Abstract
The overall conformation of M1 RNA, the catalytic RNA subunit of RNase P in Escherichia coli, was analyzed in vivo and, in the presence of the C5 protein subunit, in vitro by lead(II) acetate probing. The partial cleavage patterns obtained are congruent with previous structure mapping performed in vitro. Most of the known major and minor cleavages in M1 RNA were supported and could be mapped onto a secondary structure model. The data obtained indicate that C5 has only minor effects on the overall structure of the RNA subunit. The similar cleavage patterns obtained in vitro and in vivo furthermore suggest that the intracellular environment does not greatly alter the overall conformation of M1 RNA within the holoenzyme complex. Moreover, our data indicate that M1 RNA in vivo is present in at least two states—the major fraction is bound to tRNA substrates and a minor fraction is substrate free. Finally, both in this and previous work we found that lead(II) probing data from in vivo experiments conducted on longer RNAs (tmRNA and M1 RNA) generally gives superior resolution compared to parallel in vitro experiments. This may reflect the absence of alternative conformers present in vitro and the more natural state of these RNAs in the cell due to proper, co-transcriptional folding pathways and possibly the presence of RNA chaperones.
Keywords: RNase P RNA, lead(II) cleavage, in vivo mapping, RNA structure
INTRODUCTION
The ubiquitous ribonuclease P (RNase P) is the endoribonuclease responsible for generating the mature 5′ end of tRNAs by a single endonucleolytic cleavage of their precursors. In Escherichia coli, the enzyme is composed of a 377-nt RNA (M1 RNA)—the catalytic subunit—and a smaller, 119-amino-acid protein subunit—the C5 protein. The protein subunit is required for activity in vivo, and is therefore essential for bacterial growth (Schedl and Primakoff 1973). However, M1 RNA, like other bacterial RNase P RNAs, is able to cleave its substrates in vitro in the absence of the protein (Guerrier-Takada et al. 1983).
For a better assessment of functional aspects of ribonucleoproteins such as M1 RNA/RNase P, structure determinations are often helpful. In the case of M1 RNA, a secondary structure model has been proposed based on phylogenetic comparisons (Haas et al. 1996), and the crystal structure of the specificity domain of a type A RNase P RNA, to which M1 RNA belongs, was recently reported (Krasilnikov et al. 2004). In addition, biochemical and genetic studies have been carried out; they identified regions/nucleotides within M1 RNA that are involved in interactions with either the tRNA substrate (Kirsebom and Svärd 1994; LaGrandeur et al. 1994; Oh and Pace 1994) or the protein subunit (Talbot and Altman 1994; Biswas et al. 2000; Sharkady and Nolan 2001; Tsai et al. 2003). All structure mapping of M1 RNA had, up to this point in time, been conducted by chemical or enzymatic probing in vitro, and no information was available about the folding of this RNA in vivo.
Lead(II) acetate (Pb2+ acetate) is a useful and versatile probe to extract information on RNA structures in living bacterial cells (Lindell et al. 2002; Valverde et al. 2004). This ion primarily induces highly specific cleavages at positions of tight metal ion binding sites (David et al. 2001), but cleaves RNA also within single-stranded regions, loops, and bulges (Gornicki et al. 1989; Kirsebom and Ciesiolka 2005). Cleavages in double-stranded regions are less frequent or absent. Pb2+ has previously been used in vitro to probe the structure of M1 RNA, alone (Tallsjö et al. 1993; Zito et al. 1993; Mattsson et al. 1994; Brännvall et al. 2001) or in a complex with a tRNA substrate (Ciesiolka et al. 1994; Kirsebom and Svärd 1994).
In this work, we undertook a first approach to analyze the overall conformation of E. coli RNase P by probing the native structure of M1 RNA in vivo with Pb2+. The results obtained were compared to those of in vitro probing of M1 RNA in the presence or absence of the C5 protein. The cleavage pattern of RNase P RNA in vivo was in good agreement with the secondary model proposed based on in vitro work. Positions of the Pb2+ induced cleavage sites in the structure of the specificity domain of RNase P RNA might be indicative of a metal ion core important for folding, as suggested for the P4/P6 domain of the Group I intron RNA (Strobel and Doudna 1997). Moreover, the observed cleavages suggest that M1 RNA is present in two main conformational states, one being characteristic of free RNase P and one of an RNase P-tRNA complex. The similarities observed in a comparison of in vitro Pb2+ cleavage patterns in the presence or absence of C5 indicate that major structural changes are not induced by the protein.
RESULTS AND DISCUSSION
Pb2+-induced cleavage of M1 RNA in vivo indicates a tRNA substrate-bound form
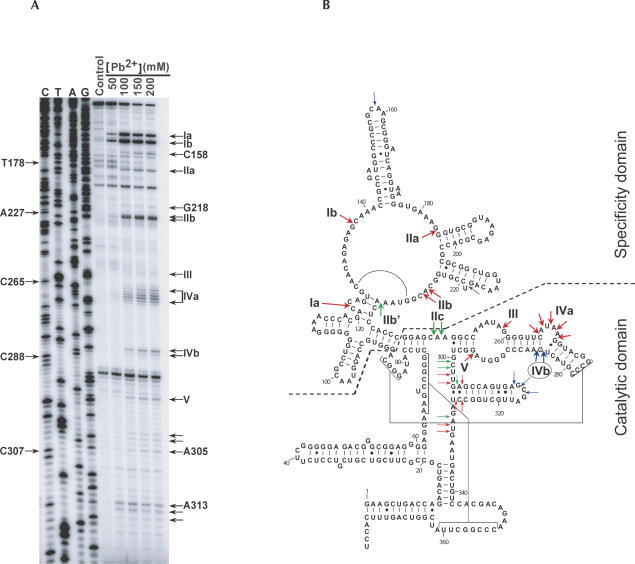
Pb2+-induced cleavage can be used to map RNA conformations and to follow discrete structural changes (e.g., Brännvall et al. 2001). Here we investigated the cleavage pattern of M1 RNA in vivo and compared it to results obtained in vitro. Partial Pb2+ cleavages of M1 RNA were obtained by treating exponentially growing E. coli cells with 50–200 mM Pb2+ acetate (Lindell et al. 2002; Materials and Methods). Primer extension analysis was carried out on total extracted RNA, and cleavage positions were determined by comparison to a sequencing reaction performed with the same primer. As a control, RNA from mock-treated cells was analyzed in parallel. Figure 1A shows a representative autoradiogram of such an analysis. The Pb2+-dependent cleavage positions that were reproducibly found in several independent experiments were mapped onto the secondary structure model proposed by Haas et al. (1996), using the denotation according to Ciesiolka et al. (1994) (Fig. 1B). Primer extension stops also seen in the control lane (absence of Pb2+) were not considered. The results obtained can also be compared to previously published in vitro Pb2+- induced cleavages (see Tallsjö et al. 1993; Zito et al. 1993; Ciesiolka et al. 1994). Pb2+ cleavages denoted by Roman numerals are strong and specific, and occur at sites at which Mg2+ ions are most likely displaced from their normal binding pocket in the RNA structure; they can be competed by increasing concentrations of Mg2+ (Ciesiolka et al. 1994; Brännvall et al. 2001). Moreover, the inference that Pb2+ and Mg2+ bind to the same or at least overlapping sites is also supported by the fact that several of the Pb2+-induced cleavage sites are also subjected to cleavage in the presence of Mg2+ (Kazakov and Altman 1991). A second, weaker type of cleavage, also detected in this experiment, is located in interhelical regions, loops, and bulges. Overall, the pattern obtained in Figure 1 is remarkably similar to those previously seen in vitro in that most major and minor cleavages were obtained. Specifically, two major site-specific cuts were detected in vivo. One is located between C122 and C123 (denoted Ia in Fig. 1A,B; see also Fig. 3 below for better resolution of the larger fragment range), in a region suggested to interact with the T-loop/stem domain of pre-tRNA; the other is between G137 and C138 (Ib). Several cleavages occurred in single-stranded segments, for example, in the bulge between C265 and G270 (IVa) and in the terminal loop between C158 and A159. Weak cleavages were seen between G218 and U219, U302 and G304, A305 and G306, and A313 and G316. In contrast to the single-stranded segments, fully helical regions were resistant to Pb2+-induced cleavages. Several minor site-specific cleavages were also observed; cleavage site IIa lies between G183 and G184 and IIb between C226 and A227, as reported earlier (Ciesiolka et al. 1994). An additional cleavage site was mapped to the vicinity of IIb (A227↓C228), and we also observed that cleavage at site III occurred between A258 and G259. Cleavages at formerly reported sites IIb′ and IIc were not seen. The apparent absence of cleavage at these two sites, and the weak cleavage at site III, suggests that the local structure of M1 RNA in vivo differs from that obtained in vitro in the absence of substrate. Furthermore, cleavage at site IIb′ was previously shown to be indicative of non-completely folded M1 RNA at low Pb2+ concentrations, whereas cleavage at site IIb occurred in the catalytically active form (Brännvall et al. 2001). Since an earlier study reported that Pb2+-induced cleavage of M1 RNA in vitro at sites IIb′-III was reduced when tRNAGly was present, this indicates a structural change in this part of the RNA as a result of substrate binding (Ciesiolka et al. 1994). Furthermore, IVb, which is only cleaved when M1 RNA is in complex with tRNA (Ciesiolka et al. 1994; Kirsebom and Svärd 1994), was also detected in vivo (Fig. 1A, U284↓A286). Thus, the results in Figure 1 tentatively suggest that a major fraction of M1 RNA in the cell is in complex with tRNA substrates. Conversely, a minor fraction of M1 RNA shows the hallmark of being free of substrate. Figure 1A shows weak but significant cleavage at site V (A295↓G296). Cuts at this site have only been seen in the absence of tRNA (Ciesiolka et al. 1994). Altogether, this suggests that we mapped at least two subpopulations of M1 RNA in living cells, one with pre-tRNA/tRNA bound (cleavage at site IVb) and one free of tRNA (cleavage at site V).
FIGURE 1.
Pb2+ cleavage analysis of M1 RNA in vivo. (A) Total RNA was extracted and used for primer extension analysis with a radiolabeled primer, FP0251. The same primer was also used to generate a DNA sequencing ladder, which in the figure is denoted according to the M1 RNA sequence (C, T, A, G). Reverse transcription products were run on an 8% denaturing gel and subsequently autoradiographed. Pb2+ concentrations used are shown. Control refers to a primer extension performed on RNA from mock-treated cells, to correct for reverse transcription stops due to structure. Site-specific Pb2+-induced cleavage sites are denoted by Roman numerals and are indicated to the right of the autoradiogram. Some additional cleavage sites, along with their assigned nucleotide positions, are indicated as well. (B) The cleavage positions that were reproducibly found in several independent experiments with two different radiolabeled primers (M1–2 and FP0251) are represented on the secondary structure model of M1 RNA (Haas et al. 1996). Red arrows indicate Pb2+ cleavages identified both in vivo and in vitro. Pb2+ cleavages found only either in vivo or in vitro are indicated by blue and green arrows, respectively. The circled site-specific Pb2+-induced cleavage at site IVb is indicative of tRNA substrate binding. Cleavages downstream of C338 were not detectable due to the location of the primer binding sites. Specificity domain and catalytic domain of M1 RNA are indicated on the secondary structure model.
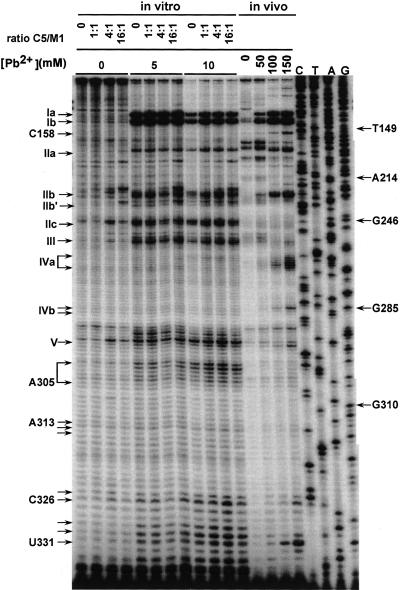
FIGURE 3.
Pb2+ cleavage analysis of M1 RNA in vitro ± C5 protein. Pb2+ concentrations used and protein:RNA molar ratios are indicated. The final concentrations of M1 RNA and C5 protein were 8 nM and 0–112 nM, respectively. Sites of Pb2+ cleavage are denoted as in Figure 1, and are indicated on the left side of the autoradiogram. Primer extensions of an in vivo Pb2+ cleavage analysis using the same radiolabeled primer, M1–2, were run on the same polyacrylamide gel for comparison.
The accessibility of M1 RNA to Pb2+ is not significantly altered by C5 protein
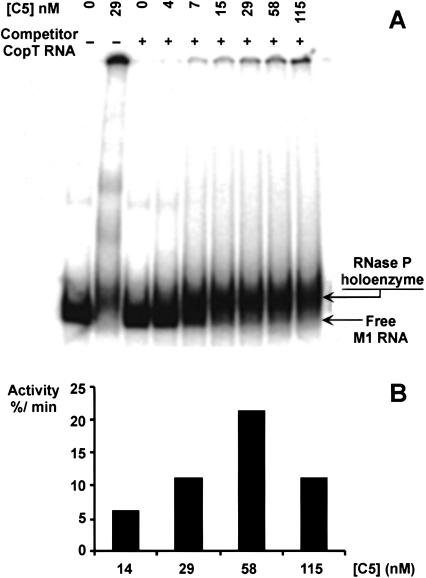
Comparing Pb2+-induced cleavage of M1 RNA in vivo with the cleavage pattern generated on naked M1 RNA in vitro did not reveal any obvious differences, apart from the possible local structural changes discussed above. Given that M1 RNA in the cell is believed to be in complex with C5, this would argue that the protein subunit does not change the overall conformation of M1 RNA significantly. To further investigate this, we conducted Pb2+-induced cleavage of M1 RNA in vitro in the presence of C5. RNase P holoenzyme was reconstituted from purified C5 protein and in vitro transcribed M1 RNA. Lack of contamination of the C5 protein preparation by M1 RNA was confirmed by primer extension and 3′-32P-end labeling (data not shown). Earlier work had shown that reconstitution of the RNase P holoenzyme required an excess of C5 protein over M1 RNA (see Kirsebom 2001 and references therein). A gel shift assay was performed at different C5 protein/M1 RNA molar ratios according to Talbot and Altman (1994). The concentration of M1 RNA was kept at 8 nM, and that of the C5 protein varied between 1.15 and 115 nM. A shift to a slower migrating form of labeled M1 RNA was observed as a function of C5 protein addition (Fig. 2A). At high C5 concentrations, the accumulation of presumed aggregates was observed, in keeping with a previous report (Talbot and Altman 1994). To assess the activity of the reconstituted holoenzyme, a cleavage assay was carried out with a previously described model substrate (Brännvall and Kirsebom 1999; Brännvall et al. 1998, 2001). Activity was at maximum between 50 and 80 nM C5 protein, in the presence of 8 nM M1 RNA (Fig. 2B; see below), using the same buffer conditions as for the Pb2+ cleavage reaction. In this assay, cleavage rates were decreased significantly at C5 concentrations that exceeded 290 nM. Cleavages occurred neither in the absence of C5 nor in the C5 alone control (data not shown).
FIGURE 2.
Gel mobility shift assay. (A) M1 RNA and C5 protein were allowed to form complexes under conditions similar to the ones used for Pb2+-induced cleavage but supplementing with 0.05% (w/v) Nonidet P40 and 5% (v/v) glycerol. Concentrations of C5 protein are given. M1 RNA was preincubated for 7 min at 37°C prior to mixing with freshly diluted C5 protein. The RNA–protein mixture was incubated for 10 more minutes before electrophoresis (see Materials and Methods). (B) RNase P holoenzyme activity test. Increasing concentrations of C5 protein were added at a constant concentration of M1 RNA (8 nM), with 300 nM of substrate (pATSerUG). Activity is expressed as the percentage of substrate cleaved/minute at steady state.
In vitro Pb2+ cleavage assays were performed in the same buffer as the one used for the gel shift (except for the absence of glycerol and NP40; see Materials and Methods), with increasing molar ratios of C5 protein/M1 RNA. The cleavage pattern in the presence of C5 protein was very similar to that in its absence. All major and minor site-specific Pb2+-induced cuts reported previously were detected (Fig. 3). Cleavage at site IVa occurred at a low frequency and was difficult to quantify due to background fragmentation, but was nevertheless confirmed in several independent experiments. These findings are in keeping with the in vivo results and confirm that the C5 protein does not greatly affect the Pb2+-induced cleavage pattern of M1 RNA. This in turn indicates that C5 does not change the overall RNA conformation to a significant extent.
Footprinting studies conducted earlier on the E. coli holoenzyme with RNase T1 or dimethyl sulfate (DMS) gave protection from cleavages in several regions (Vioque et al. 1988; Talbot and Altman 1994). Additional information about C5 protein contacts on M1 RNA came from experiments in which a Fe-EDTA cleavage compound was tethered to the protein (Biswas et al. 2000; Tsai et al. 2003) and from crosslinking studies (Sharkady and Nolan 2001). Taken together, several potential protein contact sites appear to be spread out over a rather extended RNA surface. Nevertheless, Figure 3 (in vitro, ±C5) indicates no significant C5-dependent protection from Pb2+-induced cleavage. Several things might account for this. First, Pb2+ preferentially induces cuts at positions spatially close to metal ion binding sites. Thus, only a limited number of sites are easily accessible and can be detected in the primer extension reaction. Second, protection from site-specific cleavage requires a change in the positioning (or a replacement) of the Pb2+ ion such that it no longer can promote cleavage at that particular site. This makes it impossible to detect structural changes that fail to alter the coordination of Pb2+ at these sites. Third, interactions between RNA and protein, in particular the direct contacts between the protein and nucleotides in the RNA, may be weak. The small size of the Pb2+ ion might therefore enable its entry at sites of weak interaction, resulting in cleavage. In contrast, the much larger RNase T1 is expected to be more sensitive to spatial interference. We can conclude that Pb2+ is, in this case, not suitable to map protein–RNA contacts (see, however, Valverde et al. 2004). A further limitation that Pb2+ shares with several other probing techniques lies in its preference for single-stranded regions. Thus, protein binding sites in double-stranded regions will escape detection.
In general, protection of Pb2+ cleavage sites in an RNA, dependent on a binding protein, does not necessarily reflect direct contacts, but could rather be due to induced conformational alterations. Pb2+ acetate has been shown to be a sensitive probe to map even small structure changes in RNAs (Gornicki et al. 1989). In the case of this study, Pb2+ has been unable to detect C5 binding to M1 RNA, both in vivo and in vitro (cf. Fig. 3). Based on the similarity of the cleavage patterns of M1 RNA in vitro, in absence or presence of the C5 protein, and the in vivo pattern, we can, however, conclude that no major structural rearrangements are induced in M1 RNA upon C5 protein binding.
A metal ion core in the specificity domain of RNase P RNA?
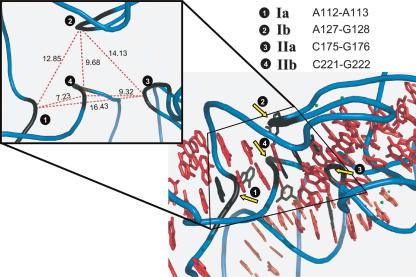
A crystal structure of a complete RNase P RNA is presently not available. However, Krasilnikov and coworkers (2004) have recently solved the crystal structure of a domain of a type A RNase P RNA. Hence, based on our present findings suggesting that the conformation of M1 RNA is not significantly different in vitro (± the C5 protein) and in vivo, and that M1 RNA belongs to type A, we felt encouraged to map the Pb2+-induced cleavage sites detected (Figs. 1, 3) onto the structure of the specificity domain of Thermus thermophilus RNase P RNA. The corresponding domain of M1 RNA is indicated in Figure 1. Figure 4 shows clustering of the cleavage sites in the center of the specificity domain. Metal ion induced cleavage of the RNA backbone results from activation of a neighboring 2′OH-group to act as a nucleophile (Kirsebom and Ciesiolka 2005). Note also that Mg2+ induces cleavage at sites I and II (Kazakov and Altman 1991). Thus, this raises the interesting possibility that the center of the specificity domain constitutes a metal ion core that is important for folding of the RNA, in a fashion similar to that proposed for the P4/P6 domain of the Group I intron (Strobel and Doudna 1997). Binding of metal ions in this region of the specificity domain is further supported by the positioning of Ba2+ ions in the crystal structure, near the Pb2+-induced cleavage sites, and the fact that Ba2+ (as well as other divalent metal ions) can suppress Pb2+- induced cleavage at these sites (Brännvall et al. 2001).
FIGURE 4.
Pb2+-induced cleavage sites mapped onto the structure of the specificity domain of RNase P RNA. Pb2+-induced cleavage sites Ia, Ib, IIa, and IIb were mapped onto the structure of the specificity domain of Thermus thermophilus RNase P RNA. The cleavage positions within the backbone are indicated in black (yellow arrows). (Inset) Distances in A° between the Pb2+ induced cleavage sites Ia, Ib, IIa, and IIb in the specificity domain (see Fig. 1 and text for details).
Pb2+ cleavages in vivo are often more distinct than those in vitro
An incidental observation both in previous work (Lindell et al. 2002; M. Lindell, unpubl.) and here (Fig. 3) concerns the more distinct bands obtained in in vivo Pb2+ probing experiments, compared to those seen in in vitro experiments conducted in parallel. Generally, we obtain much sharper cleavage sites and less background (noise) bands, even though downstream analysis is carried out by the same primer extension technique, using identical protocols and primers. This conclusion is valid for longer RNAs such as tmRNA (Lindell et al. 2002; data not shown) and M1 RNA (present work). At first, this seems surprising, since extracted total RNA contains a vast molar excess of competing RNA species that would be expected to rather increase noise. In contrast, in vitro synthesized and purified RNA would be expected to yield superior mapping data. Thus, in view of the fact that we have repeatedly observed better cleavage patterns in vivo, we tentatively conclude that this may reflect the more “natural” setting of the cellular environment, with the presence of RNA chaperones and an ordered cotranscriptional folding pathway as likely important factors. If this is so, then the less distinct patterns obtained frequently on longer RNAs in vitro may reflect the presence of different conformers, possible arising from improper folding. In addition, minor fragmentation of in vitro transcribed M1 RNA might also contribute to the background bands observed after reverse transcription.
In conclusion, we have shown that the in vivo structure of the M1 RNA subunit of RNase P can be assessed by probing with Pb2+ acetate. Our data suggest that protection by the C5 protein cannot be detected (Figs. 1, 3) and that the overall conformation of M1 RNA is not significantly changed upon protein binding (Fig. 3). The simultaneous presence of several hallmark cleavages, and their relative intensities, furthermore indicates that RNase P in the cell is predominantly bound by substrate, whereas a minor fraction is in its free form.
MATERIALS AND METHODS
Oligodeoxyribonucleotides
DNA templates used for sequencing reactions were generated by colony-PCR using the following primer pair: AV-22 (5′-AGGT GAAACTGACCGATAAG) and FP0016 (5′-CGTCCATTCGCCA TTCAGGCTGCGC). For primer extension and sequencing reactions, the following primers were used: M1–2 (5′-GCCGGGTT CTGTCGTGGACAG; +338 to +358 of rnpB) and FP0251 (5′-GT TCTGTCGTGGACAGTCATTCATCTAGG; +325 to +353 of rnpB).
In vivo Pb2+ acetate cleavage of RNA
The protocol used here has essentially been described (Lindell et al. 2002). E. coli cells were grown at 37°C in L Broth (LB). Overnight cultures were diluted 100× in LB and grown to mid-log phase (OD600 = 0.5). Immediately before treatment, Pb2+ solutions were prepared by mixing 3 volumes of a lead solution (appropriately diluted 1 M Pb2+ acetate (Merck) in H2O) with 1 volume of prewarmed 4× concentrated LB. Unless otherwise stated, 8 mL of this Pb2+ acetate/LB solution were added to 20 mL of culture to give a final Pb2+ concentration of 100 mM. Cultures were further incubated with vigorous shaking at 37°C for 7 min. Reactions were stopped by addition of a 1.5-fold molar excess of EDTA and immediately put on ice. Total RNA was extracted by the hot phenol method (Blomberg et al. 1990) and RNA samples were further treated with DNase I (Amersham Biosciences) to eliminate DNA.
Pb2+ cleavages were identified by primer extension. The 5′-32P-end- labeled primers (M1–2 or FP0251) were annealed to 10 μg of total RNA samples (incubations: 90°C for 1 min, 1 min on ice, and 5 min at 20°C), and primer extension was performed at 45°C for 30 min using 200 units of Superscript II (Invitrogen) in a total reaction volume of 15 μL, containing 50 mM Tris-HCl (pH 8.5), 6 mM MgCl2, 40 mM KCl and dNTPs (1.0 mM each). Reactions were terminated by adding 20 μL of stop buffer (50 mM Tris-HCl at pH 7.5, 0.1% SDS) and 3.5 μL of 3 M KOH. After 3 min at 90°C, and subsequently 3 h at 37°C, 6 μL of 3 M acetic acid were added, and the cDNA was precipitated with ethanol. Reverse transcription products were run on 8% polyacrylamide gels containing 7 M urea. Cleavage positions were identified by comparison to sequencing reactions, generated from suitable PCR-generated DNA templates using the same 5′ end-labeled primers, run in parallel. The Circumvent thermal cycle dideoxy DNA sequencing kit (New England Biolabs) was used according to the manufacturer’s protocol.
In vitro Pb2+ acetate cleavage of M1 RNA
3′-32P-end-labeled M1 RNA (0.24 pmol) was preincubated for 10 min without or with C5 protein (final concentration of C5 protein as indicated in the figures) at 37°C in 50 mM Tris-OAc (pH 7.6), 100 mM NH4OAc, 10 mM Mg(OAc)2, 0.5 μM CopT RNA. CopT RNA was added as a carrier. Cleavage was initiated by the addition of Pb(OAc)2 to a final concentration of 5–10 mM (final reaction volume was 30 μL). The reaction was allowed to proceed for 5 min at 37°C, and was then stopped by the addition of 5 μL 0.1 M EDTA and 75 μL 99% ethanol. The precipitated RNA was vacuum-dried and dissolved in water prior to analysis by primer extension.
Gel mobility shift assay
3′-32P-end-labeled M1 RNA was preincubated in 50 mM Tris-OAc (pH 7.6), 100 mM NH4OAc, 10 mM Mg(OAc)2, 0.05% (w/v) NP40, 5% glycerol, and 1 μM CopT RNA for 7 min. An equal volume of C5 protein diluted in the same buffer (but without CopT RNA) was added to give a final concentration of 8 nM M1 RNA and 0.5 μM CopT RNA (see figure legends for the concentrations of C5 protein). The mixture was incubated for 10 min at 37°C and then immediately loaded onto 5% polyacrylamide gels in 25 mM Tris-OAc (pH 8.0), 0.5 mM Mg(OAc)2 and 0.005% NP40. The gel was run in a cold room at 5 W (≈300 V) and subsequently dried and analyzed using a PhosphorImager (Molecular Dynamics).
Activity assay
The activity of the RNase P holoenzyme was measured in the buffer used for the Pb2+ induced cleavage, but without CopT RNA. Final concentrations of M1 RNA and substrate (pATSerUG) were 8 nM and 300 nM, respectively, and C5 protein concentrations ranged from 0 to 580 nM. M1 RNA/C5 protein solution and 5′-32P-end-labeled substrate solution were preincubated separately at 37°C for 7 and 2 min, respectively, before mixing equal volumes to start the reaction. Time point aliquots were withdrawn and immediately mixed with 3 volumes of 10 M urea/15 mM EDTA to stop the reaction. Reaction products were separated by denaturing PAGE and analyzed as above. The percentage of cleaved substrate was plotted against time, and the activity was calculated from the initial rates.
Acknowledgments
The authors acknowledge support from The Swedish Research Council, Wallenberg Consortium North, and the Foundation of Strategic Research. We thank Nils E. Mikkelsen for providing the RNA structure model in Figure 4.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2590605.
REFERENCES
- Biswas, R., Ledman, D.W., Fox, R.O., Altman, S., and Gopalan, V. 2000. Mapping RNA–protein interactions in ribonuclease P from Escherichia coli using disulfide-linked EDTA-Fe. J. Mol. Biol. 296: 19–31. [DOI] [PubMed] [Google Scholar]
- Blomberg, P., Wagner, E.G.H., and Nordström, K. 1990. Control of replication of plasmid R1: The duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 9: 2331–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brännvall, M. and Kirsebom, L.A. 1999. Manganese ions induce mis-cleavage in the Escherichia coli RNase P RNA-catalyzed reaction. J. Mol. Biol. 292: 53–63. [DOI] [PubMed] [Google Scholar]
- Brännvall, M., Mattsson, J.G., Svärd, S.G., and Kirsebom, L.A. 1998. RNase P RNA structure and cleavage reflect the primary structure of tRNA genes. J. Mol. Biol. 283: 771–783. [DOI] [PubMed] [Google Scholar]
- Brännvall, M., Mikkelsen, N.E., and Kirsebom, L.A. 2001. Monitoring the structure of Escherichia coli RNase P RNA in the presence of various divalent metal ions. Nucleic Acids Res. 29: 1426–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesiolka, J., Hardt, W.D., Schlegl, J., Erdmann, V.A., and Hartmann, R.K. 1994. Lead-ion-induced cleavage of RNase P RNA. Eur. J. Biochem. 219: 49–56. [DOI] [PubMed] [Google Scholar]
- David, L., Lambert, D., Gendron, P., and Major, F. 2001. Leadzyme. Methods Enzymol. 341: 518–540. [DOI] [PubMed] [Google Scholar]
- Gornicki, P., Baudin, F., Romby, P., Wiewiorowski, M., Kryzosiak, W., Ebel, J.P., Ehresmann, C., and Ehresmann, B. 1989. Use of lead(II) to probe the structure of large RNAs. Conformation of the 3′ terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J. Biomol. Struct. Dyn. 6: 971–984. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N., and Altman, S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. [DOI] [PubMed] [Google Scholar]
- Haas, E.S., Banta, A.B., Harris, J.K., Pace, N.R., and Brown, J.W. 1996. Structure and evolution of ribonuclease P RNA in Gram-positive bacteria. Nucleic Acids Res. 24: 4775–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov, S. and Altman, S. 1991. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli. Proc. Natl. Acad. Sci. 88: 9193–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom, L.A. 2001. Escherichia coli ribonuclease P. Methods Enzymol. 342: 77–92. [DOI] [PubMed] [Google Scholar]
- Kirsebom, L.A. and Ciesiolka, J. 2005. Pb2+-induced cleavage of RNA. In Handbook of RNA biochemistry (eds. Hartmann R.K. et al.), pp. 214–228. Wiley-VCH Verlag GmbH & Co., Weinheim.
- Kirsebom, L.A. and Svärd, S.G. 1994. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 13: 4870–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov, A.S., Xiao, Y., Pan, T., and Mondragón, A. 2004. Basis for structural diversity in homologous RNAs. Science 306: 104–107. [DOI] [PubMed] [Google Scholar]
- LaGrandeur, T.E., Hüttenhofer, A., Noller, H.F., and Pace, N.R. 1994. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 13: 3945–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell, M., Romby, P., and Wagner, E.G.H. 2002. Lead(II) as a probe for investigating RNA structure in vivo. RNA 8: 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J.G., Svärd, S.G., and Kirsebom, L.A. 1994. Characterization of the Borrelia burgdorferi RNase P RNA gene reveals a novel tertiary interaction. J. Mol. Biol. 241: 1–6. [DOI] [PubMed] [Google Scholar]
- Oh, B.K. and Pace, N.R. 1994. Interaction of the 3′-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 22: 4087–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl, P. and Primakoff, P. 1973. Mutants of Escherichia coli thermo-sensitive for the synthesis of transfer RNA. Proc. Natl. Acad. Sci. 70: 2091–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel, S.A. and Doudna, J.A. 1997. RNA seeing double: Close-packing of helices in RNA tertiary structure. Trends Biochem. Sci. 22: 262–266. [DOI] [PubMed] [Google Scholar]
- Talbot, S.J. and Altman, S. 1994. Gel retardation analysis of the interaction between C5 protein and M1 RNA in the formation of the ribonuclease P holoenzyme from Escherichia coli. Biochemistry 33: 1399–1405. [DOI] [PubMed] [Google Scholar]
- Tallsjö, A., Svärd, S.G., Kufel, J., and Kirsebom, L.A. 1993. A novel tertiary interaction in M1 RNA, the catalytic subunit of Escherichia coli RNase P. Nucleic Acids Res. 21: 3927–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H.Y., Masquida, B., Biswas, R., Westhof, E., and Gopalan, V. 2003. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol. 325: 661–675. [DOI] [PubMed] [Google Scholar]
- Valverde, C., Lindell, M., Wagner, E.G.H., and Haas, D. 2004. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J. Biol. Chem. 279: 25066–25074. [DOI] [PubMed] [Google Scholar]
- Vioque, A., Arnez, J., and Altman, S. 1988. Protein–RNA interactions in the RNase P holoenzyme from Escherichia coli. J. Mol. Biol. 202: 835–848. [DOI] [PubMed] [Google Scholar]
- Zito, K., Hüttenhofer, A., and Pace, N.R. 1993. Lead-catalyzed cleavage of ribonuclease P RNA as a probe for integrity of tertiary structure. Nucleic Acids Res. 21: 5916–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]