Abstract
Protein synthesis is initiated on ribosomal subunits. However, it is not known how 70S ribosomes are dissociated into small and large subunits. Here we show that 70S ribosomes, as well as the model post-termination complexes, are dissociated into stable subunits by cooperative action of three translation factors: ribosome recycling factor (RRF), elongation factor G (EF-G), and initiation factor 3 (IF3). The subunit dissociation is stable enough to be detected by conventional sucrose density gradient centrifugation (SDGC). GTP, but not nonhydrolyzable GTP analog, is essential in this process. We found that RRF and EF-G alone transiently dissociate 70S ribosomes. However, the transient dissociation cannot be detected by SDGC. IF3 stabilizes the dissociation by binding to the transiently formed 30S subunits, preventing re-association back to 70S ribosomes. The three-factor-dependent stable dissociation of ribosomes into subunits completes the ribosome cycle and the resulting subunits are ready for the next round of translation.
Keywords: protein synthesis, sucrose density gradient centrifugation, elongation factor G, initiation factor 3
INTRODUCTION
Protein synthesis consists of initiation, elongation, termination, and recycling of spent ribosomes for the next round of translation. The last step is catalyzed by ribosome recycling factor (RRF) and elongation factor G (EF-G). This step disassembles the model post-termination complexes (PoTCs, consisting of ribosomes, unesterified tRNA, and mRNA) into mRNA, tRNA, and ribosomes (for review, see Kaji et al. 2001). The 70S ribosomes, the working form of ribosomes, have to be dissociated into subunits for the initiation of protein synthesis (Guthrie and Nomura 1968). However, the mechanism of this crucial step is not known.
Historically, this step has been a source of heated discussion and has never been resolved. One point of dispute is whether the dissociation of ribosomes into subunits occurs simultaneously with the release from mRNA (Kaempfer 1970) or subsequent to the release of 70S ribosomes from mRNA (Subramanian and Davis 1973; for a summary of this dispute, see p. 154 of the review by Janosi et al. 1996). Another point of dispute is that Davis and his colleagues believed that initiation factor 3 (IF3) is a dissociation factor that dissociates 70S ribosomes into subunits (Subramanian and Davis 1970), while Kaempfer (1972) claimed that IF3 is an anti-association factor.
The exact role of initiation factors in the subunit dissociation process has not been so clear either. Although IF3 and IF1 were reported to dissociate 70S ribosomes into subunits (Godefroy-Colburn et al. 1975), whether this is the true mechanism by which ribosomes are dissociated into subunits is questionable, based on the following reasons. The ribosome dissociation activity of IF3 by itself can be observed only under nonphysiological conditions such as a high concentration of IF3 or a low Mg2+ ion concentration (Subramanian and Davis 1970; Kaempfer 1972). Indeed, at 7 mM Mg2+ and in the absence of polyamines (and with 70 mM NH4+ and 30 mM K+), 3 μM IF3 and IF1 dissociate the 70S ribosome almost completely (Vila-Sanjurjo et al. 2004). However, in the presence of polyamines (6 mM Mg2+, 2 mM spermidine, 0.05 mM spermine, and 60 mM NH4+), these two factors were not sufficient to dissociate 70S ribosomes completely (Vila-Sanjurjo et al. 2004). We also observed that 4 μM IF3 (in vivo concentration of IF3 in growing Escherichia coli; Howe and Hershey 1983) does not dissociate 70S ribosomes under our conventional buffer conditions (8.2 mM Mg2+, no polyamines, and 80 mM NH4+) or in a buffer containing polyamines (5 mM Mg2+, 8 mM putrescine, 1 mM spermidine, 5 mM NH4+, 95 mM K+) which is optimized for in vitro translation (Jelenc and Kurland 1979; Karimi et al. 1999). Furthermore, the possible role of IF1 in this process has been questioned (Kaempfer 1972).
On the other hand, a model has been proposed suggesting that RRF, EF-G, and GTP dissociate 50S subunits from the PoTC, leaving 30S subunits and bound tRNA on mRNA (Karimi et al. 1999). This model does not explain how mRNA is released from the PoTC. For further details of this model, see the Discussion section.
The data presented below show that 70S ribosomes are transiently dissociated into subunits by RRF and EF-G, and that the subunits are then stabilized by IF3. In the presence of IF3, the model PoTC is disassembled into stable subunits by EF-G and RRF. In this paper, we define “stable subunit dissociation” as “dissociation into subunits detectable by sucrose density gradient centrifugation (SDGC)” and “transient subunit dissociation” as “dissociation into subunits detectable by the light scattering decrease but not by SDGC”. When we refer to simply “subunit dissociation”, this means “stable subunit dissociation”. The stable subunit dissociation by the cooperative action of three factors provides a solution to the longstanding fundamental question of how ribosomal subunits are provided for the initiation step of protein synthesis.
RESULTS
Ribosome preparations used
For this study, we used the following 70S ribosome preparations and ribosomal complexes: (1) washed 70S ribosomes (w-ribosomes; this is conventional high salt-washed ribosomes); (2) tight-coupled 70S ribosomes (tc-ribosomes); (3) re-associated 70S ribosomes (ra-ribosomes); and (4) model post-termination complexes (PoTCs). We used these different preparations of ribosomes because we wished to demonstrate that the three-factor-dependent subunit dissociation is valid regardless of the various ribosome preparations used among the researchers in this field. W-ribosomes were isolated by washing crude ribosomes with 0.5 M salt, which releases most of the ribosome-bound factors. Some of the w-ribosomes may contain deacylated tRNA in the E-site, as observed in recent cryo-electron microscopy studies (Agrawal et al. 2004). Tc-ribosomes were isolated based on the observation that they remain as 70S ribosomes at 4 mM Mg2+ (Agrawal and Burma 1996). The ra-ribosomes are free of possible contamination by tRNA or mRNA. PoTCs were isolated by treating the naturally occurring polysomes with puromycin. The resulting ribosomal complexes contain mRNA and deacylated tRNA in the P/E-site position (Lancaster et al. 2002; Noller et al. 2002), but the A-site is empty.
RRF, EF-G, and IF3 are required for the dissociation of 70S ribosomes into subunits detectable by the sucrose density gradient centrifugation technique
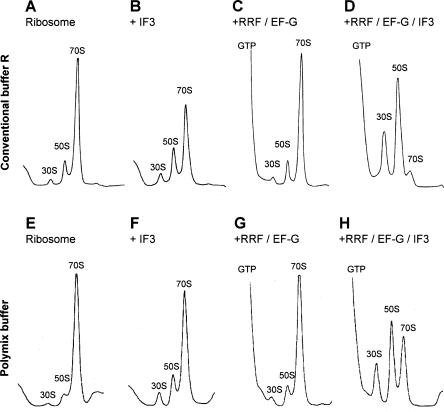
As described in Figure 1 and Table 1, 70S ribosomes were incubated with various factors and the conversion into subunits was analyzed by SDGC. The analysis of the sedimentation behavior of ribosomes through the sucrose gradient detects subunits if the 70S ribosomes are dissociated and formed subunits are stable enough to withstand re-association during centrifugation. When 70S w-ribosomes were incubated with 4.5 μM IF3 alone, only 12% of 70S ribosomes were dissociated into their subunits (Table 1, line a; Fig. 1B). Only when there are very high concentrations of IF3 does complete dissociation occur, as shown in a later section of this paper. This is consistent with the notion that IF3 is an anti-association factor but not a dissociation factor (Kaempfer 1972). IF1 (4.5 μM) alone had no effect (Table 1, line b), but, combined with IF3, had some activity (line c), confirming work by Godefroy-Colburn et al. (1975). Importantly, however, most of the ribosomes still remained as 70S particles even under these conditions. This suggests that these two factors are not sufficient for dissociation of 70S ribosomes into subunits in vivo. Similarly in the absence of initiation factors, 20 μM RRF (in vivo concentration of RRF; Andersen et al. 1999), 20 μM EF-G (in vivo concentration of EF-G; Caldas et al. 2000), and GTP also did not stably dissociate 70S ribosomes into subunits (Table 1, line i; Fig. 1C). This confirms the original observation that ribosomes released from mRNA by these two factors are detected as 70S ribosomes, not subunits, by SDGC (Hirashima and Kaji 1970; Hirokawa et al. 2002). It should be noted that RRF, EF-G, and GTP did not dissociate 70S ribosomes into stable subunits even at 5 or 3.5 mM Mg2+ conditions (data not shown).
FIGURE 1.
In vivo concentrations of RRF, EF-G, and IF3 dissociate 70S ribosomes into subunits almost completely. W-ribosomes (0.07 μM) were incubated alone (A,E), with 4.5 μM IF3 (B,F), with 20 μM RRF, 20 μM EF-G, and 0.36 mM GTP (C,G), and with 20 μM RRF, 20 μM EF-G, 0.36 mM GTP, and 4.5 μM IF3 (D,H) at 30°C for 15 min in conventional buffer R (A–D) or in the polymix buffer (E–H). Sedimentation behavior of ribosomes was analyzed as described in Materials and Methods. The sedimentation is from left to right. The peaks of GTP, 30S, 50S, and 70S ribosome are indicated.
TABLE 1.
Cooperative action of RRF, EF-G, and IF3 for converting 70S ribosomes into subunitsa (IF1 cannot replace IF3)
| Ribosome Incubation with | 70S converted into subunits (%) |
| Washed 70S ribosome (W-ribosome) | |
| a) IF3 (4.5 μM) | 12.0 |
| b) IF1 (4.5 μM) | 0.9 |
| c) IF3 (4.5 μM), IF1 (4.5 μM) | 24.7 |
| d) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), IF3 (4.5 μM) | 68.4 |
| e) RRF (20 μM), EF-G (20 μM), GTP (0.36 mM), IF3 (4.5 μM) | 91.7 |
| f) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), IF1 (4.5 μM) | 2.2 |
| g) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), IF3 (4.5 μM), IF1 (4.5 μM) | 78.2 |
| h) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM) | 4.1 |
| i) RRF (20 μM), EF-G (20 μM), GTP (0.36 mM) | 6.5 |
| j) RRF (1 μM), GTP (0.36 mM), IF3 (4.5μM) | 10.0 |
| k) EF-G (1 μM), GTP (0.36 mM), IF3 (4.5 μM) | 11.6 |
| Tight-coupled 70S ribosome (tc-ribosome) | |
| l) IF3 (4.5 μM) | 6.7 |
| m) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), IF3 (4.5 μM) | 41.2 |
| n) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM) | 5.6 |
| Re-associated 70S ribosome (ra-ribosomes) | |
| o) IF3 (4.5 μM) | 5.1 |
| p) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), IF3 (4.5 μM) | 48.5 |
| q) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM) | 8.1 |
aConversion of w-ribosomes, tc-ribosomes and ra-ribosomes into subunits by indicated factors (at 8.2 mM Mg2+) was analyzed as described in Materials and Methods.
However, when w-ribosomes were incubated with RRF, EF-G, GTP, and IF3 together, significantly more 70S ribosomes were stably dissociated into subunits (68.4% of 70S ribosomes; Table 1, line d). Furthermore, in the presence of in vivo concentrations of these factors (20 μM RRF, 20 μM EF-G, and 4.5 μM IF3) and GTP, most (91.7%) of the 70S w-ribosomes dissociated into subunits (Table 1, line e; Fig. 1D). Omitting EF-G (Table 1, line j), RRF (line k), or IF3 (line h) from the reaction resulted in only 10%, 11.6%, or 4.1% subunit dissociation, respectively. IF1 (4.5 μM) increased conversion by only ~10% when combined with all of the other three factors (Table 1, line g). More importantly, combinations of IF1, RRF, and EF-G did not change the sedimentation profile at all (Table 1, line f). Taken together, these data indicate that RRF, EF-G, and IF3 cooperatively enable the stable dissociation of 70S ribosomes into subunits. IF1 has only a slight effect, if any, on this three-factor-dependent reaction.
One may argue that the buffer condition used (8 mM Mg2+, 80 mM NH4+, and no polyamines) may be such that association of subunits is favored and only under such conditions may factor dependency be observed. It is known that the bond between ribosomal subunits is largely affected by Mg2+ ion, monovalent ions, and polyamines. It is possible that under optimal in vitro protein synthesis conditions, RRF may not be needed for the subunit dissociation. To examine this possibility, the experiment shown in Figure 1E–H was performed in the “polymix” buffer, which was carefully adjusted for the optimum in vitro translation of natural mRNA (Jelenc and Kurland 1979). The polymix buffer contains 5 mM Mg2+, 5 mM K+, 95 mM NH4+, 8 mM putrescine, and 1 mM spermidine. As shown here, even in this buffer, the combination of the three factors was required to stably dissociate 70S ribosomes into subunits. It should be noted that, as shown in Figure 1G, RRF and EF-G did not dissociate 70S ribosomes into subunits detectable by SDGC in this buffer. This does not agree with the statement of Karimi et al. (1999) that “when both EF-G and RRF were added together the profile (sedimentation by sucrose gradient) was significantly shifted toward free subunits” in the polymix buffer (p. 605). Since the difference between their system and ours is only the mRNA context, this result suggests that the mRNA context may affect the extent of the stable subunit dissociation (see Discussion).
In Table 1, lines l–n, we show that similar results were obtained with tc-ribosomes. We tested tc-ribosomes because they are not dissociated at 4 mM Mg2+ concentration (Agrawal and Burma 1996). The data show that these three factors can disrupt even the bonds between the tightly associated subunits.
To examine whether traces of mRNA and tRNA remaining on the tc-ribosomes may influence the factor-dependent dissociation of subunits, we exposed tc-ribosomes to 1 mM Mg2+, which releases residual mRNA and tRNA, if any. Ribosomes were then re-associated by raising the Mg2+ ion concentration. As shown in this table, the same requirements for the three factors were observed for the stable dissociation of these ra-ribosomes into subunits (Table 1, lines o–q).
Presence of RRF, EF-G, and IF3 together is necessary for the stable dissociation into subunits
How do the three factors function to achieve the stable dissociation? To answer this question, we examined whether one or two of these factors would change the 70S ribosomes to isolatable “stable intermediate ribosomes” ready to be dissociated into stable subunits by the addition of the other factors. In the experiment shown in Table 2, the 70S w-ribosomes were treated first with one or two factors, isolated, and washed to remove the factors. The isolated hypothetical “stable intermediate ribosomes” were then treated with the factor(s) not used in the first treatment. For example, 70S ribosomes were pretreated with RRF, EF-G, and GTP, washed, and isolated (Table 2, lines a–c). The isolated ribosomes were then examined for conversion into subunits by addition of IF3 (line b).
TABLE 2.
Pre-incubation of ribosomes with factors does not function; requires the presence of three factors together for conversion of 70S into subunits
| Pre-incubation witha | Incubation with | 70S converted into subunits (%) |
| a) RRF, EF-G, GTP | - | 3.8 |
| b) RRF, EF-G, GTP | IF3 | 14.5 |
| c) RRF, EF-G, GTP | RRF, EF-G, GTP, IF3 | 69.2 |
| d) RRF, IF3 | - | 8.8 |
| e) RRF, IF3 | EF-G, GTP | 20.4 |
| f) RRF, IF3 | RRF, EF-G, GTP, IF3 | 75.8 |
| g) EF-G, GTP, IF3 | - | 15.5 |
| h) EF-G, GTP, IF3 | RRF | 18.1 |
| i) EF-G, GTP, IF3 | RRF, EF-G, GTP, IF3 | 73.8 |
| j) EF-G, GTP | - | 5.6 |
| k) EF-G, GTP | RRF, IF3 | 29.5 |
| l) EF-G, GTP | RRF, EF-G, GTP, IF3 | 79.6 |
| m) IF3 | - | 12.8 |
| n) IF3 | RRF, EF-G, GTP | 25.9 |
| o) IF3 | RRF, EF-G, GTP, IF3 | 77.5 |
aW-ribosomes were pre-incubated with factors at 8.2 mM Mg2+ as indicated in the first column, washed and isolated by microfiltration. The isolated ribosomes were then further incubated at 8.2 mM Mg2+ with other factors as indicated in the second column, and the ribosomal sedimentation was analyzed as described in Materials and Methods. Both in the pre-incubation and the incubation periods, RRF, EF-G, GTP, and IF3 concentrations were 1 μM, 1 μM, 0.36 mM, and 4.5 μM, respectively.
Note that since the ribosome-bound RRF is difficult to remove by the microfiltration procedure (Kiel et al. 2003), pre-incubation with RRF was not performed.
As shown in Table 2, no evidence of “stable intermediate w-ribosomes” was obtained under various combinations of factors. Without any exception, when three factors were simultaneously present with 70S w-ribosomes, the stable dissociation was observed. As discussed later, this experiment does not detect transient change, which may be caused by one of the factors. The data clearly show however that no change on the ribosome by treatment with one or two of the factors took place lasting long enough to withstand washing and the period between additions of other factors. We should mention that a well accepted requirement for the simultaneous presence of EF-G and RRF for the disassembly of the PoTC was established in this manner (Hirashima and Kaji 1973). We conclude from these data that the maximum dissociation was observed when all three factors were present together.
Three-factor-dependent stable dissociation of ribosome is strictly dependent on GTP; nonhydrolyzable GTP analog does not function
As shown in Table 3, it is clear that GTP is required for the three-factor-dependent stable dissociation of ribosomes into subunits. Neither GDP, GMPPCP (a nonhydrolyzable GTP analog), nor ATP functioned. This strict requirement for GTP and the inability of GMPPCP to cause subunit dissociation is reminiscent of the same requirements for the disassembly of PoTCs (Hirashima and Kaji 1973; Karimi et al. 1999; Hirokawa et al. 2002). This is in contrast to the conventional translocation reaction, which can take place with GMPPCP at a reduced rate (Rodnina et al. 1997). It should be noted however that a partial reaction of the disassembly by RRF and EF-G, namely the release of tRNA from the model PoTC, was observed with GMPPCP (Hirokawa et al. 2002). It appears that the release of tRNA by RRF and EF-G (Hirashima and Kaji 1973; Hirokawa et al. 2002) is very similar to the translocation of tRNA by EF-G, while the release of mRNA and the subunit formation by EF-G and RRF (and IF3) are distinctly different from the tRNA release reaction and the usual translocation reaction.
TABLE 3.
Nucleotide specificity for three-factor-dependent subunits formationa
| Nucleotides | 70S converted to subunits (%) | Activity (%) |
| a) GTP | 68.4 | 100.0 |
| b) No nucleotide | 14.2 | 20.7 |
| c) GDP | 20.4 | 29.9 |
| d) GMPPCP | 17.2 | 25.1 |
| e) ATP | 18.9 | 27.7 |
aW-ribosomes were incubated with RRF (1 μM), EF-G (1 μM), and IF3 (4.5 μM) in the presence of 0.36 mM GTP or GDP or GMPPCP, or ATP at 30°C for 15 min in 275 μL of conventional buffer R, and sedimentation behavior of ribosomes was analyzed as in Table 1.
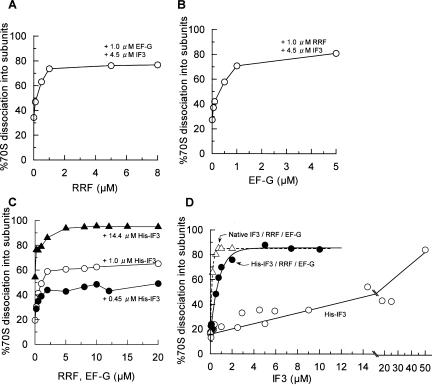
Quantitative relationship among IF3, RRF, and EF-G
In Figure 2A or 2B, a dose-response curve for RRF or EF-G in the presence of fixed amounts of EF-G (1 μM)/IF3 (4.5 μM) or RRF (1 μM)/IF3 (4.5 μM) is noted, respectively. As shown, the stable dissociation of 70S w-ribosomes into subunits was maximized when equal molar amounts of RRF and EF-G were present. Since the maximum disassembly of the model PoTC was observed with equal molar amounts of RRF and EF-G (Hirashima and Kaji 1972b), these data suggest that the stable dissociation of 70S ribosomes into subunits is similar to the disassembly of the model PoTC into mRNA, tRNA, and ribosomes observed by SDGC.
FIGURE 2.
Dose-response curve of factors for the dissociation of 70S ribosomes into subunits. W-ribosomes (0.07 μM) were incubated with factors at 30°C in conventional buffer R. Sedimentation of ribosomes was analyzed as in Figure 1, and the amounts of 70S ribosomes converted into subunits are expressed as percentages. (A) RRF dose-response curve with 1 μM EF-G, 4.5 μM His-IF3, and 0.36 mM GTP. (B) EF-G dose-response curve with 1 μM RRF, 4.5 μM His-IF3, and 0.36 mM GTP. (C) RRF/EF-G dose-response curve. (•) 0.45 μM His-IF3, various amounts of RRF, EF-G, and 0.36 mM GTP. (○) As above except for 1 μM His-IF3. (▴) 14.4 μM His-IF3. (D) IF3 dose-response curve. ○, His-IF3; •, His-IF3, 5 μM RRF, 5 μM EF-G and 0.36 mM GTP; ▵ and dotted line, as above except that native IF3 was used instead of His-IF3.
We then kept the molar ratio of RRF and EF-G at 1, and dose-response curves of these factors in the presence of three different concentrations of IF3 were performed (Fig. 2C). As shown in this figure, the final extent of subunit formation was determined by the concentration of IF3, and excess amounts of EF-G and RRF did not compensate for IF3. It should be noted that with all concentrations of IF3 tested, the dissociation did not increase beyond 5 μM RRF and EF-G.
As shown in Figure 2D, we then examined the dose-response curve of IF3 in the presence of saturating conditions of RRF (5 μM) and EF-G (5 μM). Under this experimental condition, the subunit dissociation was maximized with 2 μM histidine-tagged IF3 (His-IF3). Since modified IF3 may not behave exactly like the native IF3, we also constructed a dose-response curve of native IF3. It is clear from Figure 2D that native IF3 was more active than His-IF3, and that the subunit dissociation is already maximized at 0.5 μM IF3. It is noted that, without RRF and EF-G, a very high concentration of His-IF3 (50 μM) was required to dissociate 70S ribosomes into subunits under this buffer condition.
Stable dissociation from model post-termination complexes
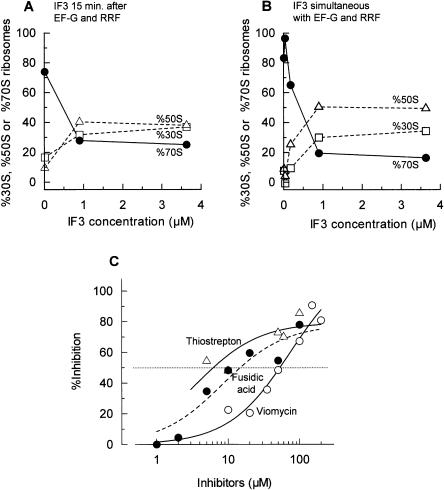
In order to further explore whether the three-factor-dependent stable dissociation from 70S ribosomes into subunits described above is important in the ribosome cycle, we examined the stable subunit dissociation from the model PoTC. Based on the finding that 70S ribosomes are dissociated into subunits by the three factors, we expected that PoTCs would be dissociated into stable subunits by RRF, EF-G, and IF3. We show just that in Figure 3A. In this experiment, the model PoTC was first treated with RRF and EF-G for 10 min. As described in Figure 4 and the Discussion, the observed 70S ribosomes in Figure 3 are re-associated ribosomes after they are dissociated into subunits by RRF and EF-G. We then added various amounts of IF3. As expected, the stable subunit dissociation took place upon addition of IF3. In the presence of 3.6 μM IF3, most (80%) of the ribosomes were dissociated. The amount of subunit dissociation was dependent on the amount of IF3 added, as in the case with w-ribosomes (Fig. 2C,D).
FIGURE 3.
Subunits dissociation from the model post-termination complexes by three factors. (A,B) IF3 dose-response curve. (A) Polysomes (0.6 A260 units) were incubated with puromycin (50 μM), RRF (1 μM), EF-G (1 μM), and GTP (0.36 mM) at 30°C for 10 min in conventional buffer R. Then, IF3 (various amounts) were added and further incubated at 30°C for 5 min. (B) Polysomes (0.6 A260 units) were incubated with puromycin (50 μM), RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), and IF3 (various amounts) at 30°C for 15 min in conventional buffer R. Sedimentation behavior of ribosomes was analyzed as in Figure 1. The percentages of 30S subunits (□, dotted line), 50S subunits (▵, dotted line), and 70S ribosomes (•, solid line) are plotted against added IF3 concentrations. (C) Effects of EF-G inhibitors on the subunit dissociation from the model PoTC by RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), and IF3 (4.5 μM) in conventional buffer R were examined as in B. ○ and solid line, viomycin; ▵ and solid line, thiostrepton; • and dotted line, fusidic acid. Percent inhibition of the increase of subunits was calculated and is plotted against the concentrations of inhibitors.
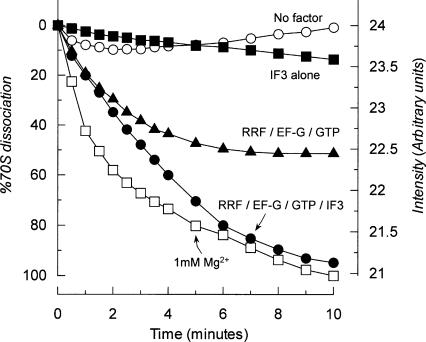
FIGURE 4.
70S ribosomes are transiently dissociated by RRF and EF-G alone. (•) w-ribosomes (0.16 μM) were incubated with RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), and IF3 (4.5 μM) at 30°C in buffer U, and the dissociation was observed following the decrease of the light scattering as described in Materials and Methods. (▴) With RRF (1 μM), EF-G (1 μM), and GTP (0.36 mM). (▪) With IF3 (4.5 μM) alone; ○ with no factors. (□) W-ribosomes (0.16 μM) were dissociated by placing into buffer S (1 mM Mg2+). The data are calculated as percentages of 70S ribosomes dissociated using the dissociation at 1 mM Mg2+ as 100%.
We then investigated whether pre-incubation with RRF and EF-G is necessary. As shown in Figure 3B, almost identical results were obtained when three factors were added simultaneously. We therefore could not determine whether or not subunit dissociation takes place simultaneously with the release of mRNA. However, these experiments established that the subunit dissociation can indeed occur during the disassembly of the model PoTC.
Effects of EF-G inhibitors on the stable dissociation of subunits
Since the involvement of EF-G in subunit dissociation has not been studied in detail, we wanted to establish the importance of the involvement of EF-G in this reaction. In the experiment shown in Figure 3C, we examined the inhibitors of EF-G on the stable dissociation into subunits from the model PoTC. We found that all of the EF-G inhibitors tested had inhibitory effects. The most efficient inhibitors were thiostrepton (50% at 6 μM), followed by fusidic acid (15 μM) and viomycin (50% at 50 μM; see Table 4A). Similar results were obtained even when the addition of IF3 was delayed (Table 4B). These concentrations of the inhibitors are known to inhibit the EF-G reaction (Bodley et al. 1970; Rodnina et al. 1997). These data indicate that EF-G indeed participates in the dissociation of 70S ribosome into subunits.
TABLE 4.
Comparison of concentrations of inhibitors required for 50% inhibition of stable subunits formation from the model PoTC by RRF/EF-G/IF3 under two different conditions
| Concentration required for 50% inhibition of | ||
| Inhibitors | (A) Subunits formation from PoTC by RRF/EF-G/IF3 simultaneouslya | (B) Subunits formation from PoTC by pre- incubation with RRF/EF-G, then IF3b |
| Thiostrepton | 6.0 μM | 6.5 μM |
| Fusidic acid | 15.0 μM | 20.0 μM |
| Viomycin | 50.0 μM | Not determined |
aEstimated from Figure 4.
bModel PoTC was first incubated with RRF (1 μM), EF-G (1 μM), and GTP (0.36 mM) for 15 min at 30°C. Then inhibitors and IF3 (4.5 μM) were added.
We have so far found three disassembly reactions involving EF-G. They are (1) disassembly of the model PoTC into mRNA, tRNA, and ribosomes by RRF and EF-G (Hirashima and Kaji 1973; Hirokawa et al. 2002); (2) stable dissociation of 70S ribosomes into subunits by RRF, EF-G, and IF3 (Fig. 1); and (3) the stable dissociation of the model PoTC into subunits and other components by RRF, EF-G, and IF3 (Fig. 3A,B). We suspect that they are all the same reaction, only observed under different conditions. If so, we would expect that the inhibitory concentrations of the EF-G inhibitors on these reactions would be similar. Table 4 shows that this notion is indeed true with cases (1) and (3). Two inhibitors, fusidic acid and thiostrepton inhibit reaction (3) under two different conditions at almost the same concentrations (shown in Table 4). The effective inhibitory concentrations of these inhibitors match the effective concentrations of reaction (1) (Hirokawa et al. 2002). It appears that EF-G participates in these disassembly reactions in a similar manner.
Three-factor-dependent stable dissociation does not take place at high Mg2+
It is well known that ribosomal subunits are tightly bound to each other at higher Mg2+ concentrations. We therefore examined whether the three-factor-dependent subunit formation would take place even at high Mg2+ concentrations. As shown in Table 5, very little dissociation took place at 14 mM Mg2+, if any at all. No dissociation was observed at 20 mM Mg2+. Since the disassembly of the model PoTC is not observed at 15 mM Mg2+ (Hirashima and Kaji 1973), these data further support the notion that the disassembly of PoTC by EF-G and RRF is a reaction similar to the three-factor-dependent dissociation of 70S ribosomes into subunits described in this paper.
TABLE 5.
High Mg2+ concentrations inhibit three-factor-dependent conversion of 70S ribosomes into subunitsa
| Mg2+ concentration Incubation with | 70S converted into subunits (%) |
| 14 mM Mg2+ | |
| a) - | - |
| b) IF3 (4.5 μM) | 1.9 |
| c) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), IF3 (4.5 μM) | 15.9 |
| 20 mM Mg2+ | |
| a) - | - |
| b) IF3 (4.5 μM) | 3.8 |
| c) RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), IF3 (4.5 μM) | 3.2 |
aConversion of w-ribosomes into subunits at 14 or 20 mM Mg2+ was examined as described in Table 1 except that buffer V (10 mM Tris-Cl, pH 7.4, 14 mM MgSO4 , 80 mM NH4Cl, 0.2 mM DTT) or buffer W (10 mM Tris-Cl, pH 7.4, 20 mM MgSO4 , 80 mM NH4Cl, 0.2 mM DTT) was used instead of conventional buffer R.
70S ribosomes are transiently dissociated by RRF and EF-G
All of the experiments described above depended on the dissociation of subunits that were stable enough to be detected by the SDGC technique. We then wondered whether RRF and EF-G alone may transiently dissociate the 70S ribosomes into subunits but not be detectable by SDGC. Such a transient dissociation must be detectable by the decrease of light scattering by 70S ribosomes upon dissociation into subunits (Godefroy-Colburn et al. 1975; Goss et al. 1980). As shown in Figure 4, such experiments yielded evidence of transient dissociation of 70S ribosomes into subunits by RRF and EF-G without IF3.
In Figure 4, open squares (□) show the real-time dissociation kinetics of 0.16 μM 70S w-ribosomes upon exposure to 1 mM Mg2+. The rate constant for dissociation was 0.013 ± 0.001 s−1, and this agrees with the published value (Goss et al. 1980). Then, the 70S w-ribosomes were incubated with factors at 5 mM Mg2+. In the presence of 1 μM RRF, 1 μM EF-G and 4.5 μM IF3, almost all of the ribosomes were dissociated after 10 min of incubation (closed circles,•.). Importantly, even in the absence of IF3, significant dissociation (~50%) by RRF and EF-G was observed (closed triangles, ▴). This shows that 70S ribosomes are indeed dissociated by RRF and EF-G alone. IF3 (4.5 μM) alone did not decrease the light scattering significantly (closed squares, ▪).
It is noted in Figure 4 that in the presence of IF3, the extent of the dissociation is increased (cf. ▴, •.). This suggests that equilibrium between subunits and the 70S ribosome is shifted to dissociation by IF3, because IF3 removes available 30S subunits from this equilibrium between 70S ribosomes and subunits (see the next section).
It would have been ideal to examine the transient dissociation under ionic conditions identical to those of the preceding figures (8 mM Mg2+ in conventional buffer). However, due to the abnormal increase of the scattering of 70S ribosomes alone under these conditions, we could not get reliable values. Under the ionic condition used in Figure 4 (5 mM Mg2+ in conventional buffer), the transient dissociation by EF-G and RRF was not observed by the SDGC technique (data not shown).
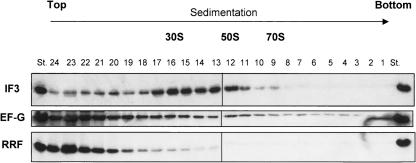
IF3 stabilizes the transiently dissociated 30S subunits
We then tested whether IF3 stays on the 30S subunit after the stable subunit dissociation from 70S ribosomes by the three factors. As shown in Figure 5, 70S w-ribosomes were incubated with RRF, EF-G, GTP, and IF3, and then subjected to SDGC. Fractions were taken from the bottom of the tube, and then RRF, EF-G, or IF3 was detected by Western blotting. As can be seen from this figure, most of the IF3 was detected on 30S subunits, whereas RRF and EF-G were detected mostly in the supernatant fractions. The IF3–30S interaction is stable enough to withstand centrifugation through sucrose solution. The data suggest that, after the transient dissociation of 70S ribosomes into subunits by RRF and EF-G, IF3 stabilizes subunits as an anti-association factor (Kaempfer 1972). Therefore, the subunits are stable enough to be detected by SDGC.
FIGURE 5.
IF3, not RRF/EF-G, stays on subunits after the subunit formation by three factors. W-ribosomes (0.07 μM) were dissociated into subunits by 1 μM RRF, 1 μM EF-G, 0.36 mM GTP, and 4.5 μM IF3 in conventional buffer R, then sedimented through a 15%–30% sucrose gradient. Fractions were taken from the bottom of the sucrose gradient (10 drops per fraction, 24 fractions), and IF3, EF-G, and RRF were detected by Western blotting. Positions of 30S, 50S, and 70S are indicated. St (standard), purified IF3, EF-G, and RRF as controls.
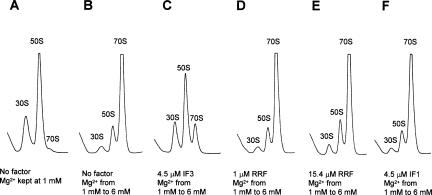
RRF does not have anti-association activity
Our finding that RRF, together with EF-G, has ribosome dissociation activity prompted us to examine whether RRF by itself has anti-association activity like IF3. This is a reasonable question, because RRF binds to 50S subunits (Ishino et al. 2000; Hirokawa et al. 2002) at the intersubunit face (Lancaster et al. 2002; Agrawal et al. 2004; Wilson et al. 2004), and RRF bound to 50S subunits cannot be released by EF-G (Kiel et al. 2003). Ribosomes were exposed to a solution of 1 mM Mg2+ (Fig. 6A) and then to 6 mM Mg2+ (Fig. 6B) in the presence of IF3 (Fig. 6C), RRF (Fig. 6D,E), or IF1 (Fig. 6F). It is clear that either 1 or 15 μM RRF (Fig. 6, cf. D and E with B) did not prevent subunits from associating in the presence of 6 mM Mg2+. In contrast, a control experiment with 4.5 μM IF3 did prevent significant amounts of subunits from associating into 70S ribosomes (Fig. 6C). This is consistent with the observation that the three factors (including IF3) are necessary to form stable subunits. As expected, IF1 alone did not have any anti-association activity (Fig. 6F).
FIGURE 6.
RRF does not have anti-association activity. (A,B) W-ribosomes (0.07 μM) were incubated alone, (C) with 4.5 μM IF3, (D) with 1 μM RRF, (E) with 15.4 μM RRF, and (F) with 4.5 μM IF1, at 30°C for 5 min in buffer S (containing 1 mM MgSO4). Then, the MgSO4 concentration of B–F, but not of A, was raised to 6 mM and further incubated at 30°C for 10 min. Ribosomes were sedimented through a 15%–30% sucrose density gradient in buffer S (A) or buffer T (containing 6 mM MgSO4; B–F) and analyzed as in Figure 1. The sedimentation is from left to right. The peaks of 30S, 50S, and 70S ribosome are indicated.
DISCUSSION
The most important point in this paper is that RRF, EF-G, and IF3 together caused the stable dissociation of 70S ribosomes and the model PoTC into subunits (Fig. 7A–D). This finding is important because this fills the long-standing historical gap between the termination (Capecchi 1967; Caskey et al. 1968) and the initiation steps (Guthrie and Nomura 1968). In support of this finding, there are some recent observations consistent with the notion that RRF may be involved in the dissociation of 70S ribosomes into subunits in vivo. Inactivation of RRF in vivo leads to accumulation of 70S ribosomes. These 70S ribosomes can still translate poly(U) or leaderless mRNA, but they cannot translate conventional mRNA with canonical initiation sequences (Hirokawa et al. 2004; Moll et al. 2004). Furthermore, our present observation of the active involvement of IF3 in the stable dissociation of subunits is consistent with the report that in vivo inactivation of IF3 results in accumulation of 70S ribosomes (Olsson et al. 1996).
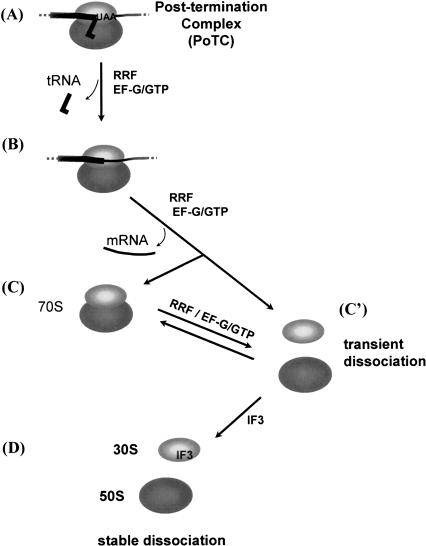
FIGURE 7.
Model of ribosomal subunit formation from a post-termination complex. (A) Post-termination complex with bound tRNA remained after releasing the nascent peptide. (B) Complex without tRNA due to the action of EF-G and RRF (Hirashima and Kaji 1973; Hirokawa et al. 2002). (C) 70S ribosome is free of mRNA and tRNA; (C′) transiently dissociated subunits by RRF and EF-G without IF3. (D) Stable subunits with IF3.
Figure 7 shows our hypothesized series of events during ribosome recycling. Our hypothesis is based upon the data presented in this study and those published. Complex A is the post-termination complex, which has mRNA and deacylated tRNA. In the first step, deacylated tRNA is released from the complex A by RRF and EF-G (Hirashima and Kaji 1973; Hirokawa et al. 2002), resulting in the intermediate complex, complex B. Recent experiments suggested that RRF moves to the second site on the ribosome during this process from A to B in a fashion analogous to that of tRNA movement during the translocation step (Hirokawa et al. 2002; Kiel et al. 2003; Raj et al. 2005). The next step is still uncertain. There are two possible pathways from the intermediate complex B leading to the transiently dissociated form of subunits (C′): (1) B → C (70S ribosome) → C′, or (2) B → C′ directly. Ever since the discovery of RRF by using the SDGC technique, we observed conversion of puromycin-treated polysomes to monosomes by RRF and EF-G, leading us to believe that mRNA is released from the intermediate complex B to form C (Hirashima and Kaji 1973). We now realize that the real product, produced from the model PoTC by interaction with these two factors (RRF and EF-G), is not the 70S ribosome (C) but the transiently dissociated form of ribosome (C′). The transiently dissociated form of ribosome (C′) is then stabilized by IF3. The transiently dissociated subunits (C′) associate back to 70S ribosomes during SDGC because of the absence of RRF and EF-G in the sucrose gradient and are detected as C.
In light of this new finding, the intermediate formation of 70S ribosome (C) may not be necessary during the ribosome recycling process. Rather, it is more reasonable to assume that the 70S ribosome is transiently dissociated by RRF and EF-G at the same time that mRNA is detached from the ribosome by these factors (step from B to C′ directly). Otherwise, two RRF molecules (one for A through B to C and the other for C to C′) would be required for the recycling of one ribosome, because the release of the initially bound RRF from the ribosome is necessary to release mRNA (Raj et al. 2005). The dissociation of 70S ribosomes into subunits and the release of ribosomes from mRNA by RRF and EF-G measured by SDGC appear to share the same mechanism, as suggested by their equal sensitivity to the EF-G inhibitors (Table 4; Fig. 3C). This is consistent with the notion that a direct pathway from B to C′ takes place.
Despite this evidence, we hasten to point out that we still do not have compelling direct evidence to disprove the existence of the intermediate 70S ribosome (C in Fig. 7) during the recycling process. It is possible that the intermediate C is formed under physiological conditions where rapid ribosome recycling is not necessary. Indeed, it is well known that 70S ribosomes are accumulated in the cell by the energy source shift-down (Ruscetti and Jacobson 1972), and that during the stationary phase, most ribosomes exist as 100S ribosomes (the dimerized form of 70S ribosomes) (Wada et al. 1990). Although the possibility of the intermediate 70S ribosome during recycling remains, we must state clearly that our previous conclusion that the final products of the disassembly reaction of the PoTC by RRF and EF-G are 70S ribosomes (Kaji et al. 2001) has to be modified, because RRF and EF-G do indeed transiently dissociate 70S ribosomes into subunits.
The concept that the dissociation of 70S ribosomes into subunits by RRF and EF-G alone (C to C′) is transient is supported by the following observations. (1) The light scattering measurement of ribosomes (Fig. 4) but not the SDGC technique detected the dissociation by RRF and EF-G (Fig.1; Table 1; Hirashima and Kaji 1972a; Kaji et al. 2001; Hirokawa et al. 2002). (2) The simultaneous presence of RRF, EF-G, and IF3 was required to detect the stable dissociation of 70S ribosomes into subunits (Table 2), and the amount of IF3 determined the final extent of subunit dissociation (Fig. 2C). Thus, the transient intermediate (C′ in Fig. 7) rapidly goes back to 70S ribosomes (C) during SDGC in the absence of IF3. (3) RRF or EF-G does not have an anti-association property detectable by the SDGC technique (Fig. 6).
How do RRF and EF-G transiently dissociate 70S ribosomes into subunits? Domain I of RRF is known to interact with helices 69 and 71 of 23S rRNA (Lancaster et al. 2002; Agrawal et al. 2004; Wilson et al. 2004). Since these helices 69 and 71 of 23S rRNA interact with helix 44 of 16S rRNA forming the intersubunit bridges B2a and B3, respectively (Yusupov et al. 2001), the possible breakage of bridges B2a and B3 by the EF-G-dependent movement of RRF (Hirokawa et al. 2002; Kiel et al. 2003; Raj et al. 2005) may cause the transient dissociation into subunits. However, we believe that the possible disruption of B2a and B3 by RRF is not enough to cause the dissociation of ribosomes into subunits. This disruption may prepare the ribosome for the next step where the actual dissociation takes place. We postulate that the disruption of some other intersubunit bridge(s) such as B2b and B2c by the EF-G-dependent motion of RRF, resulting in its release from the E-site of the ribosome (Raj et al. 2005), is required for the dissociation of the 70S ribosomes into subunits. This is based on the observation that the complete disassembly of the model PoTC requires the EF-G-dependent release of RRF from the site where RRF is moved by EF-G (Raj et al. 2005). This site to which RRF is moved overlaps with the E-site (V.S. Raj, H. Kaji, and A. Kaji, unpubl.). The transient dissociation of subunits by RRF and EF-G makes it possible for IF3 to enter the intersubunit space to convert the transiently dissociated form (C′) to the stable form (D).
Consistent with the above hypothesis, IF3 is found on the 30S subunits formed by the three-factor-dependent dissociation of 70S ribosomes into subunits (Fig. 5). The C-terminal domain of IF3, which is important for the anti-association or dissociation activity of IF3 (Petrelli et al. 2001), was shown to be localized at the interface side of the 30S subunit (McCutcheon et al. 1999), particularly helices 23, 24, and 45 of 16S rRNA (Dallas and Noller 2001). The interaction between helices 24 and 45 of 16S rRNA and helices 67 and 69 of 23S rRNA comprises the intersubunit bridge B2b (Yusupov et al. 2001), which may be disrupted during the release of RRF from the ribosome as discussed above. Due to its anti-association property (Kaempfer 1972; Gualerzi et al. 2001), IF3 interacting with the bridge B2b can now stably keep the subunits separated to produce D of Figure 7, which can be detected by the SDGC technique. It is noted that, for the IF3 binding, deacylated tRNA at the P/E site (Lancaster et al. 2002) has to be removed first (A, B in Fig. 7) because the binding site of IF3 may overlap with the E-site tRNA (Dallas and Noller 2001).
As mentioned in the Introduction, Karimi and colleagues proposed a different scheme based on their data (Karimi et al. 1999). In their scheme, the 50S subunit is first dissociated from the PoTC by RRF and EF-G prior to the release of tRNA. This is not transient dissociation, because they were able to detect this dissociation by the SDGC technique. After the dissociation of the 50S subunit, the complex of tRNA, mRNA, and 30S subunits remained. Then, tRNA is released from the complex by IF3, in analogy with the removal of aminoacyl tRNA from the complex of the 30S subunit and mRNA by IF3 reported earlier (Gualerzi et al. 1971). This is different from what we observed with our systems of natural MS2 phage mRNA (Ogawa and Kaji 1975) as well as with the model PoTC (Hirashima and Kaji 1972a). In our system, both tRNA and mRNA are released from the ribosome by the action of RRF and EF-G. IF3 is necessary for the stable dissociation of subunits. The differences between our conditions and theirs are the buffers used and the sequence of mRNAs. However, under the buffer conditions identical to those used in the Karimi experiment, we could not demonstrate the stable subunit dissociation of 70S ribosomes by EF-G and RRF (Fig. 1G). Thus, the difference in the behavior of ribosomes in these two systems must be due to the difference in the mRNA sequences. Actually, this is not unexpected, because it is known that the behavior of ribosomes at the termination codon is strongly influenced by the mRNA sequence surrounding the termination codon. For example, not all the ribosomes that complete one round of translation leave mRNA at the border of two translationally coupled open reading frames (ORFs) (Inokuchi et al. 2000).
Our model PoTC system is different from natural PoTC because the A-site codon is not the termination codon. On the other hand, the Karimi system has the A-site termination codon, but their short mRNA has a strong SD sequence and 3′ poly(A) tails very close to the termination codon. Thus, both systems do not truly represent a typical naturally occurring PoTC. It is necessary to carry out more defined in vitro experiments using natural mRNA.
MATERIALS AND METHODS
Buffers
Conventional buffer R: 10 mM Tris-Cl, pH 7.4, 8.2 mM MgSO4, 80 mM NH4Cl, 0.2 mM DTT.
Polymix buffer (Karimi et al. 1999): 5 mM Mg(OAc)2, 0.5 mM CaCl2, 5 mM NH4Cl, 95 mM KCl, 8 mM putrescine, 1 mM spermidine, 5 mM potassium phosphate, pH 7.3, 1 mM DTT.
Buffer S: 10 mM Tris-Cl, pH 7.4, 1 mM MgSO4, 80 mM NH4Cl, 0.2 mM DTT.
Buffer T: 10 mM Tris-Cl, pH 7.4, 6 mM MgSO4, 80 mM NH4Cl, 0.2 mM DTT.
Buffer U: 10 mM Tris-Cl, pH 7.4, 5 mM MgSO4, 80 mM NH4Cl, 0.2 mM DTT.
Ribosomes and factors
NH4Cl-washed ribosomes (w-ribosomes) and tight-coupled ribosomes (tc-ribosomes) were prepared from E. coli MRE600 cells (purchased from the University of Alabama Fermentation Facility) as described in Kiel et al. (2003) and Makhno et al. (1988), respectively. RRF and EF-G were purified as described (Hirashima and Kaji 1972b; Kiel et al. 2003). N-terminal His-tagged IF1 and IF3 (Shimizu et al. 2001) were purified from XL1-Blue cells harboring plasmids expressing His-IF1 and His-IF3 (obtained from Dr. T. Ueda, Tokyo University). Native IF3 was obtained from Dr. C.O. Gualerzi, University of Camerino. His-IF3 was used in most of the experiments unless otherwise mentioned.
Preparation of re-associated ribosomes (ra-ribosomes)
Tc-ribosomes were incubated in buffer S at 30°C for 5 min, and the resulting ribosomal subunits were isolated. The subunits were then re-associated by exposing them to the buffer containing 20 mM MgSO4 at 30°C for 7 min.
Model termination complexes and model post-termination complexes (PoTC)
Polysomes with peptidyl tRNA were prepared from E. coli Q13 cells as described previously (Hirashima and Kaji 1972a). The PoTC was formed by treating the polysomes with puromycin (see methods below for the dissociation assay).
Ribosome dissociation assay by sedimentation through the sucrose gradient
W-ribosomes (0.07 μM), tc-ribosomes (0.05 μM), and ra-ribosomes (0.05 μM) were incubated with factors as specified in the table and figure legends at 30°C for 15 min in 275 μL of conventional buffer R or in a polymix buffer. For production of subunits from the model post-termination complexes, isolated polysomes (0.6 A260 units) were incubated with factors at 30°C for 15 min in 275 μL of conventional buffer R containing 50 μM puromycin. Ribosomes were sedimented through a 15%–30% sucrose density gradient in the same buffer by ultracentrifugation (Beckman SW50.1 rotor, 40,000 rpm, 2.5 h at 4°C), and the sedimentation behavior was monitored by an ISCO UA-6 spectrophotometer.
Pre-incubation of ribosomes with factors
W-ribosomes (0.35 μM) were pre-incubated with factors for 15 min in 275 μL of conventional buffer R, and then isolated by microfiltration (Nanosep 300K, Pall Life Sciences) at 3000g followed by six washings with 300 μL of conventional buffer R. The isolated ribosomes (0.07 μM) were then used for dissociation into subunits.
Dissociation of ribosomes measured by light scattering decrease
Light scattering experiments were performed at 30°C with the Perkin-Elmer 650–10S Fluorescent Spectrophotometer (excitation, 2 mm slit; emission, 5 mm slit; wave-length at 436 nm at 90° angle). Mixture A (1.35 mL) containing factors in buffer U as specified in the legend of Figure 4 was pre-incubated at 30°C for 2 min. Then mixture B (150 μL, pre-incubated at 30°C) containing 1.6 μM w-ribosomes in the same buffer was added to mixture A and mixed with a magnetic stirrer. Recording of the scattering signal started 2 sec after the addition. The signal corresponding to 100% dissociation was determined by measuring the scattering of w-ribosomes (0.16 μM) in buffer S (1 μM Mg2+).
Western blotting of RRF, EF-G, and IF3
W-ribosomes (0.07 μM) were incubated with 1 μM RRF, 1 μM EF-G, 0.36 μM GTP, and 4.5 μM IF3 in conventional buffer R at 30°C for 15 min, then layered onto a 15%–30% sucrose gradient in the same buffer. After the ultracentrifugation (Beckman SW50.1, 2.5 h, 4°C), fractions were taken from the bottom of the tube (10 drops per fraction), and the A260 units were measured to determine the positions of 30S, 50S, and 70S. Proteins in each fraction were precipitated by adding 20 μL of 100% TCA, washed by a mixture of ether/ethanol (50% each), and subjected to 12% SDS-PAGE. RRF, EF-G, and IF3 were detected by rabbit antiserum against RRF (1:12,500 dilution), against EF-G (1:12,500 dilution), and mouse monoclonal antibody against histidine-tag (0.4 μg/mL, QIAGEN), respectively.
Anti-association analysis
W-ribosomes (0.07 μM) were incubated at 30°C for 5 min in 275 μL of buffer S. Then, the MgSO4 concentration was raised to 6 mM both with and without factors and then further incubated at 30°C for 10 min. Ribosomes were sedimented through a 15%–30% sucrose density gradient in buffer T by ultracentrifugation (Beckman SW50.1 rotor, 40,000 rpm, 2.5 h at 4°C), and the sedimentation behavior was monitored with the ISCO UA-6 spectrophotometer.
Acknowledgments
We thank Dr. Michael C. Kiel (University of Pennsylvania) and Dr. Anna Maria Giuliodori (University of Camerino) for helpful discussions and critical comments; Dr. Takuya Ueda (University of Tokyo) for kindly providing the expression plasmids for IF3 and IF1; Dr. Claudio O. Gualerzi (University of Camerino) for kindly providing the purified native IF3; and Drs. Jane Vanderkooi, Tomoko Ohnishi, and Takahiro Yano (University of Pennsylvania) for the use of the light scattering measuring device. This work was supported by NIH grant GM60429, The Creative Biomedical Research Institute, the Nippon Paint Research Fund, and a grant-in-aid from the Japan Society for the Promotion of Science (JSPS). G.H. is a JSPS Research Fellow.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2520405.
REFERENCES
- Agrawal, R.K. and Burma, D.P. 1996. Sites of ribosomal RNAs involved in the subunit association of tight and loose couple ribosomes. J. Biol. Chem. 271: 21285–21291. [DOI] [PubMed] [Google Scholar]
- Agrawal, R.K., Sharma, M.R., Kiel, M.C., Hirokawa, G., Booth, T.M., Spahn, C.M.T., Grassucci, R.A., Kaji, A., and Frank, J. 2004. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc. Natl. Acad. Sci. 101: 8900–8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, L.D., Moreno, J.M.D., Clark, B.F.C., Mortensen, K.K., and Sperling-Petersen, H.U. 1999. Immunochemical determination of cellular content of translation release factor RF4 in Escherichia coli. IUBMB Life 48: 283–286. [DOI] [PubMed] [Google Scholar]
- Bodley, J.W., Zieve, F.J., Lin, L., and Zieve, S.T. 1970. Studies on translocation III. Conditions necessary for the formation and detection of a stable ribosome-G factor-guanosine diphosphate complex in the presence of fusidic acid. J. Biol. Chem. 245: 5656–5661. [PubMed] [Google Scholar]
- Caldas, T., Binet, E., Bouloc, P., and Richarme, G. 2000. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 271: 714–718. [DOI] [PubMed] [Google Scholar]
- Capecchi, M.R. 1967. Polypeptide chain termination in vitro: Isolation of a release factor. Proc. Natl. Acad. Sci. 58: 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey, C.T., Tompkins, R., Scolnick, E., Caryk, T., and Nierenberg, M. 1968. Sequential translation of trinucleotide codons for initiation and termination of protein synthesis. Science 162: 135–138. [DOI] [PubMed] [Google Scholar]
- Dallas, A. and Noller, H.F. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8: 855–864. [DOI] [PubMed] [Google Scholar]
- Godefroy-Colburn, T., Wolfe, A.D., Dondon, J., and Grunberg-Manago, M. 1975. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J. Mol. Biol. 94: 461–478. [DOI] [PubMed] [Google Scholar]
- Goss, D.J., Parkhurst, L.J., and Wahba, A.J. 1980. Kinetics of ribosome dissociation and subunit association. The role of initiation factor IF3 as an effector. J. Biol. Chem. 255: 225–229. [PubMed] [Google Scholar]
- Gualerzi, C., Pon, C.L., and Kaji, A. 1971. Initiation factor dependent release of aminoacyl-tRNAs from complexes of 30S ribosomal subunits, synthetic polynucleotide and aminoacyl tRNA. Biochem. Biophys. Res. Commun. 45: 1312–1319. [DOI] [PubMed] [Google Scholar]
- Gualerzi, C.O., Brandi, L., Caserta, E., Garofalo, C., Lammi, M., La Teana, A., Petrelli, D., Spurio, R., Tomsic, J., and Pon, C.L. 2001. Initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harb. Symp. Quant. Biol. 66: 363–376. [DOI] [PubMed] [Google Scholar]
- Guthrie, C. and Nomura, M. 1968. Initiation of protein synthesis: A critical test of the 30S subunit model. Nature 219: 232–235. [DOI] [PubMed] [Google Scholar]
- Hirashima, A. and Kaji, A. 1970. Factor dependent breakdown of polysomes. Biochem. Biophys. Res. Commun. 41: 877–883. [DOI] [PubMed] [Google Scholar]
- ———. 1972a. Factor-dependent release of ribosomes from messenger RNA—Requirement for two heat-stable factors. J. Mol. Biol. 65: 43–58. [DOI] [PubMed] [Google Scholar]
- ———. 1972b. Purification and properties of ribosome-releasing factor. Biochemistry 11: 4037–4044. [DOI] [PubMed] [Google Scholar]
- ———. 1973. Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J. Biol. Chem. 248: 7580–7587. [PubMed] [Google Scholar]
- Hirokawa, G., Kiel, M.C., Muto, A., Selmer, M., Raj, V.S., Liljas, A., Igarashi, K., Kaji, H., and Kaji, A. 2002. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 21: 2272–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, G., Inokuchi, H., Kaji, H., Igarashi, K., and Kaji, A. 2004. In vivo effect of inactivation of ribosome recycling factor—Fate of ribosomes after unscheduled translation downstream of open reading frame. Mol. Microbiol. 54: 1011–1021. [DOI] [PubMed] [Google Scholar]
- Howe, J.G. and Hershey, J.W. 1983. Initiation factor and ribosome levels are coordinately controlled in Escherichia coli growing at different rates. J. Biol. Chem. 258: 1954–1959. [PubMed] [Google Scholar]
- Inokuchi, Y., Hirashima, A., Sekine, Y., Janosi, L., and Kaji, A. 2000. Role of ribosome recycling factor (RRF) in translational coupling. EMBO J. 19: 3788–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino, T., Atarashi, K., Uchiyama, S., Yamami, T., Saihara, Y., Yoshida, T., Hara, H., Yokose, K., Kobayashi, Y., and Nakamura, Y. 2000. Interaction of ribosome recycling factor and elongation factor EF-G with Escherichia coli ribosomes studied by the surface plasmon resonance technique. Genes Cells 5: 953–963. [DOI] [PubMed] [Google Scholar]
- Janosi, L., Hara, H., Zhang, S., and Kaji, A. 1996. Ribosome recycling by ribosome recycling factor (RRF)-An important but overlooked step of protein biosynthesis. Adv. Biophys. 32: 121–201. [DOI] [PubMed] [Google Scholar]
- Jelenc, P.C. and Kurland, C.G. 1979. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl. Acad. Sci. 76: 3174–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaempfer, R. 1970. Dissociation of ribosomes on polypeptide chain termination and origin of single ribosomes. Nature 228: 534–537. [DOI] [PubMed] [Google Scholar]
- ———. 1972. Initiation factor IF-3: A specific inhibitor of ribosomal subunit association. J. Mol. Biol. 71: 583–598. [DOI] [PubMed] [Google Scholar]
- Kaji, A., Kiel, M.C., Hirokawa, G., Muto, A., Inokuchi, Y., and Kaji, H. 2001. The fourth step of protein synthesis: Disassembly of the posttermination complex is catalyzed by elongation factor G and ribosome recycling factor, a near-perfect mimic of tRNA. Cold Spring Harb. Symp. Quant. Biol. 66: 515–529. [DOI] [PubMed] [Google Scholar]
- Karimi, R., Pavlov, M.Y., Buckingham, R.H., and Ehrenberg, M. 1999. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell 3: 601–609. [DOI] [PubMed] [Google Scholar]
- Kiel, M.C., Raj, V.S., Kaji, H., and Kaji, A. 2003. Release of ribosome-bound ribosome recycling factor by elongation factor G. J. Biol. Chem. 278: 48041–48050. [DOI] [PubMed] [Google Scholar]
- Lancaster, L., Kiel, M.C., Kaji, A., and Noller, H.F. 2002. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell 111: 129–140. [DOI] [PubMed] [Google Scholar]
- Makhno, V.I., Peshin, N.N., Semenkov, Y.P., and Kirillov, S.V. 1988. Modified method of producing “tight” 70S ribosomes from Escherichia coli, highly active in individual stages of the elongation cycle. Mol. Biol. 22: 670–679. [PubMed] [Google Scholar]
- McCutcheon, J.P., Agrawal, R.K., Philips, S.M., Grassucci, R.A., Gerchman, S.E., Clemons Jr., W.M., Ramakrishnan, V., and Frank, J. 1999. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc.Natl. Acad. Sci. 96: 4301–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, I., Hirokawa, G., Kiel, M.C., Kaji, A., and Blasi, U. 2004. Translation initiation with 70S ribosomes: An alternative pathway for leaderless mRNAs. Nucleic Acids Res. 32: 3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller, H.F., Yusupov, M.M., Yusupova, G.Z., Baucom, A., and Cate, J.H. 2002. Translocation of tRNA during protein synthesis. FEBS Lett. 514: 11–16. [DOI] [PubMed] [Google Scholar]
- Ogawa, K. and Kaji, A. 1975. Requirement for ribosome-releasing factor for the release of ribosomes at the termination codon. Eur. J. Biochem. 58: 411–419. [DOI] [PubMed] [Google Scholar]
- Olsson, C.L., Graffe,M., Springer, M., andHershey, J.W.B. 1996. Physiological effects of translation initiation factor IF3 and ribosomal protein L20 limitation in Escherichia coli. Mol. Gen. Genet. 250: 705–714. [DOI] [PubMed] [Google Scholar]
- Petrelli, D., LaTeana, A., Garofalo, C., Spurio, R., Pon, C.L., and Gualerzi, C.O. 2001. Translation initiation factor IF3: Two domains, five functions, one mechanism? EMBO J. 20: 4560–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, V.S., Kaji, H., and Kaji, A. 2005. Interaction of RRF and EF-G from Escherichia coli and T. thermophilus with ribosomes from both origins—insight into the mechanism of the ribosome recycling step. RNA 11: 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina, M.V., Savelsbergh, A., Katunin, V.I., and Wintermeyer, W. 1997. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385: 37–41. [DOI] [PubMed] [Google Scholar]
- Ruscetti, F.W. and Jacobson, L.A. 1972. Accumulation of 70S mono-ribosomes in Escherichia coli after energy source shift-down. J. Bacteriol. 111: 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K., and Ueda, T. 2001. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19: 751–755. [DOI] [PubMed] [Google Scholar]
- Subramanian, A.R. and Davis, B.D. 1970. Activity of initiation factor F3 in dissociating Escherichia coli ribosomes. Nature 228: 1273–1275. [DOI] [PubMed] [Google Scholar]
- ———. 1973. Release of 70S ribosomes from polysomes in Escherichia coli. J. Mol. Biol. 74: 45–56. [DOI] [PubMed] [Google Scholar]
- Vila-Sanjurjo, A., Schuwirth, B.S., Hau, C.W., and Cate, J.H. 2004. Structural basis for the control of translation initiation during stress. Nat. Struct. Mol. Biol. 11: 1054–1059. [DOI] [PubMed] [Google Scholar]
- Wada, A., Yamazaki, Y., Fujita, N., and Ishihama, A. 1990. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc. Natl. Acad. Sci. 87: 2657–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, D.N., Schluenzen, F., Harms, J.M., Yoshida, T., Ohkubo, T., Albrecht, R., Buerger, J., Kobayashi, Y., and Fucini, P. 2004. X-ray crystallography study on ribosome recycling: The mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 24: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]