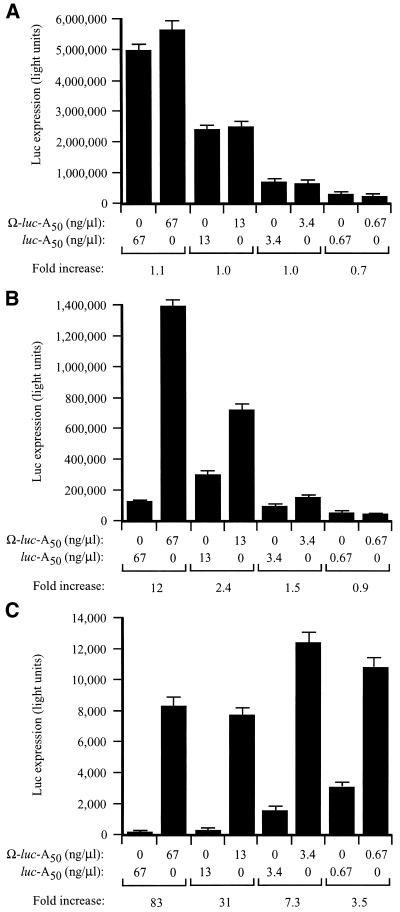
To examine whether eIF4G is necessary for the function of Ω, the functional requirements of Ω in stimulating translation were investigated using fractionated and unfractionated wheat germ lysates in which capped-luc-A50 and Ω-luc-A50 mRNAs were translated at different concentrations. Ω conferred no translational advantage at any concentration in unfractionated lysate (Fig. 6A). This is consistent with the previous observation that a 5′-cap does not confer a substantial increase in translation in unfractionated lysate and supports the observation that wheat germ lysate is highly message dependent due to a low concentration of endogenous transcript and the high level of unengaged translational machinery. As a consequence, those features of an mRNA that increase its ability to recruit translational machinery, such as a cap or Ω, would not be expected to confer a translational advantage under conditions (e.g. unfractionated lysate) in which the translational machinery is readily available. Accordingly, Ω-stimulated translation should be possible following the removal of the excess capacity of those factors most important for Ω function.
Figure 6.
Ω confers a translational advantage in vitro when the level of eIF4F, eIFiso4F or PABP is limiting. Unfractionated (A), eIF4F/eIFiso4F-depleted (B) or PABP-depleted (C) wheat germ lysate was programmed with capped Ω-luc-A50 or luc-A50 mRNAs at the concentration indicated below the histograms. The degree to which each mRNA was translated was determined by luciferase assays. Luciferase activity is indicated as the average (from 2 µl of lysate) of three translation reactions with the standard deviation for each construct shown. The degree to which the presence of Ω increased translation relative to the control (i.e. fold increase) is indicated below each pair of mRNAs for each concentration tested.
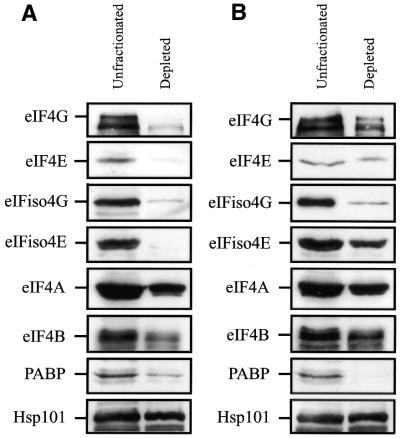
A fractionated eIF4G-dependent, eIFiso4G-dependent lysate was described recently (31,32). Because eIF4F (in which eIF4G and eIF4E are subunits) and eIFiso4F (in which eIFiso4G and eIFiso4E are subunits) bind m7GTP, the level of both factors in wheat germ lysate could be readily reduced through their binding to m7GTP-Sepharose (24,25). Likewise, the level of PABP could be significantly reduced by its binding to poly(A)–agarose. Western analysis confirmed that the level of eIF4E and eIFiso4E was substantially reduced as was eIF4G and eIFiso4G in the lysate depleted for eIF4F and eIFiso4F (Fig. 7). The level of eIF4A and eIF4B, factors known to associate with eIF4F and eIFiso4F, was reduced slightly as was PABP, which also physically interacts with eIF4G and eIFiso4G (2,3). No reduction in the level of HSP101 was observed. Following binding to poly(A)– agarose, the level of PABP was substantially reduced (Fig. 6) and the levels of eIF4G and eIFiso4G were also partially reduced. Little to no reduction was observed in the levels of eIF4E, eIFiso4E, eIF4A, eIF4B or HSP101.
Figure 7.
Depletion of eIF4F, eIFiso4F and PABP from wheat germ lysate. Wheat germ lysate was incubated with (A) m7GTP-Sepharose or (B) poly(A)-Sepharose for 30 min. Western analysis was performed to determine the level of eIF4G, eIF4E, eIFiso4G, eIFiso4E, eIF4A, eIF4B, PABP and Hsp101 in depleted lysate relative to unfractionated lysate.
Following depletion, the fractionated lysates were used for the analysis of Ω function by examining the extent of translation from capped-luc-A50 and Ω-luc-A50 mRNAs translated at different concentrations. In eIF4F/eIFiso4F-depleted lysate, Ω-luc-A50 mRNA was translated 12-fold higher than the control mRNA at the highest concentration tested and decreased with reduced RNA concentrations (Fig. 6B). The depletion of PABP and the reduction in eIF4G and eIFiso4G results in a greater level of cap-dependent translation than that observed in the eIF4G/eIFiso4G-dependent lysate supporting the notion that high levels of unengaged PABP can promote translation non-specifically (24). In this lysate, Ω conferred a translational advantage at all RNA concentrations but increased with the concentration of RNA (Fig. 6C).
Because Ω stimulated translation to the greatest extent in the PABP-depleted and eIF4G/eIFiso4G-reduced lysate, we examined whether the function of Ω requires eIF4F or eIFiso4F. The observation that the translation of an mRNA in PABP-depleted, eIF4F/eIFiso4F-reduced lysate improves substantially if it contains Ω suggests that this sequence promotes the recruitment of a factor that is required for translation initiation but whose availability is limited. Consequently, restoring the factor in question to the lysate would be expected to increase expression from the mRNA lacking Ω to a greater extent than that from the mRNA-containing Ω, thereby reducing the translational advantage conferred by Ω to a level similar to that observed in unfractionated lysate as seen in Figure 6A. Such a strategy was employed recently to investigate the function of the internal ribosome entry site (IRES) present in the 5′-leader of tobacco etch virus genomic mRNA that confers cap-independent translation (25).
To assay the requirement of eIF4F or eIFiso4F for Ω function, capped-luc-A50 and Ω-luc-A50 mRNAs were translated in PABP-depleted, eIF4F/eIFiso4F-reduced lysate supplemented with either eIF4F or eIFiso4F and their effect on translation and the translational advantage conferred by Ω determined. A similar approach was used to demonstrate the requirement for eIF4F and eIFiso4F to mediate the function of the 5′-cap (24). The presence of a 5′-cap stimulated translation to a greater extent in the depleted lysate demonstrating that the importance of the 5′-cap increased when the level of eIF4F and eIFiso4F had been reduced. Supplementation of the depleted lysate with either eIF4F or eIFiso4F increased expression from the control construct disproportionately, thereby reducing the translational advantage afforded by the 5′-cap (24).
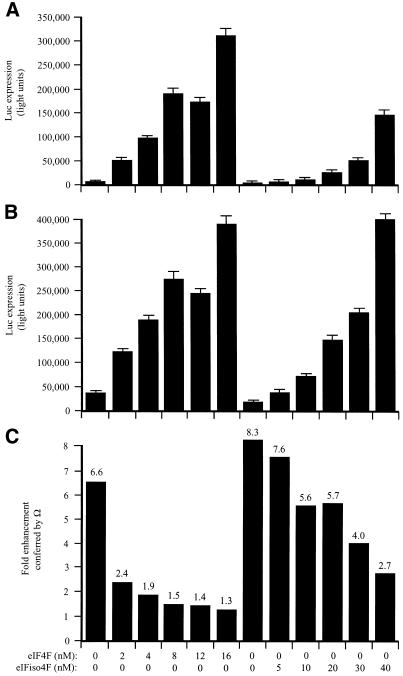
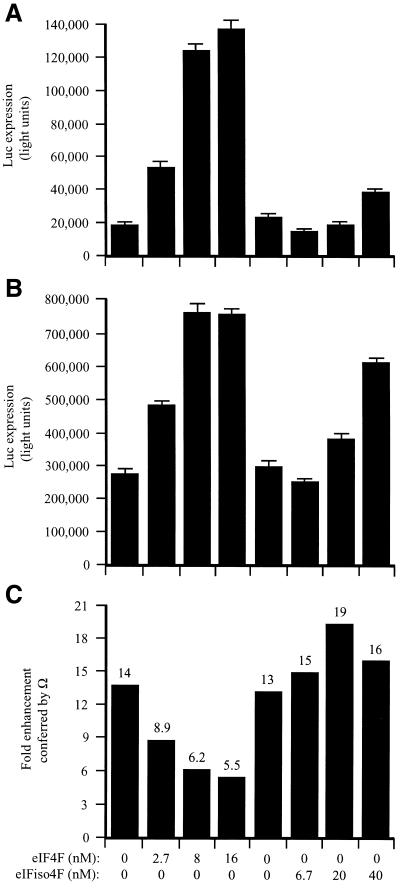
In the fractionated lysate, Ω increased translation 6.6-fold (Fig. 8C). The addition of eIF4F increased expression from the control (Fig. 8A) and Ω-containing (Fig. 8B) constructs, demonstrating that the fractionated lysate was limiting for these factors. Expression from the control construct increased disproportionately as the limiting amount of eIF4F was relieved (Fig. 8C). Consequently, the translational advantage conferred by Ω was reduced in a dose-dependent manner (Fig. 8C). Supplementation with eIFiso4F also reduced the degree to which Ω increased expression but required substantially greater molar amounts of the factor to do so and was not as effective as supplementation with eIF4F even at the highest degree of supplementation tested (i.e. 20-fold greater than eIF4F). The data suggest that Ω functions by recruiting eIF4F preferentially but not exclusively to a capped mRNA when the factor is present in limiting amounts but that the translational advantage conferred by Ω is lost when the concentration of eIF4F is no longer limiting.
Figure 8.
eIF4F but not eIFiso4F mediates Ω function in capped mRNAs. PABP-depleted, eIF4F/eIFiso4F-reduced wheat germ lysate was programmed with capped luc-A50 mRNA (A) or capped Ω-luc-A50 (B) and was supplemented with the indicated amounts of eIF4F or eIFiso4F (each including eIF4A). mRNA constructs were translated in triplicate and the average value and standard deviation for each construct is reported. Luciferase expression is indicated as light units from 2 µl of translation lysate. The degree to which Ω stimulated translation relative to the control construct is indicated in (C).
The results in Figures 1–3 suggested that Ω and the 5′-cap each function to recruit eIF4F. In order to investigate whether Ω alone exhibits a preference for eIF4F or eIFiso4F, uncapped mRNAs were tested so that the 5′-cap would not be present to complicate the analysis. Uncapped luc-A50 and Ω-luc-A50 constructs were translated in the PABP-depleted, eIF4F/eIFiso4F-reduced lysate supplemented with either eIF4F or eIFiso4F. Ω increased translation from uncapped mRNAs (Fig. 9C) as it had for capped mRNAs. Because the translational advantage conferred by Ω in the depleted lysate would be to promote recruitment of a factor that is required for translation initiation but whose availability is limited, restoring the factor in question would be expected to preferentially increase translation from the control mRNA and thus reduce the observed translational advantage conferred by Ω. This prediction was borne out by the observation that, as for capped mRNAs, the addition of eIF4F increased expression from the control (Fig. 9A) and Ω-containing (Fig. 9B) constructs, however, expression from the control construct increased disproportionately as the concentration of eIF4F was increased. Therefore, the translational advantage conferred by Ω was reduced in a dose-dependent manner (Fig. 9C). In contrast, although supplementation with eIFiso4F increased expression from both mRNAs, it did so equally for both (Fig. 9A and B) and therefore was not specifically required for Ω function (Fig. 9C). The data suggest that in the absence of a cap, Ω exclusively recruits eIF4F in order to enhance translation.
Figure 9.
eIF4F but not eIFiso4F mediates Ω function in uncapped mRNAs. PABP-depleted, eIF4F/eIFiso4F-reduced wheat germ lysate was programmed with uncapped luc-A50 mRNA (A) or uncapped Ω-luc-A50 (B) and was supplemented with the indicated amounts of eIF4F or eIFiso4F (each including eIF4A). mRNA constructs were translated in triplicate and the average value and standard deviation for each construct is reported. Luciferase expression is indicated as light units from 2 µl of translation lysate. The degree to which Ω stimulated translation relative to the control construct is indicated in (C).
DISCUSSION
The mechanism by which the TMV 5′-leader, Ω, enhances translation in plants has been suggested to involve HSP101-mediated recruitment of eIF4G (18). In this study, we have shown that Ω functionally recruits eIF4G to an mRNA, is more effective following a heat shock, and overlaps functionally with a 5′-cap and poly(A) tail during translation. We observed that the presence of Ω reduced the translational stimulatory function of the 5′-cap and the poly(A) tail by 3–4-fold. Similarly, the presence of a 5′-cap reduced the function of Ω by 3-fold. In contrast, the presence of a poly(A) tail had little effect on Ω function in the absence of a 5′-cap, suggesting that the effect of the poly(A) tail on Ω function is mediated through the cap. Moreover, the effect on Ω function by the poly(A) tail was specific to this 3′ regulatory element as the translational enhancer within the TMV 3′-UTR did not have the same effect on Ω. These observations suggest that the function of Ω in promoting translation initiation overlaps with that of the cap and the poly(A) tail in a way that reduces their effectiveness in promoting translation initiation.
The 5′-cap and poly(A) tail have been shown to recruit eIF4G (as part of eIF4F) to an mRNA. The 5′-cap contributes to eIF4G recruitment by serving as the binding site for eIF4E which binds directly to eIF4G whereas the poly(A) tail promotes eIF4G recruitment by serving as the binding site for PABP which also contacts eIF4G directly (2,3). In the absence of the 5′-cap, a poly(A) tail does not stimulate translation significantly in plants and the 5′-cap only moderately increases the translation of an mRNA lacking a poly(A) tail or a functionally equivalent element such as the TMV 3′-UTR (12,13). Therefore, although the cap and poly(A) tail are each involved in the recruitment of eIF4G they work together synergistically. This synergy is consistent with the fact that eIF4E and PABP bind to eIF4G at distinct and separate regions. However, Ω does not interact synergistically with either the 5′-cap or poly(A) tail but instead overlaps with their function. In contrast, the TEV IRES functions synergistically with the poly(A) tail to increase translation in the absence of a 5′-cap (25,33,34). The TEV IRES is functionally equivalent to the 5′-cap and acts to recruit eIF4G (25). The TEV IRES is similar to Ω in that its presence reduces the function of the 5′-cap (25) but differs from Ω in that its function is enhanced by the presence of the poly(A) tail (33). The similarities and differences between Ω and the TEV IRES in their interactions with the cap and poly(A) tail reveal substantial differences in the functional interactions for these two viral translational enhancers.
Plants, as with other eukaryotes, contain two distinct eIF4G proteins (20). The two plant eIF4G proteins, referred to as eIF4G and eIFiso4G, differ in size (165 and 86 kDa, respectively). eIF4F or eIFiso4F support the translation of an unstructured mRNA but eIF4F supports internal initiation and translation from uncapped mRNAs to a greater extent than eIFiso4F (24). Moreover, eIF4F is responsible for the cap-independent translation conferred by the TEV IRES (25). These observations suggest that eIF4F and eIFiso4F have undergone functional specialization that allows them to discriminate between mRNAs. Ω preferentially recruited eIF4F to an mRNA over eIFiso4F whether the mRNA was capped or not. eIF4F is present in plants at just 11% the level of eIFiso4F (35). Because the low concentration of eIF4F is considered rate limiting, the ability of Ω to recruit this factor, which is at least 20–30-fold more active on a molar basis in stimulating translation than is eIFiso4F (24), may explain the basis of the translational enhancement associated with this leader.
Ω serves to recruit HSP101, which in turn requires eIF4G to mediate the translational enhancement (18). Consistent with the involvement of this heat shock protein, the function of Ω increased following exposure to heat shock, which elevates expression of HSP101. The increase in Ω function following a heat shock is notable in that thermal stress results in a loss in cap function and a loss in the functional and physical interaction between the 5′-cap and poly(A) tail (5,30). The loss in cap function following a heat shock correlates with the loss of eIF4G from the cap-binding complex (5). The ability of Ω to recruit HSP101 and function following a heat shock suggests that its ability to recruit eIF4F is not affected by a heat shock in the way the ability of the 5′-cap to recruit the factor is. This observation suggests that although Ω and the 5′-cap each function to recruit eIF4G, they may differ in how each recruits the factor.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the USDA (NRICGP 99-35301-7866 and 00-35301-9086).
REFERENCES
- 1.Sachs A.B. (2000) Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY., pp. 447–465.
- 2.Tarun S.Z. and Sachs,A.B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 3.Le H., Tanguay,R.L., Balasta,M.L., Wei,C.-C., Browning,K.S., Metz,A.M., Goss,D.J. and Gallie,D.R. (1997) Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem., 272, 16247–16255. [DOI] [PubMed] [Google Scholar]
- 4.Wei C.-C., Balasta,M.L., Ren,J. and Goss,D.J. (1998) Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry, 37, 1910–1916. [DOI] [PubMed] [Google Scholar]
- 5.Le H., Browning,K.S. and Gallie,D.R. (2000) The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F and eIF4B. J. Biol. Chem., 275, 17452–17462. [DOI] [PubMed] [Google Scholar]
- 6.Bushell M., Wood,W., Carpenter,G., Pain,V.M., Morley,S.J. and Clemens,M.J. (2001) Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J. Biol. Chem., 276, 23922–23928. [DOI] [PubMed] [Google Scholar]
- 7.Wilson T.M.A. (1984) Contranslational disassembly to tobacco mosaic virus in vitro. Virology, 137, 255–265. [DOI] [PubMed] [Google Scholar]
- 8.Wilson T.M.A. (1984) Contranslational disassembly increases the efficiency of expression of TMV RNA in wheat germ cell-free extracts. Virology, 138, 353–356. [DOI] [PubMed] [Google Scholar]
- 9.Shaw J.G., Plaskitt,K.A. and Wilson,T.M.A. (1986) Evidence that tobacco mosaic virus particles disassemble contranslationally in vivo. Virology, 148, 326–336. [DOI] [PubMed] [Google Scholar]
- 10.Mundry K.W., Watkins,P.A., Ashfield,T., Plaskitt,K.A., Eisele-Walter,S. and Wilson,T.M. (1991) Complete uncoating of the 5′ leader sequence of tobacco mosaic virus RNA occurs rapidly and is required to initiate cotranslational virus disassembly in vitro. J. Gen. Virol., 72, 769–777. [DOI] [PubMed] [Google Scholar]
- 11.Wu X., Xu,Z. and Shaw,J.G. (1994) Uncoating of tobacco mosaic virus RNA in protoplasts. Virology, 200, 256–262. [DOI] [PubMed] [Google Scholar]
- 12.Gallie D.R. and Walbot,V. (1990) RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev., 4, 1149–1157. [DOI] [PubMed] [Google Scholar]
- 13.Leathers V., Tanguay,R., Kobayashi,M. and Gallie,D.R. (1993) A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol. Cell. Biol., 13, 5331–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallie D.R., Sleat,D.E., Watts,J.W., Turner,P.C. and Wilson,T.M.A. (1987) The 5′-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res., 15, 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallie D.R., Sleat,D.E., Watts,J.W., Turner,P.C. and Wilson,T.M.A. (1988) Mutational analysis of the tobacco mosaic virus 5′-leader for altered ability to enhance translation. Nucleic Acids Res., 16, 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallie D.R. and Walbot,V. (1992) Identification of the motifs within the tobacco mosaic virus 5′ leader responsible for enhancing translation. Nucleic Acids Res., 20, 4631–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallie D.R., Sleat,D.E., Watts,J.W., Turner,P.C. and Wilson,T.M.A. (1987) In vivo uncoating and efficient transient expression of recombinant RNA packaged into pseudovirus particles. Science, 236, 1122–1124. [DOI] [PubMed] [Google Scholar]
- 18.Wells D.R., Tanguay,R.L., Le,H. and Gallie,D.R. (1998) HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Genes Dev., 12, 3236–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yisraeli J.K. and Melton,D.A. (1989) Synthesis of long, capped transcripts in vitro by SP6 and T7 RNA polymerases. Methods Enzymol., 180, 42–50. [DOI] [PubMed] [Google Scholar]
- 20.Browning K.S., Webster,C., Roberts,J.K. and Ravel,J.M. (1992) Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J. Biol. Chem., 267, 10096–10100. [PubMed] [Google Scholar]
- 21.Browning K.S., Maia,D.M., Lax,S.R. and Ravel,J.M. (1987) Identification of a new protein synthesis initiation factor from wheat germ. J. Biol. Chem., 262, 538–541. [PubMed] [Google Scholar]
- 22.Lax S.R., Lauer,S.J., Browning,K.S. and Ravel,J.M. (1986) Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol., 118, 109–128. [DOI] [PubMed] [Google Scholar]
- 23.van Heerden A. and Browning,K.S. (1994) Expression in Escherichia coli of the two subunits of the isozyme form of wheat germ protein synthesis initiation factor 4F. Purification of the subunits and formation of an enzymatically active complex. J. Biol. Chem., 269, 17454–17457. [PubMed] [Google Scholar]
- 24.Gallie D.R. and Browning,K.S. (2001) eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation and translation of structured mRNAs. J. Biol. Chem., 276, 36951–36960. [DOI] [PubMed] [Google Scholar]
- 25.Gallie D.R. (2001) Cap-independent translation conferred by the 5′-leader of tobacco etch virus is eIF4G-dependent. J. Virol., 75, 12141–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallie D.R. (1991) The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev., 5, 2108–2116. [DOI] [PubMed] [Google Scholar]
- 27.Kepes A. (1963) Kinetics of induced enzyme synthesis determination of the mean life of galactosidase-specific messenger RNA. Biochim. Biophys. Acta, 76, 293–309. [PubMed] [Google Scholar]
- 28.Pedersen S. and Reeh,S. (1978) Functional mRNA half lives in E.coli. Mol. Gen. Genet., 166, 329–336. [DOI] [PubMed] [Google Scholar]
- 29.Palmiter R.D. (1975) Quantitation of parameters that determine the rate of ovalbumin synthesis. Cell, 4, 189–197. [DOI] [PubMed] [Google Scholar]
- 30.Gallie D.R., Caldwell,C. and Pitto,L. (1995) Heat shock disrupts cap and poly(A) tail function during translation and increases mRNA stability of introduced reporter mRNA. Plant Physiol., 108, 1703–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling J., Wells,D.R., Tanguay,R.L., Dickey,L.F., Thompson,W.F. and Gallie,D.R. (2000) Heat shock protein HSP101 binds to the Fed-1 internal light regulator y element and mediates its high translational activity. Plant Cell, 12, 1213–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young T.E., Ling,J., Geisler-Lee,C.J., Tanguay,R.L. Caldwell,C. and Gallie,D.R. (2001) Developmental and thermal regulation of the maize heat shock protein, Hsp101. Plant Physiol., 127, 777–791. [PMC free article] [PubMed] [Google Scholar]
- 33.Gallie D.R., Tanguay,R.L. and Leathers,V. (1995) The tobacco etch viral 5′ leader and poly(A) tail are synergistic regulators of translation. Gene, 165, 233–238. [DOI] [PubMed] [Google Scholar]
- 34.Niepel M. and Gallie,D.R. (1999) Identification and characterization of the functional elements within the tobacco etch viral 5′-leader required for cap-independent translation. J. Virol., 73, 9080–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Browning K.S., Humphreys,J., Hobbs,W., Smith,G.B. and Ravel,J.M. (1990) Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J. Biol. Chem., 265, 17967–17973. [PubMed] [Google Scholar]