Abstract
In Saccharomyces cerevisiae, nonsense-mediated mRNA decay (NMD) requires Upf1p, Upf2p, and Upf3p to accelerate the decay rate of two unique classes of transcripts: (1) nonsense mRNAs that arise through errors in gene expression, and (2) naturally occurring transcripts that lack coding errors but have built-in features that target them for accelerated decay (error-free mRNAs). NMD can trigger decay during any round of translation and can target Cbc-bound or eIF-4E-bound transcripts. Extremely low concentrations of the Upf proteins relative to the total pool of transcripts make it difficult to understand how nonsense transcripts are selectively recruited. To stimulate debate, we propose two alternative mechanisms for selecting nonsense transcripts for NMD and for assembling components of the surveillance complex, one for the first (pioneer) round of translation, called “nuclear marking,” and the other for subsequent rounds, called “reverse assembly.” The model is designed to accommodate (1) the low abundance of NMD factors, (2) the role of nucleocytoplasmic shuttling proteins in NMD, (3) the independent and nonobligate order of assembly of two different subcomplexes of NMD factors, and (4) the ability of NMD to simultaneously reduce or eliminate the synthesis of truncated proteins produced by nonsense transcripts while down-regulating but not completely eliminating functional proteins produced from error-free NMD-sensitive transcripts.
Keywords: translation, RNA transcript degradation, RNA binding proteins, messenger ribonucleoprotein, nucleocytoplasmic transport
NMD in yeast and mammals
A great deal of attention has been paid to the question of where the steps in nonsense-mediated mRNA decay (NMD) take place: in the nucleus, during export to the cytoplasm, or in the cytoplasm. In mammals, there is good evidence that nonsense transcripts are degraded by NMD during the first round of translation, called the pioneer round, when they are bound to the CBC20/CBC80 (Cbc) cap binding complex and the exon junction complex (EJC), both of which associate with transcripts in the nucleus. In contrast, nonsense transcripts undergoing bulk translation in the cytoplasm that are bound to the cytoplasmic cap binding protein eIF-4E are reportedly immune to NMD or, at the very least, are not targeted by a mechanism directly related to the EJC (Ishigaki et al. 2001). These results suggest that the major mechanism for NMD in mammals occurs during the first round of translation, which could take place concomitantly with mRNA export from the nucleus.
The ideas presented below were initially prompted by a recent report in RNA (Kuperwasser et al. 2004) claiming important differences in the mechanism of NMD between mammals and the yeast Saccharomyces cerevisiae. The authors presented evidence that nonsense transcripts are insensitive to NMD when restricted to the nucleus, due to a block in nuclear export. In addition, analysis of a nonsense reporter transcript indicated that it was insensitive to NMD when bound to the nuclear cap binding complex (Cbc1p/Cbc2p). They concluded that nonsense transcripts are not degraded directly in the nucleus and that there is no special pioneer round of translation in yeast. NMD must therefore occur during bulk translation of eIF-4E-bound transcripts in the cytoplasm.
These conclusions were quickly challenged (Gao et al. 2005). Examining six NMD-sensitive transcripts, those authors found that all of them were susceptible to NMD regardless of whether they were bound to Cbc- or eIF-4E cap binding complexes. This evidence supports the existence of a special pioneer round of translation. Furthermore, the results are compatible with other evidence showing that NMD can occur during any round of translation (Maderazo et al. 2003; Keeling et al. 2004). The finding that the Upf proteins associate with polyribosomes and are not restricted to 80S monosomes (Atkin et al. 1997) is another indication that NMD is not limited to Cbc-bound transcripts. Considering all of these observations, the most reasonable premise is that yeast lacks a Upf-dependent mechanism for degrading nonsense transcripts trapped in the nucleus but retains a special pioneer round of translation. Nonsense transcripts can be degraded during the pioneer round or during bulk cytoplasmic translation.
Recruitment of the factors required for NMD in yeast
An unanswered question remains, however, as to the mechanism by which appropriate transcripts are selectively recruited for NMD. Recruitment for the pioneer round could depend on Upf3p (Shirley et al. 2002) or Hrp1p (Gonzalez et al. 2000). Both proteins shuttle between the nucleus and the cytoplasm. Upf3p has been studied extensively, and there is no doubt that it is required for NMD. Hrp1p binds to AU-rich RNAs, is essential for growth, and performs functions outside of NMD. A role in NMD was suggested by the finding that it interacts with Upf1p. Also, a temperature-sensitive hrp1 allele stabilized a nonsense reporter transcript in a temperature- shift experiment, but evidence of similar effects on other transcripts has not been reported. As proposed in an earlier model (Culbertson 1999; Culbertson and Leeds 2003), shuttling proteins like Upf3p or Hrp1p could mark Cbc-bound transcripts in the nucleus for potential decay during pioneer translation by setting the stage for the recruitment of other factors needed for NMD. However, this does not readily explain how the same factors are recruited to trigger NMD during subsequent rounds of translation in the cytoplasm.
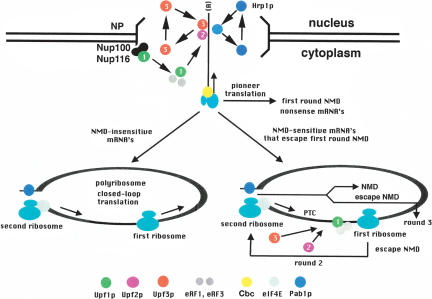
To stimulate debate about how recruitment might work for an NMD pathway that can occur during any round of translation, we propose two mechanisms: “nuclear marking” for the pioneer round of translation, and an alternate mechanism called “reverse assembly” for subsequent rounds of translation. Figure 1 illustrates both mechanisms. The cartoon depicts Cbc-bound mRNPs exiting the nucleus to engage in a pioneer round of translation. This aspect of the model deserves further explanation, because the current claim is that a pioneer ribosome can translate either Cbc-bound or eIF-4E-bound transcripts. This could be true, but there is a caveat to consider.
FIGURE 1.
Nuclear marking/reverse assembly model for yeast NMD during pioneer translation and subsequent rounds of translation. According to the model, the two sub-complexes of NMD factors assemble sequentially but in a nonobligate order. Nuclear marking: The Upf3p-Upf2p subcomplex assembles prior to the pioneer round of translation followed by translation, and then assembly of the Upf1p/RF subcomplex. Reverse assembly: The order of assembly of the subcomplexes is reversed during subsequent rounds of translation. Regardless of the order of assembly, a structurally and functionally identical surveillance complex is formed, leading to translation termination, decapping, Xrn1p-mediated 5′ decay, and exosome-mediated 3′ decay.
eIF4E-bound PGK1 mRNA is targeted by NMD in a cbc1-Δ strain, which is surprisingly viable (Gao et al. 2005). Due to the absence of Cbc1p and because NMD requires translation, it was argued that pioneer translation must occur on eIF-4E-bound transcripts in this abnormal circumstance. Furthermore, in wild-type strains, intron-containing pre-mRNAs are bound to either Cbc or eIF-4E. Since nuclear splicing produces mature mRNAs with both kinds of cap binding complexes before any translation is possible, it was argued that some mRNAs that undergo pioneer translation must have eIF-4E bound to the cap. The percentage of transcripts falling into this category is substantial. Although only 264 of ~5800 genes contain introns, they are concentrated in the most highly expressed genes. Moreover, 50% of all mRNAs are derived from pre-mRNAs (Li et al. 1999).
However, pre-mRNAs (by virtue of intronic stop codons) must be exported to the cytoplasm to be degraded by NMD. From that location, they are no longer precursors to mature mRNA because the splicing machinery is in the nucleus. If some of the pre-mRNAs escape NMD during pioneer translation, eIF-4E could replace Cbc before the next round of translation, and that could be why Gao et al. (2005) detected eIF-4E-bound pre-mRNAs. The only way their model for pioneer translation of eIF-4E-bound pre-mRNAs holds up is if some eIF-4E is nuclear. Because it is only 24.3 kDa in size, eIF-4E could import by passive diffusion (Pante and Aebi 1995), but this has not been tested. When cells over-express eIF-4E at 100-fold above normal (1.42 × 106 molecules per cell), some nuclear accumulation occurs (Lang et al. 1994; Ghaemmaghami et al. 2003), but mislocalization at this level of expression is common. Lang et al. (1994) also claimed that eIF-4E can be detected in the nucleus when expressed at the normal level, but the data was not shown. This point is so critical to the conclusion that eIF-4E-bound transcripts undergo pioneer translation that we should not incorporate this into the dogma of NMD until better evidence for nuclear eIF-4E is published.
As illustrated in Figure 1, nuclear marking posits that shuttling proteins function in NMD by exporting from the nucleus in association with every mRNP (mRNA/protein) complex. During or following export, the pioneer ribosome displaces the transcript-bound marker proteins, after which they shuttle back into the nucleus for another round of mRNP-associated export. However, when translation terminates prematurely, the marker proteins remain associated with the transcript, followed by assembly of a “surveillance” complex. The complete NMD-competent complex, which contains the factors required for translation termination and NMD (eRF1, eRF3, Upf1p, Upf2p, Upf3p), triggers termination of translation, decapping, and 5′ decay-mediated by the Xrn1p exonuclease. There is also a link between NMD and exosomal 3′ decay mediated by an interaction between Upf1p and Ski7p (Mitchell and Tollervey 2003; Takahashi et al. 2003). Thus, nonsense transcripts appear to be degraded from both ends.
Full assembly of the surveillance complex is most likely preceded by the prior assembly of two subcomplexes (Atkin et al. 1997). One contains Upf2p and Upf3p, and the other contains Upf1p, eRF1, and eRF3 (Upf1p/RF). Staged assembly of the two subcomplexes is supported by two observations. (1) Upf2p fails to associate with translating ribosomes in the absence of Upf3p. (2) Upf1p associates with translating ribosomes in the absence of Upf2p and Upf3p. These and other findings suggest that the two subcomplexes are recruited separately and assembled independently. NMD only occurs when both subcomplexes are present.
We propose that the Upf3p/Upf2p subcomplex assembles on Cbc-bound transcripts before the pioneer round of translation commences. If the pioneer ribosome encounters a premature stop codon, the Upf1p/RF subcomplex is recruited, and the surveillance complex is assembled. During bulk translation of eIF-4E-bound transcripts, the subcomplexes assemble in reverse order. The structure and function of the fully assembled surveillance complex could be identical regardless of the order of assembly of the subcomplexes.
Limitations caused by the low abundance of Upf proteins
The primary impetus for suggesting reversible assembly of the subcomplexes is that it helps explain how nonabundant Upf proteins, one of which shuttles between the nucleus and the cytoplasm as part of its function in NMD, can selectively form surveillance complexes on nonsense mRNAs during both pioneer translation and subsequent rounds of translation. In mammals the Upf proteins are present at tens of thousands to millions of copies per cell (Maquat and Serin 2001), but in S. cerevisiae the Upf proteins are present at very low intracellular concentrations (Table 1). The estimates for protein abundance vary (Atkin et al. 1997; Li et al. 1999; Maderazo et al. 2000; Ghaemmaghami et al. 2003), but they are consistently much lower than cellular transcripts (~15,000/cell), ribosomes (~200,000/cell), and release factors (variable but as high as 79,000/cell), and they consistently point to Upf3p as the least abundant of the three proteins. A major challenge is to understand how a small number of Upf proteins can locate and assemble on nonsense transcripts in the total pool of transcripts. We argue that prior to the pioneer round of translation, nuclear marking simplifies the identification of minority populations of nonsense transcripts in the relatively small pool of exporting transcripts compared to the larger pool of cytoplasmic transcripts.
TABLE 1.
Estimates for haploid intracellular concentrations of yeast Upf proteins
| Protein | Abundance (Western)a | Abundance (GFP/TAP)b |
| Upf1p | 1600 | 6091 |
| Upf2p | 160 | 1280 |
| Upf3p | 80 | 1250 |
a(Maderazo et al. 2000). Numbers indicate molecules/cell. The estimates were derived by quantitative Western blotting of proteins detected by a common monoclonal antibody to an epitope tag.
b(Ghaemmaghami et al. 2003). Numbers indicate molecules/cell. The estimates of abundance for proteins listed in the GFP protein localization database are typically much higher than estimates derived by other methods. The actual concentrations of the Upf proteins are likely to be somewhere in between the estimates shown in the table for the two methods.
To illustrate the limits imposed by low Upf protein abundance, let’s assume that the cytoplasmic mRNA pool has an average half-life of 20 min, such that nascent transcripts must export from the nucleus at a rate of 375/min (15,000/2/20) to replace degraded cytoplasmic transcripts and to maintain a constant steady-state bulk mRNA level. This and the requirement for translation in NMD imply that the minimum required abundance of nuclear marking proteins is dictated by the combined rates of shuttling and translation. Protein import from the cytoplasm to the nucleus through nuclear pores takes 1 msec (Weis 2002). In contrast, estimates of the transit time for elongation indicate that translation of the average yeast ORF requires about 10 sec. Assuming that the combined rate of shuttling and translation is 10 sec, then one marker protein molecule could shuttle six times per minute. Thus, the minimum abundance required for a marker protein to export with every mRNP is ~60 molecules/cell (375/6). This is slightly below the lowest estimate for Upf3p abundance of 80 molecules per cell and considerably less than the highest estimate (Table 1). It therefore seems plausible that shuttling proteins could mark every mRNP, leaving the nucleus even when they are present at low intracellular concentrations.
Despite the apparent feasibility of nuclear marking based on the above arguments, it would only be fair to point out a caveat of the model regarding Upf3p that needs to be addressed. Conclusions grounded in firm evidence include the following: (1) The import mechanism for Upf3p has been established—it binds to β-importin Srp1p and is excluded from the nucleus in an srp1 mutant (Shirley et al. 2002). (2) Mutations in UPF3 have been identified that block export, cause nuclear accumulation, and confer an Nmd− phenotype. (3) Upf3p associates with 80S particles and polyribosomes (Atkin et al. 1997). However, one assumption of nuclear marking has not been resolved at the molecular level. Efforts by us and by others to detect Upf3p bound to mRNA have failed. Upf3p might associate with mRNPs indirectly by binding to another mRNP protein, but besides Upf2p, no other interacting proteins have surfaced in global two-hybrid screens. These difficulties could indicate one of two possibilities: Either the nuclear marking model is wrong or the interaction of Upf3p with mRNPs is too transient or too weak to purify in immunoprecipitation experiments.
For subsequent rounds of translation in the cytoplasm, the low concentrations of the Upf proteins impose severe restrictions on any mechanism for efficient selection of NMD substrates among the 15,000 cytoplasmic mRNAs. Compared to the Upf proteins, most translation factors are highly abundant, including the release factors eRF1 and eRF3. Complexes between the release factors and Upf1p can only form with a small fraction of the release factor pool per unit time, suggesting that the Upf1p/RF complex may be restricted to aberrant termination events. Although the rapid cycling of factors must be an essential feature of shuttling proteins that function in NMD, the need for rapid cycling is even more critical in subsequent rounds of translation in the cytoplasm. The Upf proteins cannot be sequestered on any one transcript for very long. There is not enough to go around.
Upf protein localization is compatible with assembly on Cbc- or eIF-4E-bound transcripts
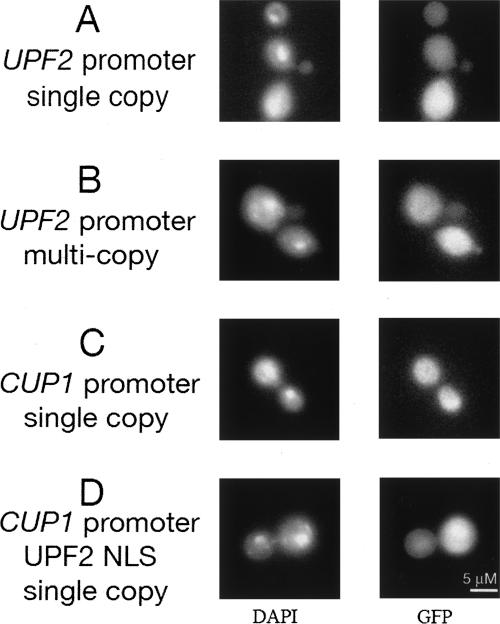
Upf3p shuttles between the nucleus and cytoplasm and accumulates in the nucleus when overexpressed or when export is blocked, but most of the protein is cytoplasmic in wild-type cells (Shirley et al. 1998, 2002). With the use of sensitive techniques such as confocal microscopy, Upf1p appears cytoplasmic and cannot be detected in the nucleus under any conditions (Atkin et al. 1995). Because the localization of Upf2p has never been reported, we determined the distribution of Upf2-GFP (Fig. 2). The fluorescent signal was predominantly cytoplasmic even when overexpressed. A putative nuclear localization signal sequence (NLS) that potentially directs Upf2p import into the nucleus failed to direct a cytoplasmic GFP reporter to the nucleus. In contrast, three similar NLSs located in Upf3p direct nuclear import through the β-importin Srp1p (Shirley et al. 1998). These results support a cytoplasmic location for Upf2p.
FIGURE 2.
Localization of GFP-Upf2p. GFP-Upf2p was localized by visualizing green fluorescent protein fusions in live cells by expressing hybrid fusions from plasmids introduced into strain NEY4 (MATαura3-52 trp1-Δ1 leu2-2 tyr7-1 can1-100 his3-Δ200 upf2Δ1::HIS3). Cells were treated with DAPI to visualize nuclear DNA. The plasmids used were (A) pNE22 (GFP-UPF2, CEN4, TRP1); (B) pNE25 (GFP-UPF2, 2 μm origin of replication, TRP1); (C) pNE85-UPF2 (CUP1-GFP-UPF2, CEN4, TRP1); (D) pNE85-UPF2(19–47) (amino acids 19–47 including the putative nuclear localization signal motif in Upf2p). Cells were grown in medium containing 25 μM Cu++ without tryptophan.
Upf1p was recently shown to interact with nuclear pore proteins on the cytoplasmic side of nuclear pores (Nazarenus et al. 2005). From this location, Upf1p could be poised to associate with Cbc-bound-mRNPs carrying prebound Upf3p. The mRNP protein Hrp1p is normally localized around the nucleus on both sides of the nuclear envelope but is not distributed throughout the cytoplasm, suggesting that whatever role it might play in NMD may be restricted to the translation of nascent transcripts emerging from nuclear pores. It is worth noting, however, that Hrp1p spreads throughout the cytoplasm during heat-shock and could influence NMD in a more general way when cells are subjected to stress (Henry et al. 2003).
Recruitment of factors for NMD following the first round of translation
All three Upf proteins are required when NMD occurs during subsequent rounds of translation following the pioneer round (Maderazo et al. 2003; Keeling et al. 2004). The rapid nucleocytoplasmic shuttling of marking proteins provides a rational way to recruit RNA substrates and assemble the surveillance complex in preparation for the pioneer round of translation, but what happens when transcripts that are targeted for NMD escape first-round decay? The two possibilities are: (1) NMD factors are retained on the transcript, or (2) they are displaced by the pioneer ribosome and must be re-assembled. We favor displacement and reassembly. These proteins cannot be sequestered during multiple rounds of translation from the pools of free Upf proteins needed to find and recruit new NMD-sensitive transcripts among the constantly emerging nascent transcripts. Rapid cycling between successive rounds of translation would be required. NMD-sensitive transcripts must be re-recruited and Upf proteins re-assembled between each round of translation. The arguments that make nuclear marking an attractive model for recruitment in preparation for the pioneer round no longer apply in subsequent rounds, because recruitment and assembly take place among the larger pool of cytoplasmic transcripts. A different mechanism seems likely.
We propose that in subsequent rounds of translation, the Upf1p/RF subcomplex is the most likely one to be recruited first when a premature termination codon is encountered and recognized as an aberrant termination codon. Recent evidence shows that aberrant termination occurs when the stop codon is followed by an imposter sequence posing as an improperly configured 3′-UTR (Amrani et al. 2004). This sequence is most likely functionally equivalent to the degenerate AU-rich downstream element (DSE) shown to be required for NMD (Zhang et al. 1995).
In the cytoplasm where the subcomplexes must find the substrates for NMD in the larger pool of total transcripts, a reverse order of assembly makes sense, but not out of necessity. It is favored by probability; Upf1p/RF is 5–20 times more abundant than Upf3p/Upf2p (Table 1). After the Upf1p/RF subcomplex is recruited, the Upf2p/Upf3p subcomplex associates through the binding domains known to tether all three proteins together in the surveillance complex (He et al. 1997). The Upf3p/Upf2p subcomplex forms its own association with RF3 (Gonzalez et al. 2001; Wang et al. 2001). As mentioned earlier, we don’t envision any necessity for the fully assembled surveillance complex to differ in structure or function between the pioneer round and subsequent rounds of translation.
Error-containing versus error-free targets of NMD
Transcripts that contain coding errors are best eliminated during the pioneer round of translation to prevent any truncated protein product from being made. However, wild-type transcripts have been identified where the turn-over rate is controlled by the Upf genes as part of the normal repertoire of gene expression (Lelivelt and Culbertson 1999; He et al. 2003). These transcripts, which code for important functional proteins, are targeted for NMD not because they contain coding errors, but for other reasons. The Upf proteins down-regulate their abundance and reduce the amount of protein product made, but without altogether eliminating their translation.
The difference between transcripts targeted to eliminate a protein product versus those targeted to reduce but not eliminate a protein product is not immediately obvious because for both of them, steady-state RNA transcript levels are not reduced by NMD to zero. Most nonsense transcripts are reduced fourfold to 10-fold. For example, met8 and pgk1 nonsense transcripts are reduced fourfold and 10-fold, respectively (Gao et al. 2005). In addition, 82% of 529 error-free transcripts that are targeted by NMD are reduced by three-fold or less, but 18% are reduced much more—as high as 11-fold (Lelivelt and Culbertson 1999).
We believe the reason that low levels of transcripts are always detected is that for all transcripts there is a protected class—those nascent transcripts that have not yet been exported from the nucleus and have therefore not been exposed to NMD through translation. Since the balance between rates of transcription and decay vary for each transcript, the steady-state percentage of protected transcripts will vary. Because of this, it is reasonable to assume that RNA surveillance, while not eliminating all detectable mRNA, may very well limit the synthesis of truncated proteins to the one made by necessity during pioneer translation to test whether the transcript can be translated full-length. This argues for very efficient RNA surveillance during the first round of translation, a level of efficiency that could be achieved by nuclear marking. In the case of error-free transcripts, the example below illustrates one way transcripts can be targeted by NMD while maintaining production of a functional protein product.
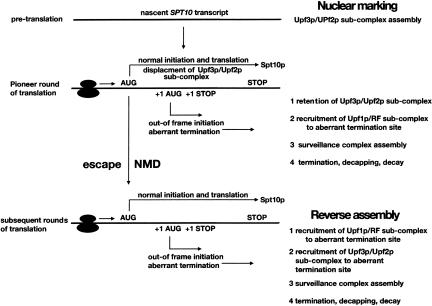
Error-free transcripts are most likely substrates for NMD during all rounds of translation. One of these, the wild-type SPT10 transcript, is three times more abundant and threefold more stable when NMD is inactivated, and is targeted for NMD by “leaky scanning” (Welch and Jacobson 1999). SPT10 is NMD-sensitive because ribosomes fail to initiate efficiently at the first AUG codon due to a suboptimal context. As a result, ribosomes frequently bypass the initiation codon and scan to a second out-of-frame AUG (Fig. 3). When out-of-frame initiation occurs, a stop codon in this alternate frame triggers NMD. SPT10 transcripts escape NMD when translation initiates at the normal AUG, but they are repeatedly exposed to the possibility of NMD, which can be triggered or bypassed in each successive round of translation. The beauty of leaky scanning is that decay rates can be reduced by a mechanism that permits synthesis of a necessary protein product. Transcripts targeted by leaky scanning most likely decay with composite rates that depend on the frequency of ribosomal scanning past the first AUG and the relative efficiencies of Upf protein recruitment during pioneer versus bulk translation.
FIGURE 3.
Translation and decay of NMD-sensitive transcripts targeted by leaky scanning. The figure depicts two mechanisms for the assembly of the surveillance complex, “nuclear marking” during the pioneer round of translation, and “reverse assembly” for subsequent rounds of translation.
Summary
Why should there be two mechanisms for transcript selection and complex assembly in NMD? We think the question can be rephrased—why should Upf3p shuttle between the nucleus and the cytoplasm if there is only one mechanism? To accommodate shuttling, we propose that the pioneer round of translation is distinguished from subsequent rounds by the order of the steps leading to NMD. For NMD to occur during the pioneer round, the Upf3p/Upf2p subcomplex is preloaded on mRNPs by nuclear marking, the pioneer round commences, and the presence of a premature stop codon leads to termination of translation concomitant with assembly of the Upf1p/RF complex. Nuclear marking in preparation for the pioneer round of translation could enhance the probability of NMD in the pioneer round.
In subsequent rounds of translation, neither subcomplex is associated with a nonsense transcript until translation comes to a premature halt, at which time the Upf1p/RF subcomplex is most often the first to associate by virtue of being more abundant. During bulk translation of eIF-4E-bound transcripts, reverse assembly enhances the probability of NMD. The efficiencies of NMD during pioneer translation and subsequent rounds of translation may not be the same.
Sufficient similarities exist between yeast and mammalian NMD to suppose that divergent pathways in these organisms represent alternative evolutionary outcomes derived from a common ancestral pathway (Culbertson and Leeds 2003). In yeast, NMD targets both Cbc-bound and eIF-4E-bound transcripts. Two alternatives may be available to assemble the full surveillance complex from two subcomplexes. By accounting for differences between rounds of translation, the combination of nuclear marking and reverse assembly explains how the majority of nonsense transcripts might be preselected by one or more shuttling proteins in preparation for degradation during the pioneer round of translation, while providing an alternate mechanism using the same factors to degrade transcripts that are targeted for NMD but escape first-round decay.
Acknowledgments
We thank Dr. Jon Warner for sharing unpublished data and ideas, Dr. Gerry Fink for his encouragement, and the members of the Culbertson lab who contributed their ideas. Support was from the University of Wisconsin College of Agricultural and Life Sciences, the University of Wisconsin Medical School, and NIH grant GM65172 (M.R.C.). This is Laboratory of Genetics paper no. 3621.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2113605.
REFERENCES
- Amrani, N., Ganesan, R., Kervestin, S., Mangus, D., Ghosh, S., and Jacobson, A. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118. [DOI] [PubMed] [Google Scholar]
- Atkin, A., Altamura, N., Leeds, P., and Culbertson, M. 1995. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Cell Biol. 6: 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin, A., Schenkman, L., Eastham, M., Dahlseid, J., Lelivelt, M., and Culbertson, M. 1997. Relationship between yeast polyribosomes and Upf proteins required for nonsense-mediated mRNA decay. J. Biol. Chem. 272: 22163–22172. [DOI] [PubMed] [Google Scholar]
- Culbertson, M. 1999. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 15: 74–80. [DOI] [PubMed] [Google Scholar]
- Culbertson, M. and Leeds, P. 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Gen. Dev. 13: 207–214. [DOI] [PubMed] [Google Scholar]
- Gao, Q., Das, B., Sherman, F., and Maquat, L. 2005. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc. Natl. Acad. Sci. 102: 4258–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami, S., Huh, W., Bower, K., Howson, R., Belle, A., Dephoure, N., O’Shea, E., and Weissman, J. 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C., Ruiz-Echevarria, M., Vasudevan, S., Henry, M., and Peltz, S. 2000. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 5: 489–499. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C.I., Bhattacharya, A., Wang, W., and Peltz, S.W. 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274: 15–25. [DOI] [PubMed] [Google Scholar]
- He, F., Brown, A., and Jacobson, A. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17: 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F., Li, X., Spatrick, P., Casillo, R., Dong, S., and Jacobson, A. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12: 1439–1452. [DOI] [PubMed] [Google Scholar]
- Henry, M., Mandel, D., Routson, V., and Henry, P. 2003. The yeast hnRNP-like protein Hrp1/Nab4 accumulates in the cytoplasm after hyperosmotic stress: A novel Fps1-dependent response. Mol. Biol. Cell 14: 3929–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., and Maquat, L.E. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CPB20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Keeling, K., Lanier, J., Du, M., Salas-Marco, J., Gao, L., Kaenjak-Anageletti, A., and Bedwell, D. 2004. Leaky termination at premature stop codons antagonizes nonsense mediated mRNA decay in S. cerevisiae. RNA 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser, N., Brogna, S., Dower, K., and Rosbash, M. 2004. Nonsense-mediated decay does not occur within the yeast nucleus. RNA 10: 1907–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, V., Zanchin, I., Lunsdorf, H., Tuite, M., and McCarthy, J. 1994. Initiation factor eIF4E of Saccharomyces cerevisiae. J. Biol. Chem. 269: 6117–6123. [PubMed] [Google Scholar]
- Lelivelt, M. and Culbertson, M. 1999. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 19: 6710–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Nierras, C., and Warner, J. 1999. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol. Cell. Biol. 19: 5393–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderazo, A., He, F., Mangus, D., and Jacobson, A. 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol. 20: 4591–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderazo, A., Belk, J., He, F., and Jacobson, A. 2003. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol. Cell. Biol. 23: 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L. and Serin, G. 2001. Nonsense-mediated mRNA decay: Insights into mechanism from the cellular abundance of human Upf1, Upf2, Upf3, and Upf3X proteins. Cold Spring Harbor Symp. Quant. Biol. LXVI: 313–320. [DOI] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′. 5′ degradation. Mol. Cell 11: 1405–1413. [DOI] [PubMed] [Google Scholar]
- Nazarenus, T., Cedarberg, R., Bell, R., Cheatle, J., Forch, A., Haifley, A., Hou, A., Wanja Kebaara, B., Shields, C., Stoysich, K., et al. 2005. Upf1p, a highly conserved protein required for nonsense-mediated mRNA decay, interacts with the nuclear pore proteins Nup100p and Nup116. Gene 345: 199–212. [DOI] [PubMed] [Google Scholar]
- Pante, N. and Aebi, U. 1995. Toward a molecular understanding of the structure and function of the nuclear pore complex. Int. Rev. Cytol. 162B: 225–255. [DOI] [PubMed] [Google Scholar]
- Shirley, R., Lelivelt, M., Schenkman, L., Dahlseid, J., and Culbertson, M. 1998. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci. 111: 3129–3143. [DOI] [PubMed] [Google Scholar]
- Shirley, R., Ford, A., Richards, R., Albertini, M., and Culbertson, M. 2002. Nuclear import of Upf3p is mediated by importin-α/β and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics 161: 1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S., Araki, Y., Sakuno, T., and Katada, T. 2003. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 22: 3951–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Czaplinski, K., Rao, Y., and Peltz, S.W. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20: 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, J.R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Weis, K. 2002. Nucleocytoplasmic transport: Cargo trafficking across the border. Curr. Opin. Cell Biol. 14: 328–335. [DOI] [PubMed] [Google Scholar]
- Welch, E. and Jacobson, A. 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18: 6134–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Ruiz-Echevarria, M., Quan, Y., and Peltz, S. 1995. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol. 15: 2231–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]