Abstract
Small nucleolar RNAs (snoRNAs) are an abundant group of noncoding RNAs mainly involved in the post-transcriptional modifications of rRNAs in eukaryotes. In this study, a large-scale genome-wide analysis of the two major families of snoRNA genes in the fruit fly Drosophila melanogaster has been performed using experimental and computational RNomics methods. Two hundred and twelve gene variants, encoding 56 box H/ACA and 63 box C/D snoRNAs, were identified, of which 57 novel snoRNAs have been reported for the first time. These snoRNAs were predicted to guide a total of 147 methylations and pseudouridylations on rRNAs and snRNAs, showing a more comprehensive pattern of rRNA modification in the fruit fly. With the exception of nine, all the snoRNAs identified to date in D. melanogaster are intron encoded. Remarkably, the genomic organization of the snoRNAs is characteristic of 8 dUhg genes and 17 intronic gene clusters, demonstrating that distinct organizations dominate the expression of the two families of snoRNAs in the fruit fly. Of the 267 introns in the host genes, more than half have been identified as host introns for coding of snoRNAs. In contrast to mammals, the variation in size of the host introns is mainly due to differences in the number of snoRNAs they contain. These results demonstrate the extensive utilization of introns for coding of snoRNAs in the host genes and shed light on further research of other noncoding RNA genes in the large introns of the Drosophila genome.
Keywords: snoRNA, ncRNA, intron, RNA modification, Drosophila melanogaster
INTRODUCTION
Small nucleolar RNAs (snoRNAs) represent an abundant group of noncoding RNAs (ncRNAs) mainly involved in rRNA biogenesis (Kiss 2002). With the exception of RNase MRP, all the snoRNAs fall into two major families, box C/D and box H/ACA snoRNAs, on the basis of common sequence motifs and structural features (Balakin et al. 1996). Box C/D snoRNAs share two conserved motifs, the 5′ end box C (RUGAUGA) and the 3′ end box D (CUGA), whereas the box H/ACA snoRNAs exhibit a common hairpin-hinge-hairpin-tail secondary structure with the H (ANANNA) motif in the hinge region and an ACA triplet 3 nt from the 3′ end of the molecule (Ganot et al. 1997). Several snoRNAs, such as U3, snR30, and RNase MRP, are required for specific cleavage of pre-rRNAs (Kiss 2002). However, the majority of box C/D snoRNAs function as guides for site-specific 2′-O-ribose methylation with most box H/ACA snoRNAs functioning as guides for pseudo-uridylation in the post-transcriptional processing of rRNAs (Smith and Steitz 1997; Bachellerie et al. 2000). Recent research has shown that some snoRNAs and scaRNAs participate in the modifications of snRNAs (Kiss 2002; Zhou et al. 2002; Tycowski et al. 1998, 2004) and even tRNAs by Archaea homologs (Clouet d’Orval et al. 2001). Moreover, an increasing number of orphan snoRNAs with unknown function have been identified from different eukaryotes, suggesting they play additional roles in cellular processes. The identification of snoRNA homologs in Archaea further demonstrates their ancient origin and preservation during the course of evolution (Bachellerie et al. 2002).
The Drosophila rRNAs possess numerous post-transcriptional modifications (Ofengand and Bakin 1997), which has made D. melanogaster a good model for studying the expression of snoRNA genes (Tycowski and Steitz 2001; Huang et al. 2004). A recent study of experimental Rnomics in the fruit fly identified 66 small ncRNAs, of which 35 species belonged to the two families of snoRNAs (Yuan et al. 2003). Computational analyses of conserved structural and functional motifs have also been applied to identify typical box H/ACA snoRNAs and box C/D snoRNA, resulting in a validation of 10 box H/ACA snoRNAs (Huang et al. 2004) and 26 box C/D snoRNAs from the Drosophila genome (Accardo et al. 2004; our unpubl. results). These studies, in addition to other reports (Tycowski and Steitz 2001; Zhou et al. 2002), have identified approximately one-third of the total snoRNAs from Drosophila as estimated proportionally to the modifications on rRNAs. Furthermore, some interesting features in the expression of the snoRNA genes have also been observed, such as the discovery of dUhg (Tycowski and Steitz 2001) and intronic box H/ACA snoRNA gene clusters in the fruit fly (Huang et al. 2004). These results encouraged us to perform a large-scale genome-wide analysis of the two families of snoRNAs in Drosophila using both experimental and computational Rnomics methods. In an attempt to identify all of the snoRNAs and give an overall view of their genomic organization, we have identified 212 gene variants encoding 119 snoRNAs from the Drosophila genome, including 57 novel snoRNAs. This analysis provides more comprehensive data on the pattern of rRNA modifications as well as a panorama of the snoRNA genomic organization in D. melanogaster. Moreover, a systematic study on the snoRNA host genes has revealed how extensively and efficiently intron regions have been used for ncRNA coding in the compact genome of the fruit fly.
RESULTS
Identification of 31 novel box C/D snoRNAs from Drosophila
A special cDNA library for box C/D snoRNAs was constructed with total D. melanogaster RNA. We focused on the cDNA fraction of 60–120 nt according to the size range of most box C/D snoRNAs. After screening to eliminate abundant fragments of rRNAs, 35 box C/D snoRNA variants were identified from the library. Most of the cDNA sequences were satisfactorily intact and exhibited typical features of box C/D snoRNA. Detailed examination of the functional elements of the cDNA sequences revealed 28 box C/D snoRNA species, including 12 novel snoRNAs (Table 1).
TABLE 1.
BOX C/D snoRNA genes in D. melanogaster
| Homology | |||||||||
| SnoRNA name | Iso | Len (nt) | Exp | Modification | Antisense element | Yeast | Plants | Vertebrates | Location |
| Me5.8S-G74* | 1 | 71 | N. blot | 5.8S-Gm74 | 15 nt (3′) | SnoR39bY | Z37 | CG13900 | |
| Me18S-A28a*(U27) | 3 | 72 | + | 18S-Am28 | 13 nt (5′) | SnR74(Z4) | U27 | U27 | DUHGI |
| Me18S-A425*(Dm425) | 1 | 84 | cDNA | 28S-A1385 | 10 nt (5′) | CG4863 | |||
| 18S-Am425 | 13 nt (3′) | SnR52 | SnR52Y | U83 | |||||
| Me18S-C419* | 1 | 70 | N. blot | 18S-Cm419 | 13 nt (3′) | U14 | U14 | U14 | CG17420 |
| Mel8S-A469* | 1 | 88 | N. blot | 18S-Am469 | 13 nt (3′) | SnoR17 | Z71 | CG9253 | |
| Me18S-A1061*(DmSnR54) | 1 | 76 | + | 18S-Am1061 | 14 nt (5′) | SnR54 | U59 | U59 | DUHG5 |
| Me18S-C1096* | 1 | 85 | cDNA | 18S-C1104 | 14 nt (5′) | CG4863 | |||
| 18S-Cm1096 | 11 nt (3′) | SnoR20 | |||||||
| Me18S-U1356a* | 3 | 79 | cDNA | 18S-Um1356 | 13 nt (3′) | SnR55 | U33 | U33 | DUHG8 |
| Me18S-G1358a* | 2 | 75 | N. blot | 18S-Gm1358 | 12 nt (5′) | SnR40 | SnoR21 | U32 | CG33188 |
| Me18S-C1366* | 1 | 100 | cDNA | 18S-C1366 | 12 nt (3′) | DUHG5 | |||
| Me18S-A1374*(SnoR122) | 1 | 67 | + | 18S-Am1374 | 10 nt (3′) | CG31175 | |||
| Me18S-A1576*(Z5) | 1 | 116 | cDNA | 18S-Am1576 | 15 nt (3′) | DUHG2 | |||
| Me18S-G1620*(U25) | 1 | 71 | + | 18S-Gm1620 | 11 nt (5′) | SnR56 | SnoR19 | U25 | DUHG2 |
| Me18S-A1806* | 1 | 78 | cDNA | 18S-Am1806 | 12 nt (3′) | U82 | IR | ||
| Me18S-C1831*(SnoR737) | 1 | 90 | cDNA | 18S-Cm1831 | 12 nt (5′) | SnR70 | U43 | U43 | CG15442 |
| Me18S-G1952* | 1 | 76 | N. blot | 18S-G1952 | 10 nt (5′) | DUHG5 | |||
| Me28S-G764*(U21) | 1 | 81 | cDNA | 28S-Gm764 | 14 nt (3′) | U21 | CG8385 | ||
| Me28S-A771* | 1 | 75 | N. blot | 28S-A771 | 11 nt (5′) | CG13900 | |||
| Me28S-A774a*(DmU18) | 2 | 73 | + | 28S-Am774 | 12 nt (5′) | U18 | U18 | U18 | CG11271 |
| Me28S-C788a*(DmSnR58) | 2 | 70 | + | 28S-Cm788 | 15 nt (5′) | Z12 | SnR58Y | Z12 | CG13900 |
| Me28S-G980* | 1 | 71 | N. blot | 28S-Gm980 | 11 nt (5′) | SnR39b | SnR39bY | SnR39b | CG14792 |
| Me28S-A982a*(DmSnR39/59) | 2 | 77 | + | 28S-Am982 | 13 nt (5′) | SnR39/59 | U51 | U51/U32 | DUHG4 |
| Me28S-A992* | 1 | 71 | N. blot | 28S-Am992 | 12 nt (5′) | SnR60 | U80 | U80/U77 | DUHG5 |
| Me28S-G1083a*(DmSnR60) | 4 | 84 | cDNA | 28S-Gm1083 | 12 nt (5′) | SnR60 | U80 | U80 | DUHG6 |
| 28S-Am1092 | 10 nt (3′) | SnR84 | SnoR133 | ||||||
| Me28S-A1322*(Z1) | 1 | 77 | cDNA | 28S-Am1322 | 11 nt (5′) | SnR61 | U38 | U38 | CG8280 |
| Me28S-A1666a*(U76) | 2 | 76 | + | 28S-Am1666 | 13 nt (5′) | U24 | U24 | Z20/U76 | DUHG1 |
| Me28S-U1848* | 1 | 80 | cDNA | 28S-U1848 | 13 nt (3′) | DUHG5 | |||
| Me28S-C2017* | 1 | 93 | cDNA | 28S-C2017 | 11 nt (5′) | CG4863 | |||
| 28S-A3349 | 15 nt (3′) | ||||||||
| Me28S-A2113* | 1 | 73 | N. blot | 28S-Am2113 | 14 nt (3′) | SnoR33 | Z38 | CG10652 | |
| Me28S-U2134a*(DmSnR62) | 2 | 77 | + | 28S-Um2134 | 12 nt (5′) | SnR62 | U34 | U34 | CG1475 |
| Me28S-G2173* | 1 | 67 | N. blot | 28S-Gm2173 | 12 nt (5′) | U50 | CG4046 | ||
| Me28S-A2486* | 1 | 117 | N. blot | 28S-A2486 | 12 nt (5′) | CG31647 | |||
| Me28S-A2564*(SnoR442) | 1 | 114 | cDNA | 28S-Am2564 | 12 nt (3′) | SnR63 | U40 | U40/U46 | CG7808 |
| Me28S-A2589a* | 3 | 106 | N. blot | 28S-Am2589 | 12 nt (3′) | SnR13 | U15 | U15 | CG9696 |
| Me28S-A2634a* | 3 | 73 | cDNA | 28S-Am2634 | 12 nt (5′) | SnoR44 | U79(Z22) | DUHG7 | |
| Me28S-C2645a* (DmSnR64) | 3 | 75 | + | 28S-Cm2645 | 12 nt (5′) | SnR64 | SnoR44 | U74(Z18) | DUHG3 |
| 28S-A2653 | 10 nt (3′) | ||||||||
| Me28S-G2703a* | 3 | 91 | cDNA | 28S-Gm2703 | 10 nt (3′) | SnR190 | Z63 | DUHG5 | |
| Me28S-G3081a*(U31) | 4 | 72 | + | 28S-Gm3081 | 13 nt (5′) | SnR67 | U31 | U31 | DUHG1 |
| Me28S-G3113a*(Dm3112) | 2 | 80 | cDNA | 28S-Gm3113 | 13 nt (5′) | U58 | CG13900 | ||
| Me28S-C3227a* | 2 | 80 | cDNA | 28S-C3227 | 16 nt (3′) | DUHG5 | |||
| Me28S-G3253* | 1 | 88 | N. blot | 28S-Gm3253 | 16 nt (5′) | SnR48 | SnoR1 | CG3395 | |
| Me28S-G3255a*(DmSnR48) | 2 | 85 | cDNA | 28S-Gm3255 | 16 nt (5′) | SnR48 | U60 | DUHG4 | |
| Me28S-G3277a*(SnR38) | 3 | 80 | cDNA | 28S-Gm3277 | 13 nt (5′) | SnR38 | SnR38Y | SnR38 | DUHG1 |
| Me28S-C3341a*(U49) | 2 | 80 | cDNA | 28S-Cm3341 | 13 nt (5′) | U49 | U49 | CG6253 | |
| Me28S-U3344a* | 2 | 79 | cDNA | 28S-U3344 | 13 nt (5′) | CG1518 | |||
| Me28S-C3351* | 1 | 77 | N. blot | 28S-C3351 | 13 nt (5′) | CG12740 | |||
| 18S-A613 | 13 nt (3′) | ||||||||
| Me28S-A3407a*(U29) | 4 | 90 | cDNA | 28S-Am3407 | 11 nt (5′) | SnR71 | U29 | U29 | DUHG1 |
| Me28S-C3420a* | 2 | 91 | N. blot | 28S-Cm3420 | 12 nt (5′) | SnR73 | U35 | U35 | CG8857 |
| MeU2-C28*(SnoR700) | 1 | 87 | + | U2-Cm28 | 13 nt (3′) | SnoR101 | CG1518 | ||
| MeU5-U42*(SnoR755) | 1 | 140 | + | U5-Um42 | 13 nt (5′) | CG12316 | |||
| MeU5-C46▴(U85) | 1 | 315 | + | U5-Cm46 | 13 nt (3′) | U85 | CG1142 | ||
| U5-Ψ47 | 6 + 6 nt (5′) | ||||||||
| MeU6-A47*(Z30) | 1 | 93 | N. blot | U6-Am47 | 11 nt (3′) | Z30 | Z30 | CG1666 | |
| MeU6-C68* | 1 | 99 | N. blot | U6-C68 | 11 nt (5′) | CG2173 | |||
| DmU3a▴(U3) | 2 | 136 | + | rRNA cleave | U3 | U3 | U3 | IR | |
| DmU14a*(U14) | 2 | 84 | cDNA | rRNA cleave | U14 | U14 | U14 | DUHG2 | |
| DmOr_cdI | 1 | 106 | cDNA | Unknown | CG3074 | ||||
| DmOr_cd2*(SnoR461) | 1 | 140 | cDNA | Unknown | DUHG4 | ||||
| DmOr_cd3*(SnoR229) | 1 | 152 | + | Unknown | CG9888 | ||||
| DmOr_cd4* | 1 | 78 | N. blot | Unknown | DUHG5 | ||||
| DmOr_cd5(SnoR185) | 1 | 103 | cDNA | Unknown | IR | ||||
| DmOr_cd6▴(SnoR684) | 1 | 170 | + | Unknown | CG10576 | ||||
| DmOr_cd7▴(SnoR291) | 1 | 114 | + | Unknown | CG1646 | ||||
| DmOr_cd8* | 1 | 92 | cDNA | Unknown | CG7808 | ||||
| DmOr_cd9a* | 2 | 91 | N. blot | Unknown | DUHG6 | ||||
| DmOr_cd10*(SnoR284) | 1 | 115 | cDNA | Unknown | CG3314 | ||||
| DmOr_cd11a* | 3 | 67 | N. blot | Unknown | CG12740 | ||||
| DmOr_cd12*(DmU24) | 1 | 93 | + | Unknown | CG12740 |
Only one isoform (isoform a) and the conserved guiding function(s) are given if the snoRNA has more than one variant. (Iso) numbers of isoforms; (Len) length of the SnoRNA gene; (Exp) expression situation. (cDNA, N. blot) SnoRNA was identified by a cDNA library or Northern blotting analysis in our work. (+) Confirmed expression of snoRNAs in other works (Tycowski and Steltz 2001; Yuan et al. 2003; Accardo et al. 2004). SnoRNAs with asterisks were identified by computer-assisted analysis. Those with black triangles were not detected in our work. SnoRNAs identified in previous studies (Tycowski and Steitz 2001; Zhou et al. 2002; Yuan et al. 2003; Accardo et al. 2004) are indicated by their names in parentheses. In the column location, the protein-coding host genes are denoted by their symbols. (IR) Intergenic region. In the column Modification, a nucleotide with “m” represents the rRNA methylation site that is conserved in Saccharomyces cerevisiae, plants, and/or mammals.
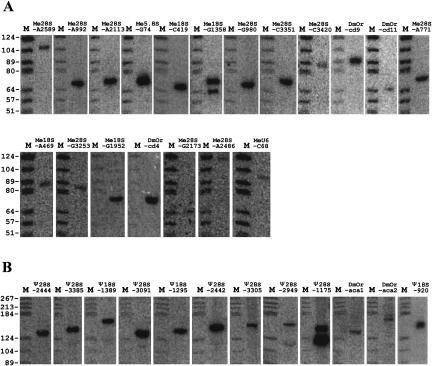
We have previously used a eukaryotic box C/D snoRNA search program to identify snoRNA genes in Oryza sativa (Chen et al. 2003). This effective computational program was applied in the present study to search the D. melanogaster genome for box C/D snoRNAs. The searches were mainly based on primary structural elements of the C/D snoRNAs and the information on rRNA methylation sites in eukaryotes such as mammals and yeast before fruit fly data were available. Computational analysis identified 97 snoRNA candidates that accounted for most of our experimental results and recently reported data (Accardo et al. 2004). Northern blotting was used to verify the novel candidates identified in silico, and 19 novel box C/D snoRNAs were validated (Fig. 1A) and added to our snoRNA database (Table 1). All snoRNA sequences were then used to find their isoforms in the Drosophila genome.
FIGURE 1.
Northern blot analyses. Aliquots of 30 μg total cellular RNA were separated on a denaturing 8% polyacrylamide gel and hybridized with the labeled oligonucleotide probes described in Materials and Methods. (Lane M) molecular weight markers (pBR322 digested with HaeIII and 5′-end labeled with [γ-32P]ATP). (A) Northern blot analyses of novel box C/D snoRNAs. (B) Northern blot analyses of novel box H/ACA snoRNAs.
In total, 102 gene variants encoding 63 box C/D snoRNAs were identified from the Drosophila genome. It has to be pointed out that a scaRNA U85 and three known snoRNAs, snoR684, snoR291, and U3, were not detected in this analysis; however these snoRNAs have been included for further analysis in the discussion. Forty-eight snoRNAs were predicted to guide 54 methylated residues in 5.8S, 18S, and 28S rRNA. Four typical snoRNAs and a scaRNA U85 were guides for internal methylations in U2, U5, and U6 snRNAs. In addition to the guide snoRNAs, 12 snoRNAs with no target in either rRNA and snRNA were termed orphan snoRNAs and their functions remain to be elucidated (Table 1; Supplementary Figure S1).
Identification of 26 novel box H/ACA snoRNAs from Drosophila
To study box H/ACA snoRNAs in Drosophila, another special cDNA library was constructed with total D. melanogaster RNA. In contrast to the box C/D snoRNA research, we focused on the cDNA fraction of 120–180 nt to evaluate the box H/ACA snoRNAs. After screening several thousand cDNA clones to eliminate heavy contamination of rRNA fragments, 31 box H/ACA candidates were identified. Sequence and structural analyses revealed that the candidates represented 29 box H/ACA snoRNA species, including 14 novel species when compared with known snoRNA data (Yuan et al. 2003; Huang et al. 2004).
Based on the conserved secondary structure and functional elements, a computational analysis for other possible box H/ACA snoRNA genes was performed on all the introns of known snoRNA host genes and ribosomal protein genes. Apart from known box H/ACA snoRNAs, 19 novel candidates were identified; 12 of them were further confirmed by the Northern blotting analyses (Fig. 1B) and added to our database.
In total, 110 gene variants encoding 56 box H/ACA snoRNAs were identified from the Drosophila genome. When compared with the published data on Drosophila, 26 novel box H/ACA snoRNAs have been first reported in this study. Fifty-one snoRNAs were predicted to guide 85 pseudouridylations in 18S and 28S rRNA. Two snoRNAs, Y28S-1232 and Y28S-3186, may guide pseudouridylations for both rRNA and U4 or U5 snRNA. Moreover, five orphan snoRNAs that lack sequence complementary to rRNA and snRNA were also determined (Table 2; Supplementary Figure S2).
TABLE 2.
Box H/ACA snoRNA genes in D. melanogaster
| Homology | |||||||||
| SnoRNA name | Iso | Len (nt) | Exp | Modification | Antisense element | Yeast | Plants | Vertebrates | Location |
| Ψ 18S-110* | 1 | 139 | cDNA | 18S-Ψ110 | 6 + 6 nt (3′) | CG13900 | |||
| Ψ 18S-176(SnoR314) | 1 | 154 | cDNA | 18S-U176 | 3 + 6 nt (5′) | IR | |||
| Ψ 18S-301 | 1 | 138 | cDNA | 18S-U301 | 7 + 5 nt (5′) | IR | |||
| 18S-Ψ363 | 4 + 5 nt (3′) | SnoR86 | U71 | ||||||
| Ψ 185-525a* | 10 | 134 | cDNA | 18S-U525 | 6 + 4 nt (5′) | CG10954 | |||
| 18S-U1313 | 3 + 10 nt (3′) | ||||||||
| Ψ 18S-531* | 1 | 152 | N. blot | 18S-Ψ531 | 5 + 10 nt (5′) | ACA42 | CG7434 | ||
| 28S-Ψ1850 | 5 + 7 nt (3′) | ||||||||
| Ψ 18S-640a* | 7 | 132 | cDNA | 18S-Ψ640 | 6 + 4 nt (5′) | SnR161 | SnoR73 | MBI-12 | CG10954 |
| Ψ 18S-841a*(SnoR66) | 4 | 144 | + | 18S-Ψ841 | 6 + 6 nt (5′) | MBI-13/ACA28 | CG9696 | ||
| Ψ 18S-920* | 1 | 141 | N. blot | 18S-U920 | 6 + 5 nt (5′) | CG7283 | |||
| Ψ 18S-996* | 1 | 153 | cDNA | 18S-Ψ996 | 5 + 8 nt (3′) | ACA14 | CG7008 | ||
| Ψ 18S-1086* | 1 | 147 | cDNA | 18S-Ψ1086 | 6 + 4 nt (5′) | SnR31 | SnoR72 | ACA8 | CG13900 |
| 28S-U2045 | 3 + 8 nt (3′) | ||||||||
| Ψ 18S-1275*(SnoR783) | 1 | 140 | + | 18S-Ψ1275 | 7 + 7 nt (5′) | SnR36 | ACA36 | CG8495 | |
| Ψ 18S-1295* | 1 | 140 | N. blot | 18S-U1295 | 6 + 3 nt (5′) | CG15693 | |||
| 18S-U740 | 3 + 8 nt (3′) | ||||||||
| 318S-1347a*(SnoR203) | 3 | 137 | cDNA | 18S-Ψ1347 | 7 + 5 nt (5′) | CG5119 | |||
| Ψ 18S-1377a*(SnoR328) | 5 | 139 | cDNA | 18S-Ψ1279 | 6 + 6 nt (5′) | SnR35 | ACA13 | CG1883 | |
| 18S-Ψ1377 | 10 + 3 nt (3′) | SnR83 | ACA4 | ||||||
| Ψ 18S-1389a* | 2 | 159 | N. blot | 18S-U1389 | 6 + 7 nt (3′) | CG9696 | |||
| Ψ 18S-1397* | 1 | 141 | cDNA | 18S-Ψ1397 | 4 + 7 nt (5′) | ACA15 | CG10576 | ||
| N. blot | |||||||||
| Ψ 18S-1820*(SnoR639) | 1 | 144 | cDNA | 18S-Ψ1820 | 7 + 3 nt (3′) | U70 | CG3333 | ||
| Ψ 18S-1854a* | 3 | 139 | cDNA | 18S-Ψ1854 | 6 + 7 nt (5′) | CG8922 | |||
| N. blot | 18S-Ψ1937 | 7 + 4 nt (3′) | |||||||
| Ψ 28S-291*(SnoR50) | 1 | 220 | cDNA | 28S-U291 | 5 + 8 nt (5′) | CG4863 | |||
| 28S-U2238 | 4 + 8 nt (3′) | ||||||||
| Ψ 28S-1060* | 1 | 150 | N. blot | 28S-Ψ1060 | 4 + 7 nt (5′) | CG3203 | |||
| Ψ 28S-1135a* | 6 | 148 | N. blot | 28S-Ψ1135 | 6 + 5 nt (3′) | SnR8 | ACA56 | CG8922 | |
| Ψ 28S-1153 | 1 | 148 | cDNA | 28S-Ψ1153 | 7 + 5 nt (5′) | IR | |||
| 18S-U1425 | 9 + 6 nt (3′) | ||||||||
| Ψ 28S-1175a* | 3 | 136 | N. blot | 28S-Ψ1175 | 6 + 6 nt (5′) | CG4046 | |||
| 28S-Ψ1311 | 6 + 3 nt (3′) | ACA32 | |||||||
| Ψ 28S-1180*(SnoR535) | 1 | 145 | + | 28S-U1930 | 5 + 8 nt (5′) | CG10897 | |||
| 28S-Ψ1180 | 12 + 4 nt (3′) | ||||||||
| Ψ 28S-1192a* | 4 | 140 | N. blot | 28S-Ψ1192 | 7 + 6 nt (5′) | SnR5 | SnoR81 | MBI-20/ACA52 | CG8922 |
| Ψ 28S-1232* | 1 | 141 | N. blot | U5-U82 | 7 + 5 nt (5′) | CG3203 | |||
| 28S-U1232 | 5 + 4 nt (3′) | ||||||||
| Ψ 28S-1837a*(SnoR14) | 3 | 137 | cDNA | 28S-Ψ1837 | 6 + 7 nt (5′) | CG10576 | |||
| Ψ 28S-2149*(SnoR143) | 1 | 144 | + | 28S-U2149 | 7 + 6 nt (5′) | CG1883 | |||
| 28S-U18 | 4 + 7 nt (3′) | ||||||||
| Ψ 28S-2442a* | 2 | 140 | N. blot | 28S-Ψ2442 | 6 + 3 nt (5′) | SnR3 | ACA6 | CG2922 | |
| 28S-Ψ2533 | 8 + 6 nt (3′) | ||||||||
| Ψ 28S-2444* | 1 | 130 | N. blot | 28S-Ψ2444 | 7 + 4 nt (5′) | SnoR87 | ACA19 | CG7726 | |
| Ψ 28S-2562* | 1 | 135 | cDNA | 28S-U925 | 6 + 4 nt (5′) | CG15693 | |||
| 28S-Ψ2562 | 7 + 6 nt (3′) | SnoR92 | MBI-1/ACA23 | ||||||
| Ψ 28S-2566*(SnoR269) | 1 | 268 | + | 28S-Ψ2566 | 5 + 5 nt (5′) | SnR191 | SnoR79 | U19/MBI-26 | CG3902 |
| 28S-Ψ2568 | 7 + 6 nt (3′) | SnR191 | U19 | U19 | |||||
| Ψ 28S-2622(SnoR3) | 1 | 140 | cDNA | 28S-U363 | 3 + 7 nt (5′) | IR | |||
| 28S-Ψ2622 | 5 + 6 nt (3′) | SnoR83 | ACA48 | ||||||
| Ψ 28S-2626* | 1 | 142 | N. blot | 28S-Ψ2626 | 6 + 4 nt (3′) | CG1883 | |||
| Ψ 28S-2648*(SnoR72) | 1 | 162 | + | 28S-Ψ2648 | 5 + 4 nt (5′) | SnR9 | MBI-12/ACA58 | CG7424 | |
| Ψ 28S-2719*(SnoR734) | 1 | 145 | cDNA | 28S-Ψ2719 | 6 + 3 nt (5′) | CG7434 | |||
| Ψ 28S-2876*(SnoR825) | 1 | 140 | + | 28S-Ψ2956 | 4 + 7 nt (5′) | CG8922 | |||
| 28S-U2876 | 9 + 3 nt (3′) | ||||||||
| Ψ 28S-2949* | 1 | 155 | N. blot | 28S-Ψ2949 | 5 + 5 nt (5′) | DUHG4 | |||
| Ψ 28S-2996* | 1 | 136 | cDNA | 28S-U2996 | 5 + 8 nt (5′) | CG17489 | |||
| Ψ 28S-3091a* | 2 | 135 | N. blot | 28S-Ψ3091 | 7 + 4 nt (5′) | SnoR78 | CG2922 | ||
| 28S-Ψ3356 | 8 + 6 nt (3′) | ACAI | |||||||
| Ψ 28S-3186*(SnoR165) | 1 | 143 | cDNA | U4-U59 | 7 + 4 nt (5′) | CG2922 | |||
| 28S-Ψ3186 | 5 + 6 nt (3′) | ||||||||
| Ψ 28S-3305a* | 3 | 150 | N. blot | 28S-Ψ3305 | 5 + 4 nt (3′) | E3 | CG9983 | ||
| Ψ 28S-3308* | 1 | 153 | cDNA | 28S-Ψ3308 | 9 + 4 nt (3′) | U68 | CG13900 | ||
| Ψ 28S-3316a* | 5 | 132 | N. blot | 28S-Ψ3316 | 6 + 3 nt (5′) | MBI-3/ACA21 | CG9696 | ||
| Ψ 28S-3327a*(SnoR586) | 3 | 140 | cDNA | 28S-Ψ3327 | 6 + 6 nt (5′) | SnR46 | ACA16 | CG11276 | |
| 18S-U1920 | 4 + 6 nt (3′) | ||||||||
| Ψ 28S-3342(SnoR644) | 1 | 164 | cDNA | 28S-Ψ3342 | 7 + 3 nt (3′) | SnR34 | U65 | U65 | IR |
| Ψ 28S-3378* | 1 | 150 | N. blot | 28S-Ψ3378 | 4 + 8 nt (5′) | CG9696 | |||
| Ψ 28S-3385a* | 2 | 150 | N. blot | 28S-Ψ3385 | 6 + 4 nt (3′) | SnR10 | SnoR74 | MBI-3/ACA21 | CG2922 |
| Ψ 28S-3405a* | 4 | 140 | cDNA | 28S-Ψ3405 | 7 + 5 nt (5′) | SnR37 | ACA10 | CG9983 | |
| 28S-U1265 | 7 + 5 nt (3′) | ||||||||
| Ψ 28S-3436a*(SnoR708) | 2 | 142 | cDNA | 28S-Ψ3436 | 6 + 5 nt (5′) | SnR42 | MBI-6/ACA27 | CG3203 | |
| Ψ 28S-3571* | 1 | 144 | cDNA | 28S-Ψ3571 | 5 + 5 nt (3′) | CG2922 | |||
| 18S-U1790 | 5 + 7 nt (5′) | ||||||||
| DmOr_aca1* | 1 | 135 | N. blot | Unknown | CG4046 | ||||
| DmOr_aca2* | 1 | 142 | N. blot | Unknown | CG10944 | ||||
| DmOr_aca3* | 1 | 145 | cDNA | Unknown | CG5502 | ||||
| DmOr_aca4 | 1 | 162 | cDNA | Unknown | Intron antisense | ||||
| DmOr_aca5*(SnoR227) | 1 | 148 | cDNA | Unknown | DUHG4 |
Only one isoform (Isoform a) and the conserved guiding function(s) are given if the snoRNA have more than one variant (Iso) numbers of Isoforms; (Len) length of the snoRNA gene; (Exp) expression situation. (cDNA, N. blot) SnoRNA was identified by a cDNA library or Northern blotting analysis in our work. (+) Confirmed expression of snoRNAs in other work (Yuan et al. 2003). SnoRNAs with stars were identified by computer-assisted analysis. SnoRNAs identified in a previous study (Yuan et al. 2003) are indicated by their names in parentheses. In the column Location, the protein-coding host genes are denoted by their symbols. (IR) Intergenic region. In the column Modification, 3 represents rRNA pseudouridine sites that are conserved in Saccharomyces cerevisiae and/or mammals, and the known rRNA pseudouridines determined. (U) Predicted pseudouridine site that has not been confirmed experimentally.
Two distinguished organizations dominate the expression of the snoRNAs in Drosophila
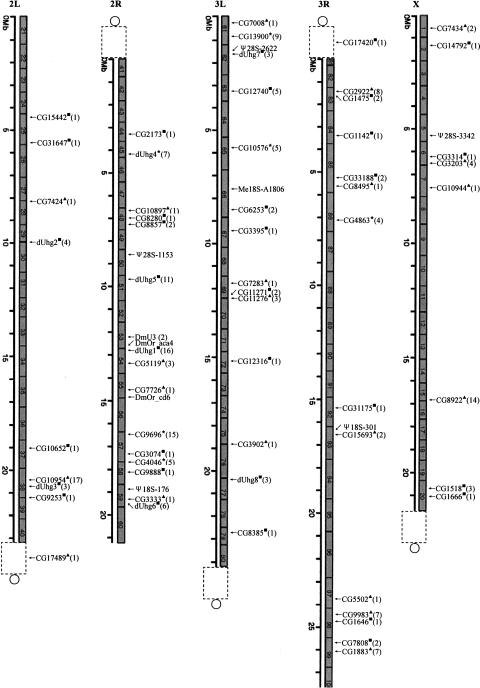
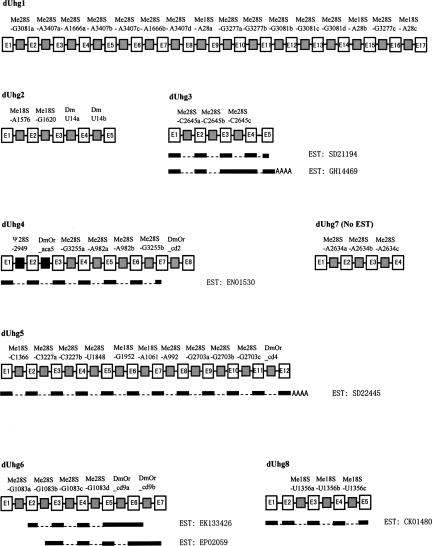
Following the identification of numerous snoRNAs, their genomic organization and expression strategy were then investigated. A large proportion of the box C/D snoRNAs were clearly located in the introns of protein-coding genes such as ribosomal proteins and proteins associated with snoRNP and transcriptional factors. However, 33 gene variants encoding 16 box C/D snoRNAs in addition to two box H/ACA snoRNAs were found clustered in six chromosomal spots that had no annotation (Fig. 2), and therefore were regarded as intergenic spacers in the Drosophila genome. Interestingly, each interval between two individual snoRNA genes was ~150–200 nt, which resembles the situations of the two dUhg genes reported previously (Tycowski and Steitz 2001). By searching typical intron splicing signals (5′GT and 3′AG with branching sequences) that flank each of the snoRNA genes, our computational analysis revealed that all the snoRNAs were indeed intron encoded (Table 3) and it was unlikely that the spliced exons of host genes encoded any protein. Therefore, six new UHG-like genes were identified and designated as dUhg 3–8. All six ncRNAs, except dUhg 7, were further supported by the presence of a perfect match with ESTs from the Flybase database (http://www.flybase.org, Database of the Drosophila Genome) (Fig. 3). As observed in the vertebrate UHG genes (Tycowski et al. 1996), the cDNA transcripts of the dUhg genes in Drosophila possessed a poly(A) tail at their 3′ end, suggesting products of RNA polymerase II (Table 3). Together with two previously identified dUhg genes (Tycowski and Steitz 2001), the eight dUhgs encode 53 snoRNA isoforms that represent half of the known box C/D snoRNA genes in Drosophila. In contrast to other eukaryotes, dUhg therefore represents a major gene organization for the expression of box C/D snoRNAs in the fruit fly. Of particular interest is that although each dUhg encodes multiple box C/D snoRNAs, the mode of one snoRNA per intron is strictly maintained.
FIGURE 2.
Chromosomal mapping of snoRNA genes in D. melanogaster. The gray and the empty boxes represent euchromatin and hetero-chromatin, respectively. Centromere is denoted by an empty circle. Intronic snoRNA gene/gene clusters are indicated by their host genes on the map. The black triangle, box, and asterisk indicate host genes encoding box H/ACA snoRNA only, box C/D snoRNA only, and both of them, respectively. The number in parentheses shows the sum of the snoRNAs encoded by the host genes.
TABLE 3.
Eight dUhg host genes in D. melanogaster
| Splice signal | ||||||||
| Name | Chr | Int. | Int. for sno. | 5′ end | 3′ end | EST record | Poly(A) tail | Accession no. |
| dUhg1 | 2R | 16 | 16 | GTAAG (7), GTAAT (3) GTGTG (2), GTGGG (1) GTTAG (1), GTAGG (1) GTATG (1) |
ACAG (6), GCAG (4) TTAG (2), CCAG (2) ATAG (1), TCAG (1) |
* | + | * |
| dUhg2 | 2L | 4 | 4 | GTAAG (2), GTAAT (2) | GCAG (2), CCAG (2) | * | + | * |
| dUhg3 | 2L | 4 | 3 | GTAAG (4) | TTAG (2), GCAG (1) ACAG (1) |
SD21194 GH14469 |
+ | AY805214 |
| dUhg4 | 2R | 7 | 7 | GTAAG (3), GTAAT (2) GTAAA (1), GTTAG (1) |
GCAG (3), TCAG (3) ACAG (1) |
EN01530 | n.d. | AY805215 |
| dUhg5 | 2R | 11 | 11 | GTAAG (4), GTAAT (2) GTATG (1), GTGAG (1) GTAGG (1), GTAAA (1) GTATA (1) |
ACAG (4), GCAG (2) TTAG (2), TCAG (1) CTAG (1), AAAG (1) |
SD22445 | + | AY805219 |
| dUhg6 | 2R | 6 | 6 | GTAAA (3), GTAAG (1) GTTAG (1), GTGAG (1) |
ACAG (3), TCAG (1) GCAG (1), GTAG (1) |
EK133426 GCAG (1), GTAG (1) |
n.d. | AY805216 |
| dUhg7 | 3L | 3 | 3 | GTGTG (2), GTAAT (1) | TCAG (2), ACAG (1) | n.d. | n.d. | AY805217 |
| dUhg8 | 3L | 4 | 3 | GTAAG (1), GTAAT (1) GTTAA (1), GTATT (1) |
ACAG (2), TCAG (1) GCAG (1) |
CK01480 | n.d. | AY805218 |
(Int) number of introns in a host gene; (Int. for sno) number of introns encoding for snoRNA. Number in the parentheses indicates the frequency of donor or acceptor sequences found in introns; (n.d.) situation is unknown; (*) dUhg1 and dUhg2 that had been experimentally verified in previous work (Tycowski and Steitz 2001).
FIGURE 3.
Schematic diagram of eight dUhg genes in D. melanogaster. Exons and introns of host genes are indicated by empty boxes and solid lines, respectively. Box H/ACA and box C/D snoRNA genes within introns are indicated by black and gray boxes, respectively. Names of host genes and snoRNA genes are also shown. The black bars indicate the cDNA and the spliced introns are indicated by dashed lines. The EST ID is also shown.
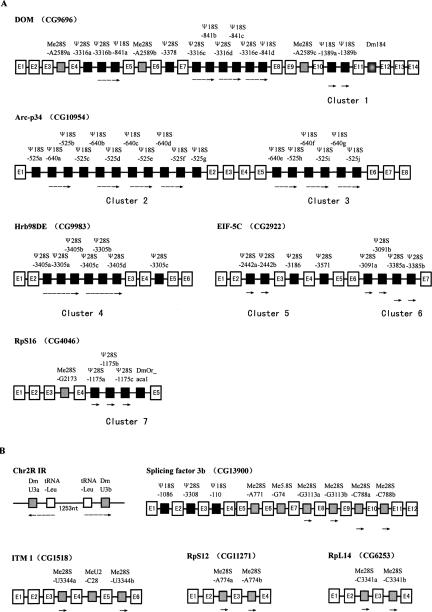
Apart from intron-encoded snoRNAs, three box C/D snoRNAs, including U3 snoRNA, which is involved in rRNA processing, are transcribed independently from intergenic regions, showing a diverse gene organization. It is also worth noting that the U3 snoRNA gene is clustered with a tRNALeu gene in an inverted repeat (Fig. 4).
FIGURE 4.
SnoRNA gene clusters and their duplication in D. melanogaster. (A) Seven novel intronic box H/ACA snoRNA gene clusters. (B) Inverted U3 snoRNA-tRNA clusters and repeated units of box C/D snoRNA gene in introns. Exons and introns of host genes are indicated by empty boxes and solid lines, respectively. Box H/ACA and box C/D snoRNA genes are indicated by black and gray boxes, respectively. Names of host genes and snoRNA genes are also shown. The repeated units of the snoRNA genes are indicated by arrows.
In terms of the box H/ACA snoRNAs, most of them are intron-encoded in protein-coding genes, especially rich in ribosomal proteins. A distinguishing characteristic of the genomic organization for this family of snoRNA is the prevalence of intronic clusters. In addition to the clusters reported previously (Huang et al. 2004), a total of 17 intronic clusters encoding 77 box H/ACA snoRNAs (70% of the gene variants identified) have been identified from the Drosophila genome (Fig. 4), highlighting the importance of this gene organization for the expression of box H/ACA snoRNAs. In general, the clusters are composed of isoforms of the same snoRNA genes, suggesting that local duplication has served as a major way to form the multiple snoRNA clusters in the introns of protein-coding genes. Interestingly, DmOr-aca4, an orphan snoRNA, was mapped entirely to an intron of a hypothetical pre-mRNA, but was transcribed in the opposite orientation of the protein-coding gene. This peculiar gene organization may suggest a possible antisense function of the orphan snoRNA in the regulation of pre-mRNA processing. In addition to being intron encoded, five box H/ACA snoRNAs appear to be transcribed independently from intergenic regions.
Sixty host genes encoding 207 snoRNAs were scattered over the two large autosomes and the X chromosome of Drosophila, but none was found in chromosome 4, which is the shortest chromosome. All the host genes were mapped in the region of euchromatin, with the exception of two located in the heterochromatin where the expression of most genes is not active (Fig. 2). The overall genome-wide analysis of snoRNA genes in Drosophila shows a preferential transcription of polycistronic snoRNA from two distinct gene organizations, first, dUhgs, which mainly encode box C/D snoRNA, and second, intronic clusters, which dominate the expression of box H/ACA snoRNA.
Extensive utilization of introns for snoRNA-coding in the Drosophila host genes
It is evident that in D. melanogaster snoRNAs can usually be found in more than one intron of a variety of host genes. Among the 267 introns in the host genes, more than half of them (145 introns) have been identified as host introns for the snoRNAs and a small ncRNA (Dm184). A higher percentage for snoRNA coding in large introns of the host genes can be attained when taking account of 85 “empty” introns that are smaller than 150 nt, the minimum length required to accommodate one box C/D snoRNA. These empty introns can therefore be easily eliminated from the list of potential host introns. For example, nine of the 13 introns of Dom gene are longer than 150 nt, and 8 turn out to encode 15 snoRNAs and a small ncRNA. In splicing factor 3b, there are nine introns longer than 150 nt, all of which are host introns for snoRNAs. Remarkably, the dUhg genes, which are unlikely to encode any protein, seem to be perfectly designed for snoRNA coding. The eight dUhg genes possess 55 introns in total, only two of which are empty (Fig. 3).
Thirty-seven empty introns larger than 150 nt were further examined in detail. Interestingly, 11 of them contained 50–120-nt-long conserved intronic sequences (LCISs) as compared to their counterparts among different species of Drosophila, suggesting the presence of putative cis-acting elements or possible small ncRNAs other than snoRNAs. LCIS was also observed in the undefined region of some large introns that were hosts for snoRNAs. Further experiments were performed to validate some of these LCISs and two of them were characterized as stable ncRNAs with unknown function (Fig. 5). The remaining 26 introns (about 10% of the total introns in this study) were devoid of any LCIS. However, 15 of the empty introns (57.7%) belonged to the first intron of ribosomal protein genes or other housekeeping genes. This suggests that instead of snoRNA coding, these introns may have a specialist function, as it has been shown that the first introns that contain binding sites for transcriptional factors are important for the expression of ribosomal protein genes in mammals (Antoine and Kiefer 1998).
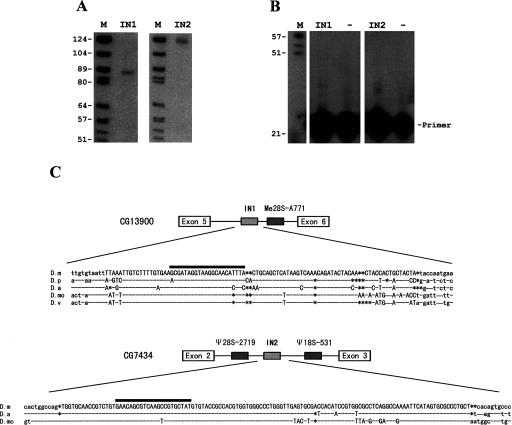
FIGURE 5.
Experimental determination of two ncRNAs encoded by LCIS. (Lane M) molecular weight markers (pBR322 digested with HaeIII and 5′-end labeled with [γ-32P]ATP). Lane IN1 and IN2 are two samples of LCIS detected by specific probes in (A) Northern blot and (B) reverse transcription. (Lane 3) negative control. (C) Alignment of the LCIS sequences from different species of Drosophila. The coding regions for intronic ncRNAs are in capital letters. Nucleotide identities among different sequences are denoted by hyphens and deletions by asterisks. D.m, D.p, D.a, D.mo, and D.v represent D. melanogaster, D. pseudo, D. ananassae, D. mojavensis, and D. virilis, respectively. Sequences of the probes or primers used in analyses are indicated by black bars above the alignment.
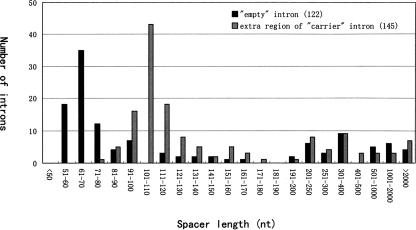
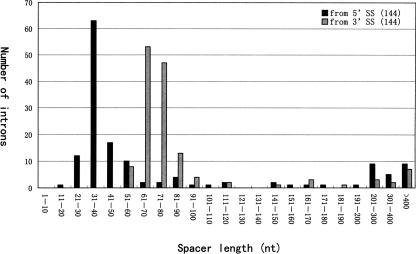
The average size of the empty introns was ~60–70 bp (Fig. 6), which corresponds perfectly to the length of the minimal intron (61 bp) in Drosophila (Yu et al. 2002). Due to snoRNA or snoRNA cluster containing, the host introns can vary in length from 150 bp to 2 kb. However, after removing all snoRNA/snoRNA clusters and ncRNA-coding regions from the 145 host introns, the size distribution of the remaining sequence in these introns was reduced to 90–120 bp, which was slightly longer than the minimal intron length (Fig. 6). Taking account of sequences necessary for snoRNA processing (Hirose and Steitz 2001), the intron hosts are only sufficiently long enough to hold a snoRNA gene or gene cluster without any redundant sequence. Interestingly, this compact structure of the host introns appears strictly maintained to encode the two families of snoRNAs in different host genes. In fact, this compact structure of host introns results mainly from parsimonious spacers from the snoRNA-coding region to the 5′ splice site, most of which were centered around 30–40 bp despite the intron sizes varying to a large extent (Fig. 7). The distance from the snoRNA gene/gene cluster to the 3′ splice sites averaged between 60 and 80 bp, which was very similar to the intronic positioning of box C/D snoRNA genes in mammals. This distance has been proven to be important for the effective processing of the snoRNAs from their host mRNA precursor (Hirose and Steitz 2001).
FIGURE 6.
Distribution of lengths of spacer sequences for 267 introns from 60 snoRNA host genes in D. melanogaster. Empty intron denotes the introns in which no intronic ncRNA was found. Extra region of “carrier” intron refers to the remaining sequences after the intronic ncRNA gene is removed from the host introns.
FIGURE 7.
Distribution of lengths of spacer sequences for 144 D. melanogaster snoRNA genes/gene clusters. The black and gray bars represent distances from 5′ and 3′ splice sites, respectively.
DISCUSSION
A more complete list of the two families of snoRNA genes in Drosophila
To obtain a comprehensive understanding of the genomic organization and expression strategy of ncRNA in D. melanogaster, we have performed a large-scale analysis of the two major families of snoRNA genes using both experimental and computational RNomics methods. In this study, 212 gene variants encoding 56 box H/ACA and 63 box C/D snoRNAs have been identified in the fruit fly. These data are consistent with previous works (Tycowski and Steitz 2001; Yuan et al. 2003; Accardo et al. 2004; Huang et al. 2004) and further includes 57 novel snoRNAs. In addition, two novel small ncRNAs other than snoRNAs were also validated. This extensive study indicates the complexity of the snoRNA gene families in the Drosophila genome and, furthermore, strengthens the analytical strategies used in this research, such as size-fractioned cloning for specialized cDNA libraries that are enriched in snoRNAs. It is also evident that the computational analysis of the Drosophila genome is complementary to the cDNA cloning approach. In D. melanogaster, a high degree of pseudouridylation on rRNA has been reported (Ofengand and Bakin 1997), while rRNA methylation sites have not yet been mapped. In this study, we have added to the long list a number of novel box C/D snoRNAs that are absent from previous works (Tycowski and Steitz 2001; Yuan et al. 2003; Accardo et al. 2004), reflecting that the level of rRNA methylation in Drosophila is not low. According to our computational analysis of the Drosophila genome, we have further predicted that the number of box C/D snoRNA for rRNA is about 97, although dozens of them remain to be confirmed by experimental detection. Interestingly, most of the box C/D snoRNAs from Drosophila possess only one antisense sequence, which is predicted to guide a single methylation site in the RNAs. In contrast, more than half of the box H/ACA snoRNAs exhibit two functional elements guiding two pseudouridylations in the RNAs (some of which are predicted to guide two pseudouridylations in rRNA and snRNA). It is not clear whether the high proportion of the bifunctional box H/ACA snoRNAs is relative to the constraint of high rRNA pseudouridylation in Drosophila or to a selective advantage in the functional evolution of these snoRNAs. From the distribution of the modified residues predicted by the snoRNAs, we estimate that more than two-thirds of the guide snoRNA for rRNA methylation and pseudouridylation in Drosophila have been described in this study. Although by no means exhaustive, the analysis has provided a more complete list of the two families of snoRNA genes in Drosophila.
Evolution of snoRNA gene organization and expression strategy
Identification of a large number of snoRNA genes provides a unique opportunity to investigate the genomic organization and expression of the ncRNAs in Drosophila. Similar to vertebrates (Maxwell and Fournier 1995), almost all snoRNAs in Drosophila are intron encoded. This organization not only emphasizes the utility of introns that have long been considered as junk DNA (Flam 1994), but also suggests an intriguing link between mRNA splicing and the expression of snoRNAs that are mainly involved in the post-transcriptional modifications of rRNAs in higher animals. In general, the host genes of snoRNAs are mostly protein-coding genes related to ribosomal biogenesis and nucleolar formation, showing expressional coordination among the various components of the protein translation machinery (Bachellerie et al. 2000). Some ncRNA genes, such as UHG, have also served as host genes specifically encoding box C/D snoRNA, which demonstrates introns, instead of exons, in an RNA precursor producing stable and functional RNAs. Since UHG was originally identified in mammals (Tycowski et al. 1996), only a few UHG-like genes have been further identified for snoRNA coding. In this study, we have shown at least eight dUhgs in the Drosophila genome. The abundance of these genes together with their powerful ability for snoRNA coding is a characteristic of snoRNA gene organization in Drosophila. Thus the importance of large ncRNAs, such as UHG, for small RNA coding, particularly in higher animals, may be underestimated because most of the noncoding genes in the mammal genome remain to be annotated (News Staff 2004). For instance, recent studies have revealed that some imprinted ncRNA genes in mammals comprise a large number of repeated tandem introns that encode box C/D snoRNAs specifically expressed in brain (Cavaille et al. 2000, 2002). It remains to be answered why in both mammals and Drosophila, the UHG or dUhg, respectively, encode mainly box C/D snoRNAs, but rarely box H/ACA snoRNAs. For example, the eight dUhgs in Drosophila contained only two box H/ACA snoRNAs compared to 51 box C/D snoRNAs.
The majority of box H/ACA snoRNAs are also intron encoded in Drosophila, but they favor another gene organization, that is, intronic cluster. SnoRNA gene clusters were originally found in higher plants (Leader et al. 1995) and the budding yeast Saccharomyces cerevisiae in which most snoRNAs are independently transcribed from singletons and five polycistronic snoRNA clusters (Lowe and Eddy 1999; Qu et al. 1999). Gene clusters have also been found in some primordial organisms, such as trypanosomes (Dunbar et al. 2000), Euglena (Russell et al. 2004) and Giardia (our unpubl. results), implying that the clusters may be an ancient gene organization conserved through the course of evolution. Remarkably, both intronic and independently transcribed polycistrons have considerably developed and become the predominant genomic organizations of snoRNAs in flowering plants (Qu et al. 2001; Liang et al. 2002; Brown et al. 2003; Chen et al. 2003). An atypical discistron, the tRNA-snoRNA gene cluster, was also identified in the rice genome (Kruszka et al. 2003). The intronic polycistron has never been reported in metazoan other than Drosophila and recognized as a predominant genomic organization for the box H/ACA snoRNAs.
Among all the organisms analyzed so far, it is only in Drosophila that the two families of snoRNAs exhibit such an important divergence in gene organization and expression strategies. This may reflect intrinsic differences in the mechanisms by which the two families of snoRNA genes evolved respectively during the speciation of the fruit fly.
It is worth noting that box H/ACA snoRNAs have many more isoforms than C/D snoRNAs in Drosophila. Unlike C/D snoRNA isoforms, which are highly conserved, mutations occurred frequently in box H/ACA snoRNA isoforms. The conserved secondary structures and box elements in H/ACA isoforms remain unchanged, which guarantees the maturation of snoRNA. Interestingly, the accumulation of mutations in the isoforms would lead to partial alternation of snoRNA’s function in loss or gain of rRNA complementary sequences (Supplementary Table S1).
Intronic regions deserve more attention in searching for ncRNAs in Drosophila
As a type of intervening sequence of genes, splicesomal introns with a large size variation are present in most (especially multicellular) eukaryotes (Logsdon 1998; Nixon et al. 2002). By comparison to the housefly, Drosophila possesses a very small genome; however, a large number of introns (about five introns per gene) have been estimated for the Drosophila genome (Yu et al. 2002). Interestingly, comparative genomic analysis has revealed similar constraints in the intergenic and intronic sequences of the Drosophila genome and about one-fourth of intronic sequences are conserved (Bergman and Kreitman 2001). A draft expression map of the Drosophila genome has also shown thousands of uncharacterized transcripts expressed from noncoding DNA in a developmentally coordinated manner (Stolc et al. 2004). Nevertheless, the variation of intron sizes and function of most introns in the Drosophila genome still remain to be elucidated.
Recently, a systematic analysis of intron size has revealed the presence of minimal introns in most multicellular (and some unicellular) eukaryotes (Yu et al. 2002). The minimal introns showed a sharp peak in the distribution of intron size in all the organisms and were not randomly distributed among genes. They were therefore suggested to function as a type of cis-element for enhancing the export of spliced mRNA from the cell nucleus (Yu et al. 2002). The minimal introns in D. melanogaster have a sharp “spike” around 60 bp, and account for about half of the total introns in the genome.
In contrast to a small genome, the majority of the remaining introns in Drosophila genes are moderately large, falling between 0.1 kb and 2.0 kb. Numerous cis-elements were also found in the large introns, which function as regulatory sequences in alternative splicing of mRNA precursor (Standiford et al. 2001) or as enhancers for gene transcription (Meredith and Storti 1993). In addition, many ncRNAs, such as miRNAs, were frequently identified from these intronic sequences (Aravin et al. 2003; Lai et al. 2003; Yuan et al. 2003). In this work, we have systematically analyzed 267 introns from the 60 host genes and found a very high proportion of the large introns for snoRNA or other small ncRNA coding. The size variations of the snoRNA-containing introns are mainly due to different numbers of snoRNAs inside them, reflecting a compact and informative intron structure. These results demonstrate the highly efficient utilization of large introns and their sequences for functional RNA coding in the host genes and provide important clues for further research of other ncRNA genes in the large introns of the Drosophila genome.
MATERIALS AND METHODS
Computational search for snoRNA genes in the Drosophila genomic database
The D. melanogaster genome scaffolds available at the Flybase database were searched for potential box C/D snoRNAs in the following ways. (1) A eukaryote snoRNA search program (Chen et al. 2003) was used to identify putative snoRNA genes with box C/D, a terminal stem with at least three base pairings and, in most cases, an rRNA complementary sequence. (2) Flanking sequences (about 1 kb) of the snoRNA candidates and all intronic sequences of the Drosophila ribosomal protein genes, which are available at the Ribosomal Protein Gene Database (http://ribosome.miyazaki-med.ac.jp), were also examined for other possible box C/D snoRNAs. The sequences were also analyzed for additional noncanonical C/D candidates. (3) BLAST (Altschul et al. 1990) and FASTA (Pearson and Lipman 1998) programs were used to find gene variants of novel snoRNA genes and establish the sum of snoRNA isoforms. Sequence alignment of snoRNA isoforms was performed with Clustal X 1.8 and DNAstar packages.
All intronic sequences of box H/ACA snoRNA host genes and ribosomal protein genes in the Drosophila genome were obtained from the Flybase database and the Ribosomal Protein Gene Database. The novel box H/ACA snoRNA candidates were identified using our computer program, which takes into account both the sequence motifs and secondary structures in the snoRNAs (Huang et al. 2004). Identification of the gene variants and sequence alignment were performed as above.
Construction and screening of cDNA libraries and RNA analyses
Fresh wild-type D. melanogaster larvae were cultured and collected for RNA extraction. Total cellular RNA was isolated and purified according to the method of guanidine thiocyanate/ phenol-chloroform (Chomczynski and Sacchi 1987).
An aliquot of 50 μg total cellular RNA was polydenylated using poly(A) polymerase (Takara). Synthesis of the first strand of cDNA was performed with 25 μg of poly(A)+-tailed RNA in a 20-μL reaction mix containing 200 U of MMLV reverse transcriptase (Promega) and 0.5 μg of primer oligodT23 for 45 min at 42°C. The reaction mixture was separated on a denaturing 8% polyacrylamide gel (8 M urea, 1× TBE buffer). cDNAs with sizes ranging from 60 to 120 nt for box C/D cDNA library and ranging from 120 to 180 nt for box H/ACA cDNA library were excised and eluted from the gel. cDNAs were tailed with poly(dG) at the 3′ end by using terminal deoxynucleotidyl transferase (Takara), and then amplified by PCR with primers Hin dIII(T)16 and Bam HI(C)16, and cloned into plasmid pTZ18 as described previously (Zhou et al. 2002). The two cDNA libraries were screened by PCR with the P47 and P48 universal primer pair. Only the recombinant plasmids carrying fragments of the expected size were selected for sequencing, which was performed with an automatic DNA sequencer (Applied Biosystems, 377) using the Big Dye Deoxy Terminator cycle-sequencing kit (Applied Biosystems).
An aliquot of 30 μg total RNA was analyzed by electrophoresis on 8% acrylamide/7 M urea gels. Electrotransfer onto nylon membrane (Hybond-N+; Amersham) was followed by UV irradiation for 5 min. Hybridization with 5′-labeled probes was performed as previously described (Zhou et al. 2002).
Oligodeoxynucleotides
Oligonucleotides were synthesized and purified by Sangon Co. The sequences of oligonucleotide probes used for Northern blotting and reverse transcription and oligonucleotide primers used for cDNA libraries construction and screening are shown in Supplementary Table S2. The primers and probes used in reverse transcription and Northern blotting were 5′-end labled with [γ-32P] ATP (Yahui Co.) and submitted to purification according to standard laboratory protocols as previously described (Sambrook et al. 1989).
Database accession codes
All novel snoRNA gene and dUHG host gene sequences identified in this study have been deposited in the EMBL and GenBank databases. Accession numbers are shown in Table 3 and Supplementary Table S1.
Supplementary materials
Supplementary materials are available upon request (send an e-mail message containing the keyword “Drosophila snoRNA” to lsbrc04@zsu.edu.cn).
Acknowledgments
We thank Xiao-Hong Chen for her technical assistance and Quan-Shen Du for the D. melanogaster culture. We thank Professor Mohsen Ghadessy and Dr. Roxana S. Ghadessy for improving the text. This research is supported by the National Natural Science Foundation of China (key project 30230200) and the Program for Changjiang Scholars and Innovative Research Team in University from the Ministry of Education of China.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2380905.
REFERENCES
- Accardo, M.C., Giordano, E., Riccardo, S., Digilio, F.A., Iazzetti, G., Calogero, R.A., and Furia, M. 2004. A computational search for box C/D snoRNA genes in the Drosophila melanogaster genome. Bioinformatics 20: 3293–3301. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Antoine, M. and Kiefer, P. 1998. Functional characterization of transcriptional regulatory elements in the upstream region and intron 1 of the human S6 ribosomal protein gene. Biochem. J. 336: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350. [DOI] [PubMed] [Google Scholar]
- Bachellerie, J.P., Cavaille, J., and Qu, L.H. 2000. Nucleotide modifications of eukaryotic rRNAs: The world of small nucleolar RNA guides revisited. In The ribosome: Structure, function, antibiotics and cellular interactions (eds. R.A. Garrett et al.), pp. 191–203. ASM Press, Washington, DC.
- Bachellerie, J.P., Cavaille, J., and Huttenhofer, A. 2002. The expanding snoRNA world. Biochimie 84: 775–790. [DOI] [PubMed] [Google Scholar]
- Balakin, A.G., Smith, L., and Fournier, M.J. 1996. The RNA world of the nucleolus: Two major families of small nucleolar RNAs defined by different box elements with related functions. Cell 86: 823–834. [DOI] [PubMed] [Google Scholar]
- Bergman, C.M. and Kreitman, M. 2001. Analysis of conserved non-coding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 11: 1335–1345. [DOI] [PubMed] [Google Scholar]
- Brown, J.W., Echeverria, M., and Qu, L.H. 2003. Plant snoRNAs: Functional evolution and new modes of gene expression. Trends Plant Sci. 8: 42–49. [DOI] [PubMed] [Google Scholar]
- Cavaille, J., Buiting, K., Kiefmann, M., Lalande, M., Brannan, C.I., Horsthemke, B., Bachellerie, J.P., Brosius, J., and Huttenhofer, A. 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. 97: 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille, J., Seitz, H., Paulsen, M., Ferguson-Smith, A.C., and Bachellerie, J.P. 2002. Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum. Mol. Genet. 11: 1527–1538. [DOI] [PubMed] [Google Scholar]
- Chen, C.L., Liang, D., Zhou, H., Zhou, M., Chen, Y.Q., and Qu, L.H. 2003. The high diversity of snoRNAs in plants: Identification and comparative study of 120 snoRNA genes from Oryza sativa. Nucleic Acids Res. 31: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P. and Sacchi, N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 732–735. [DOI] [PubMed] [Google Scholar]
- Clouet d’Orval, B., Bortolin, M.L., Gaspin, C., and Bachellerie, J.P. 2001. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 29: 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar, D.A., Chen, A.A., Wormsley, S., and Baserga, S.J. 2000. The genes for small nucleolar RNAs in Trypanosoma brucei are organized in clusters and are transcribed as a polycistronic RNA. Nucleic Acids Res. 28: 2855–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flam, F. 1994. Hints of a language in junk DNA. Science 266: 1320. [DOI] [PubMed] [Google Scholar]
- Ganot, P., Bortolin, M.L., and Kiss, T. 1997. Related site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809. [DOI] [PubMed] [Google Scholar]
- Hirose, T. and Steitz, J.A. 2001. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc. Natl. Acad. Sci. 98: 12914–12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Z.P., Zhou, H., Liang, D., and Qu, L.H. 2004. Different expression strategy: Multiple intronic gene clusters of box H/ACA snoRNA in Drosophila melanogaster. J. Mol. Biol. 341: 669–683. [DOI] [PubMed] [Google Scholar]
- Kiss, T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Kruszka, K., Barneche, F., Guyot, R., Ailhas, J., Meneau, I., Schiffer, S., Marchfelder, A., and Echeverria, M. 2003. Plant dicistronic tRNA-snoRNA genes: A new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J. 22: 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E.C., Tomancak, P., Williams, R.W., and Rubin, G.M. 2003. Computational identification of Drosophila microRNA genes. Genome Biol. 4: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader, D.J., Sanders, J.F., Turnbull-Ross, A., Waugh, R., and Brown, J.W. 1995. Genomic organisation of plant U14 snoRNA genes. Biochem. Soc. Trans. 23: 314S. [DOI] [PubMed] [Google Scholar]
- Liang, D., Zhou, H., Zhang, P., Chen, Y.Q., Chen, X., Chen, C.L., and Qu, L.H. 2002. A novel gene organization: Intronic snoRNA gene clusters from Oryza sativa. Nucleic Acids Res. 30: 3262–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon Jr., J.M. 1998. The recent origins of spliceosomal introns revisited. Curr. Opin. Genet. Dev. 8: 637–648. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171. [DOI] [PubMed] [Google Scholar]
- Maxwell, E.S. and Fournier, M.J. 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 35: 897–934. [DOI] [PubMed] [Google Scholar]
- Meredith, J. and Storti, R.V. 1993. Developmental regulation of the Drosophila tropomyosin II gene in different muscles is controlled by muscle-type-specific intron enhancer elements and distal and proximal promoter control elements. Dev. Biol. 159: 500–512. [DOI] [PubMed] [Google Scholar]
- News Staff. 2004. Breakthrough of the year: The runners-up. Science. 306: 2013–2017. [DOI] [PubMed] [Google Scholar]
- Nixon, J.E., Wang, A., Morrison, H.G., McArthur, A.G., Sogin, M.L., Loftus, B.J., and Samuelson, J.A. 2002. Spliceosomal intron in Giardia lamblia. Proc. Natl. Acad. Sci. 99: 3701–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand, J. and Bakin, A. 1997. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archae-bacteria, mitochondria and chloroplasts. J. Mol. Biol. 266: 246–268. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. and Lipman, D.J. 1998. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. 85: 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, L.H., Henras, A., Lu, Y.J., Zhou, H., Zhou, W.X., Zhu, Y.Q., Zhao, J., Henry, Y., Caizergues-Ferrer, M., and Bachellerie, J.P. 1999. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 19: 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, L.H., Meng, Q., Zhou, H., and Chen, Y.Q. 2001. Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 29: 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A.G., Schnare, M.N., and Gray, M.W. 2004. Pseudouridine-guide RNAs and other Cbf5p-associated RNAs in Euglena gracilis. RNA 10: 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2d ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Smith, C.M. and Steitz, J.A. 1997. Sno storm in the nucleolus: New roles for myriad small RNPs. Cell 89: 669–672. [DOI] [PubMed] [Google Scholar]
- Standiford, D.M., Sun, W.T., Davis, M.B., and Emerson Jr., C.P. 2001. Positive and negative intronic regulatory elements control muscle-specific alternative exon splicing of Drosophila myosin heavy chain transcripts. Genetics 157: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M.F., Rifkin, S.A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P.E., et al. 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306: 655–660. [DOI] [PubMed] [Google Scholar]
- Tycowski, K.T. and Steitz, J.A. 2001. Non-coding snoRNA host genes in Drosophila: Expression strategies for modification guide snoRNAs. Eur. J. Cell. Biol. 80: 119–125. [DOI] [PubMed] [Google Scholar]
- Tycowski, K.T., Shu, M.D., and Steitz, J.A. 1996. A mammalian gene with introns instead of exons generating stable RNA products. Nature 379: 464–466. [DOI] [PubMed] [Google Scholar]
- Tycowski, K.T., You, Z.H., Graham, P.J., and Steitz, J.A. 1998. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell 2: 629–638. [DOI] [PubMed] [Google Scholar]
- Tycowski, K.T., Alar, A., and Steitz, J.A. 2004. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. 14: 1985–1995. [DOI] [PubMed] [Google Scholar]
- Yu, J., Yang, Z., Kibukawa, M., Paddock, M., Passey, D.A., and Wong, G.K. 2002. Minimal introns are not “junk.” Genome Res. 12: 1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, G., Klambt, C., Bachellerie, J.P., Brosius, J., and Huttenhofer, A. 2003. RNomics in Drosophila melanogaster: Identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res. 31: 2495–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Chen, Y.Q., Du, Y.P., and Qu, L.H. 2002. The Schizosac-charomyces pombe mgU6–47 gene is required for 2′-O-methylation of U6 snRNA at A41. Nucleic Acids Res. 30: 894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]