Abstract
ATP-dependent nucleosome remodeling plays a central role in the regulation of access to chromatin DNA. Swi/Snf remodeling complexes characterized in yeast, Drosophila and mammals all contain a conserved set of core subunits composed of homologs of yeast SNF2-type DNA-dependent ATPase, SNF5 and SWI3 proteins. So far, no complete Swi/Snf-type complex has been characterized in plants. Arabidopsis contains a single SNF5-type gene, BSH, which has been shown to complement the yeast snf5 mutation. Here we describe the characterization of AtSWI3B, the smallest of the four Arabidopsis homologs of SWI3. The gene encoding AtSWI3B is expressed ubiquitously in the plant. AtSWI3B is localized to nuclei and is associated mostly with the chromatin and soluble protein fractions. When expressed in Saccharomyces cerevisiae, the cDNA encoding AtSWI3B partially complements the swi3 mutant phenotype. However, like BSH, AtSWI3B is unable to activate transcription in yeast when tethered to DNA. The analysis by yeast two-hybrid indicates that AtSWI3B is capable of forming homodimers and interacts with BSH as well as with two other members of the Arabidopsis SWI3 family: AtSWI3A and AtSWI3C. The results of phage display screen using recombinant protein, confirmed by direct yeast two-hybrid analyses, indicate that AtSWI3B interacts with FCA, a regulator of flowering time in Arabidopsis. This interaction is through the C-terminal region of FCA, located outside the conserved RNA- and protein-binding domains of this protein.
INTRODUCTION
In eukaryotes, chromatin remodeling is a critical step in transcriptional regulation of genes and other processes that require access to DNA. Remodeling is mediated by complexes that covalently modify the core histones (1) and by complexes using ATP to change the location or conformation of the nucleosomes (2). ATP-dependent chromatin remodeling has been characterized in such diverse organisms as yeast, Drosophila and humans. The different ATP-dependent complexes identified so far share a similar type of central subunit—a DNA (or chromatin)-dependent ATPase. These ATPases all contain the ‘SNF2_N’ domain and form a characteristic subclass of the DEAD/H superfamily of nucleic acid-stimulated ATPases (3). The current classification of the ATP-dependent complexes is based on the type of ATPase acting as the central catalytic subunit. The best known of these ATPases belong to three main subfamilies represented by the SWI2/SNF2, ISWI and Mi-2 types. While the Swi/Snf- and Mi-2-type complexes are large assemblies composed of eight or more subunits, the ISWI complexes are smaller and contain only two to five subunits. The subunit composition seems to be least variable in the case of the Swi/Snf-type complexes. The yeast, Drosophila and human complexes of this type all contain a minimal structural and functional core composed of three proteins: the SWI2/SNF2 ATPase, SNF5 and SWI3. This minimum core complex can remodel chromatin in vitro (4). The composition of the ISWI- and Mi-2-type complexes is less well conserved. In addition to the core proteins, the yeast Swi/Snf complex contains a set of subunits, which were shown by genetic analysis to be required for common functions in vivo (5).
So far, no complete chromatin-remodeling complex has been described in plants. However, examinations of the databases (6) indicate that the existence of such complexes in plants is highly probable. Our laboratory has been involved in the systematic analysis of the components of putative plant Swi/Snf-type complexes. In an earlier study we identified and characterized the Arabidopsis BSH gene and established that it encodes the functional homolog of the yeast SNF5 protein, a key subunit of the Swi/Snf-type complexes (7). An analysis of the complete Arabidopsis genome sequence revealed that BSH is the only homolog of SNF5 present (S.Kaczanowski and A.Jerzmanowski, unpublished results). Given the critical function of the SNF5 subunit in organizing the core of the Swi/Snf complex (8), this makes BSH an ideal marker of the putative plant Swi/Snf complexes. In this report we describe the characterization of a member of a small subfamily of Arabidopsis homologs of the yeast SWI3 protein, another key core subunit of the canonical Swi/Snf complex.
MATERIALS AND METHODS
Cloning of AtSWI3B cDNA
The AtSWI3B cDNA was amplified by PCR with primers SWI170NU, CAT GCC ATG GCC ATG AAA GCT CCC GAT, and SWI170N2L, GCG GAT CCT GCC CAA GCT CTT TCA GAT TC, designed on the basis of the corresponding Arabidopsis cDNA sequence found in GenBank. PCR conditions were as follows: 95°C for 5 min, 30× (56°C for 30 s, 72°C for 1 min 30 s, 95°C for 30 s), 56°C for 30 s, 72°C for 4 min. The PCR product was amplified from cDNA generated using a Gibco BRL RT SuperScript™ kit with a total RNA isolated from Arabidopsis Col-0 ecotype using the RNeasy kit (Qiagen). This product was then digested with NcoI and BamHI and inserted into the expression vector pQE60 (Qiagen). The resulting plasmid pQESWI3B was sequenced on both strands using an ALF (Pharmacia) DNA sequencer to confirm that the cloned fragment was free from errors.
Complementation of yeast swi3 mutant strain
A full-length AtSWI3B PCR product amplified from cDNA with primers SWI3Compl2U, TCC CCC GGG GAT GGC CAT GAA AGC TCC CGA, and SWI3Compl1L, CGG GAT CCT CAT TGA GTA TAA CCA TAT AAT, was digested with BamHI and SmaI and inserted into the pSI4 yeast multicopy expression vector (kindly provided by Dr Iwona Smaczynska) to produce the pSISWI3B plasmid in which the expression of the cloned gene is driven by the CTA1 inducible promoter. PCR conditions were the same as for the SWI170NU and SWI170N2L primers. The cDNA in pSISWI3B was sequenced on both strands using the ALF sequencer. The phenotype of the yeast swi3 strain CY 165 (kindly provided by Dr Craig Peterson) after transformation with the pSISWI3B, empty pSI4 or plasmid RS 313 SWI3 (kindly provided by Dr Craig Peterson), containing the yeast SWI3 gene, was tested with an assay for HO:LacZ activity using ONPG as a substrate for β-galactosidase, as described in Kaiser et al. (9).
Transcription activation assay
The full-length AtSWI3B cDNA was excised with BamHI and SmaI from pSISWI3B and inserted into pGBT9(DNA-BD) (Clontech) resulting in the pGBTSWI3B plasmid. The pGBTSWI3B plasmid was transformed into the Y190 (Clontech) yeast strain. The level of β-galactosidase activity was measured using the replica lift method as described in the Clontech yeast protocols handbook. As controls, the Y190 strain was transformed with pGBT9 alone and with plasmid pCL1 to express the full-length GAL 4 protein.
Analysis of AtSWI3B expression by RT–PCR
RNA was extracted from roots, leaves, stalks and flowers of Arabidopsis plants using the RNeasy kit and used for generation of cDNA using Gibco BRL RT kit according to the manufacturer’s instructions. The PCR was performed with SWI3Compl2U and SWI3Compl1L primers with the conditions described above. As a control, a fragment of the Arabidopsis protein actin 2 (AA2G18780) cDNA was amplified with primers ACT1, AGA GAT TCA GAT GCC CAG AAG TCT TGT TCC, and ACT2, AAG GAT TCC TGG ACC TGC CTC ATC ATA CTC, using the same conditions.
Production of anti-AtSWI3B antibodies
Recombinant AtSWI3BHis6 protein containing amino acids from 1 to 390 of AtSWI3B with six histidines fused to the C-terminus was overproduced in Escherichia coli SG13009 cells using the pQESWI3B plasmid and purified by selective binding to NiNTA (Qiagen) followed by elution according to the manufacturer’s protocol. The final preparation (∼1 mg of protein) was separated by preparative SDS–PAGE. The recombinant protein was eluted from a stained band cut from the gel, digested with trypsin and subjected to mass fingerprinting using a QTof (Micromass) spectrometer. The obtained mass fingerprint of peptides confirmed that the overexpressed protein was AtSWI3B. Gel fragments containing pure AtSWI3BHis6 were used as the source of antigen to immunize rabbits. Anti-AtSWI3BHis6 rabbit serum was prepared by Eurogentec (Seraing, Belgium).
Isolation of nuclei and extraction of nuclear proteins
Arabidopsis nuclei were isolated according to Manzara and Gruissem (10). Seventy-five grams of frozen plants was briefly pulverized in a mortar and homogenized with 300 ml of buffer containing 25 mM PIPES pH 7.0, 10 mM NaCl, 5 mM EDTA pH 8.0, 250 mM sucrose, 0.15 mM spermine, 0.5 mM spermidine, 20 mM 2-mercaptoethanol, 0.1% Nonidet P-40, 0.2 mM PMSF. The homogenate was filtered through one layer of Myracloth (Calbiochem) over three layers of Nitex (Tetco) nylon membrane (pore sizes 300, 100 and 56 µm) and centrifuged in a Sorvall SLA-1000 rotor at 6000 r.p.m. for 20 min at 4°C. After discarding the supernatant the nuclear pellet was washed three times with 20 ml of homogenization buffer by resuspension and centrifugation in a Sorvall SA-300 rotor at 4500 r.p.m. for 10 min at 4°C. Finally, the pellet was resuspended in 1 ml of a buffer containing 50 mM HEPES–KOH pH 7.6, 100 mM NaCl, 5 mM MgCl2, 10 mM KCl, 50% glycerol, 1 mM DTT and protease inhibitor cocktail (Complete EDTA-free; Boehringer, Mannheim). In order to extract nuclear proteins the suspension was incubated for 20 min at room temperature with 50 U DNase I (Roche, Mannheim) and homogenized by passing through a syringe needle.
Isolation of nuclear matrix by high- and low-salt methods
The procedures were adapted from Reyes (11). For high-salt isolation of nuclear matrix, nuclei were resuspended in CSK buffer: 10 mM PIPES pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA supplemented with protease inhibitor cocktail (Complete EDTA-free; Boehringer) at one tablet for 50 ml of buffer, 1 mM PMSF, 1 mM DTT and 0.5% (v/v) Triton X-100. After 3 min at 4°C the nuclei were separated from soluble proteins by centrifugation at 5000 g for 3 min. Chromatin was solubilized by digestion of DNA with 1 mg/ml of RNase-free DNase I (Roche) in CSK buffer for 15 min at 37°C. Ammonium sulphate was then added from a 1 M stock solution in CSK buffer to a final concentration of 0.25 M and after 5 min at 4°C, the samples were centrifuged again. The pellet was extracted three times with 2 M NaCl in CSK buffer by resuspension, incubation at 4°C for 5 min and centrifugation. The remaining pellet containing nuclear matrix was solubilized in urea buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris–HCl pH 8). For low-salt preparation of nuclear matrix, nuclei were stabilized in isolation buffer containing 3.75 mM Tris–HCl pH 7.5, 0.05 mM spermine, 0.125 mM spermidine, 0.5 mM EDTA, 5 mM MgCl2, 20 mM KCl, 1 mM DTT and protease inhibitor cocktail (Complete EDTA-free; Boehringer) for 20 min at 37°C. DNA was then digested by incubation at 37°C for a further 15 min in the presence of 1 mg/ml RNase-free DNase I. After centrifugation, nuclei were washed with isolation buffer and extracted with the same buffer supplemented with 25 mM 3,5-diiodosalicylic acid, lithium salt (LIS) for 5 min at room temperature. Chromatin-depleted nuclei were recovered by centrifugation and the pellet was solubilized in urea buffer.
Protein characterization and immunoblot analyses
The characterization of protein samples by SDS–PAGE and western blot analysis were as described previously (7). The concentration of proteins in the samples was determined using the Bradford procedure with the BioRad Protein Assay kit (BioRad Laboratories, Inc.) according to the manufacturer’s protocol.
Phage display analysis
AtSWI3BHis6 recombinant protein was refolded and used to screen for interacting heptapeptides using the Ph.D.-7™ Phage Display Peptide Library Kit (New England Biolabs). All steps were carried out according to the manufacturer’s instructions.
The DNA encoding heptapeptides in phage, which interacted with AtSWI3BHis6, was sequenced and this was used to search the SWISSPROT Database using Findpattern from the GCG package.
Analysis of AtSWI3B interactions in yeast two-hybrid system
Interactions with BSH, AtSWI3A, AtSWI3B and AtSWI3C. The pGBSH (DNA-BD) plasmid containing full-length cDNA of BSH was kindly provided by Katarzyna Olczak. The AtSWI3A cDNA was amplified by PCR with primers SWI3AU, TCC CCC GGG GGA AGC CAC TGA TCC AAG C, and SWI3AL, CGG GAT CCT TTC ACG TAC GTA TGA TCC C, designed on the basis of the corresponding Arabidopsis cDNA sequence found in GenBank. PCR conditions were as follows: 95°C for 5 min, 30× (59°C for 30 s, 72°C for 1 min 30 s, 95°C for 30 s), 59°C for 30 s, 72°C for 4 min. The AtSWI3C cDNA was amplified by PCR with primers SWI3CU, TCC CCC GGG GCC AGC TTC TGA AGA TAG AAG AG, and SWI3CL, ACG CGT CGA CTA GTT TAA GCC TAA GCC GGA, designed on the basis of the corresponding Arabidopsis cDNA sequence found in GenBank. PCR conditions were as follows: 95°C for 5 min, 30× (58°C for 30 s, 72°C for 2 min 30 s, 95°C for 30 s), 58°C for 30 s, 72°C for 4 min. The PCR products were amplified from cDNA generated using a Gibco BRL RT kit with a total RNA isolated from Arabidopsis Col-0 ecotype using the RNeasy kit (Qiagen). These products were then sub cloned using SmaI and BamHI restriction sites into vector pGAD424 (GAL4-AD) (Clontech) to produce the constructs pGADSWI3A and pGADSWI3C, respectively. The obtained constructs were sequenced on both strands using an ALF (Pharmacia) DNA sequencer to confirm that the cloned fragments were error-free. The full-length AtSWI3B cDNA was excised with BamHI and SmaI from pGBTSWI3B and inserted into pGAD424 (AD) (Clontech) resulting in the vector pGADSWI3B. The yeast strain Y190 was transformed with plasmid pairs: pGBSH/pGADSWI3B, pGBTSWI3B/pGADSWI3B, pGBTSWI3B/pGADSWI3A and pGBTSWI3B/ pGADSWI3C. As a control, Y190 yeast cells were transformed with plasmids, pGBSH, pGBTSWI3B, pGADSWI3A, pGADSWI3B, pGADSWI3C and pCL1 singly. All transformants were grown on a selection medium containing 100 mM 3-amino-1,2,4-triazole and the level of β-galactosidase activity was measured by the replica lift method described in the Clontech yeast protocols handbook.
Interactions with FCA. The cDNAs encoding the full-length and C-terminal half of FCA were subcloned from plasmids p226 and p227, respectively (kindly provided by Dr Caroline Dean laboratory) into vector pGAD424 (GAL4 AD) (Clontech) using EcoRI and SalI restriction sites, to produce the constructs pGADFCA and pGAD227, respectively. A truncated cDNA encoding FCA without the C-terminal region was obtained by PCR using plasmid p227 as template, with primers FCA-U1, CCG GAA TTC ATG AAT GGT CCC CCA GAT AG, and FCA-U2209, A CGC GTC GAC TCA TGA TAT ATC ATT C. PCR conditions were as follows: 95°C for 5 min, 30× (57.7°C for 30 s, 72°C for 2 min 30 s, 95°C for 30 s), 57.7°C for 30 s, 72°C for 5 min. Interactions between AtSWI3B and FCA were analyzed using the Matchmaker GAL4 Two-Hybrid System (Clontech) according to the manufacturer’s instructions.
The yeast strain Y190 was transformed with plasmid pairs pGADFCA/pGBTSWI3B and pGAD227/pGBTSWI3B. As a control, Y190 yeast cells were transformed with plasmids pGADFCA, pGAD227, pGBTSWI3B, pCL1 singly. All transformants were grown on a selection medium containing 100 mM 3-amino-1,2,4-triazole and the level of β-galactosidase activity was measured by the replica lift method described in the Clontech yeast protocols handbook.
Homology searches and phylogenetic analyses
The basic database used for the homology searches was a sum of the protein sequence databases of SWISSPROT, SP-TrEMBL and MIPS MATDB from March 2001. Low complexity segments in the basic database were masked using the GCG implementation of the SEG program (12). The database was searched using the GCG implementation of the SSEARCH and FASTA programs. Multiple alignments were generated using the PILEUP program from the GCG package. The alignments were analyzed and edited with the SeqLab color editor from the GCG package. The consensus maximum parsimony trees were calculated using the PROTPARTS program from the PHYLIP package (13). The maximum likelihood trees and the likelihood ratio test of the molecular clock hypothesis (14) were calculated using the PUZZLE program (15).
RESULTS
The Arabidopsis genome contains a small family of SWI3-type genes
An alignment of the sequences obtained by searching databases with yeast SWI3 as the query revealed that the majority of the SWI3 sequence is not evolutionarily conserved. The most conserved domain is located in the central region between amino acids 306 and 393. A search of the databases (see Materials and Methods for a list of data bases) with the whole yeast SWI3 sequence revealed four Arabidopsis proteins: AT2G47620, AT2G33610, AT1G21700 and AT4G34430, which we named AtSWI3A, AtSWI3B, AtSWI3C and AtSWI3D, respectively. Another search performed with just the conserved SWI3 domain as the query sequence resulted in a list containing only three proteins from Arabidopsis, AtSWI3A to AtSWI3C. This result was confirmed by a multiple alignment of the fourth putative Arabidopsis homolog, AtSWI3D, with sequences containing the main SWI3 domain. The fourth Arabidopsis sequence contained a fragment with homology to a part of the conserved sequence, but in general did not appear to be closely related to the other proteins (results not shown). However, a survey of the recent RIKEN Genomic Sciences Center cDNA sequence release from January 2002 revealed that the protein annotated as AT4G34430 in the MIPS database (AtSWI3D in this work), which was deduced from the Arabidopsis genomic sequence, is incomplete due to an error in assigning the start codon. The correct sequence is longer and contains the complete conserved domain of the SWI3-type proteins. The full AtSWI3D protein has the GenBank ID AAL67003.
An alignment of sequences corresponding to the conserved fragment between amino acids 306 and 393 of yeast SWI3 (with the variable region between amino acids 332 and 334 removed) was used to construct phylogenetic trees. Both the maximum parsimony tree (Fig. 1A) and the likelihood tree (not shown) had similar topologies. The critical statistical significance obtained in a likelihood ratio test was 5.13%, which is indicative of a molecular clock. The pairs AtSWI3A/AtSWI3B and AtSWI3C/AtSWI3D occur in separate clusters, it is however difficult to conclude which of them is more related to the branches of yeast and animal SWI3 sequences. The Arabidopsis SWI3 homologs within the clusters do not show a high degree of sequence identity, which may indicate non-redundant or only partly overlapping functions. All four potential orthologs of Saccharomyces cerevisiae SWI3 share characteristic motifs with the yeast protein, although only AtSWI3C has a similar overall size to the yeast counterpart (Fig. 1B). In an attempt to identify a homolog with a potentially novel function we decided to characterize AtSWI3B, the smallest member of the family, with a predicted molecular weight of 47 595 Da.
Figure 1.
The family of Arabidopsis proteins homologous to yeast SWI3. (A) Maximum parsimony tree for SWI3-type proteins. The tree is a consensus of 100 maximum parsimony trees based on sequences of SWI3-type proteins aligned as described in Materials and Methods. Percentage values on each branch represent the corresponding bootstrap probability values obtained in 100 replications. Arabidopsis proteins were given tentative names: AtSWI3A, AtSWI3B, AtSWI3C and AtSWI3D. (B) Distribution of conserved regions in Arabidopsis and yeast SWI3 proteins. Protein sequences were analyzed using the MEME program from the GCG package. The lengths of the proteins are indicated in parentheses.
AtSWI3B is ubiquitously expressed in Arabidopsis
To analyze the expression pattern of AtSWI3B in Arabidopsis, RNA, isolated from different organs of the plant, was used as the template for the RT–PCR with primers specific for the full-length coding sequence. PCR products of the expected size were amplified from cDNA of roots, stalks, leaves and flowers (Fig. 2), indicative of ubiquitous expression of AtSWI3B in the plant.
Figure 2.
RT–PCR analysis of AtSWI3B expression in Arabidopsis. (A) RNA was extracted from different organs of the plant and used for generation of the AtSWI3B cDNA. (B) Control showing actin gene expression, analyzed by RT–PCR, using the same preparations of RNA as in (A).
Intracellular localization of AtSWI3B protein
The cDNA of AtSWI3B (annotated in MIPS database as AT2G33610) was cloned by RT–PCR and its identity confirmed by sequencing. This cDNA was cloned into an E.coli expression vector and the recombinant protein was used to obtain anti-AtSWI3B antibodies. To determine whether AtSWI3B is a cytoplasmic or nuclear protein, protein extracts were prepared from whole cells and from isolated nuclei. Identical amounts (30 µg) of protein from each type of extract were separated by SDS–PAGE, western blotted onto a membrane and probed with anti-AtSWI3B antibodies. The detection of AtSWI3B in the nuclear—but not in the whole cell—extract (Fig. 3A) suggests that it is a nuclear protein. To investigate the possibility that a portion of the AtSWI3B pool may bind to the inner nuclear skeleton, the nuclear matrix was isolated by two different procedures. In the first, the soluble proteins were removed by extraction with the buffer containing Triton X-100 and chromatin proteins were released by DNase I digestion and extraction with 0.25 M ammonium sulphate. After washing three times with 2 M NaCl the final nuclear pellet was presumed to contain nuclear matrix proteins and the proteins tightly associated with the nuclear matrix. Supernatants from each extraction and the nuclear pellet were analyzed by SDS–PAGE and immunoblotting. As shown in Figure 3B, >50% of AtSWI3B was found in the chromatin and soluble fractions while the remaining part was bound to the nuclear matrix. To avoid the high-salt washes, which could potentially induce precipitation of some of the soluble nuclear proteins, an alternative method of nuclear fractionation was used. The nuclei were stabilized in the isolation buffer containing spermine and spermidine, digested with DNase I and washed with the digestion buffer and then extracted with the same buffer containing the detergent-like LIS (25 mM 3,5-diiodosalicylic acid, LIS). The resulting pellet was considered to contain the nuclear matrix. As shown in Figure 3C most of the AtSWI3B is found in chromatin and in the soluble fraction. No additional AtSWI3B is released upon LIS extraction. We assume that part of the AtSWI3B signal in the nuclear matrix fraction obtained by high-salt procedure (Fig. 3B) was due to precipitation of soluble proteins and conclude that the majority of AtSWI3B in the nuclei is not associated with the matrix but occurs as a soluble and chromatin-bound form. The remaining minor AtSWI3B pool is probably attached to the nuclear matrix.
Figure 3.
AtSWI3B is localized to nuclei and associates predominantly with chromatin and soluble nuclear protein fractions. (A) Identical amounts of protein extracts prepared from whole cells or isolated nuclei of A.thaliana plants were separated by SDS–PAGE and immunoblotted for detection of AtSWI3B. Note the anomalous behavior of both recombinant (lane marked AtSWI3Bhis6) and native AtSWI3B which migrate more slowly than a protein of the predicted molecular mass of 47 595 Da. (B) Arabidopsis thaliana nuclear matrix prepared by the high-salt method as described in Materials and Methods. An equivalent aliquot from each step of the extraction procedure was subjected to SDS–PAGE and immunoblotted for detection with anti-SWI3B antibody. (C) Arabidopsis thaliana nuclear matrix prepared by the LIS method as described in Materials and Methods. An equivalent aliquot from each step of the extraction procedure was subjected to SDS–PAGE and immunoblotted for detection with anti-AtSWI3B antibody.
AtSWI3B partially complements yeast swi3 mutation but is unable to activate transcription in yeast when tethered to DNA
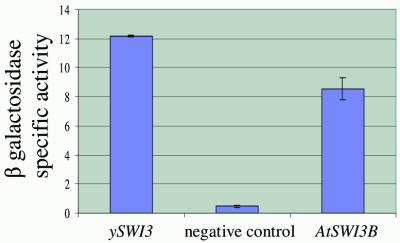
To determine whether AtSWI3B is a functional homolog of the yeast SWI3 protein, the ability of the Arabidopsis gene to complement the swi3 mutation in S.cerevisiae was tested. The characteristic phenotype caused by the swi3 mutation is loss of the HO endonuclease activity required for the mating type switch in yeast cells. In the yeast strain CY 165 carrying the swi3 mutation the HO coding sequence has been replaced with the lac Z gene encoding β-galactosidase. We transformed this strain with multicopy expression vectors bearing either the complete ySWI3 or the complete AtSWI3B. As a control of the swi3 phenotype the CY 165 yeast strain was independently transformed with the empty vector. Western blot analysis of the SDS–PAGE separated proteins confirmed that AtSWI3B is expressed in yeast (results not shown). The phenotypes were analyzed by measuring the β-galactosidase activity. As shown in Figure 4, the enzyme activity was restored in the strain transformed with the complete S.cerevisiae SWI3 gene. The level of β-galactosidase activity in this strain was taken as 100%. Compared with this, transformation with the Arabidopsis AtSWI3B gene resulted in β-galactosidase activity equivalent to 70% of the fully complemented value. These results indicate that AtSWI3B is capable of partial complementation of yeast swi3 mutation.
Figure 4.
AtSWI3B partially complements the SWI3– phenotype of the S.cerevisiae swi3 mutant. Yeast swi3 CY 165 strain was transformed with expression vectors bearing the complete yeast SWI3 (ySWI3), the complete AtSWI3B or with an empty vector (negative control). After growing the transformed cultures in liquid medium to a concentration of 1–2 × 107 cells/ml, the specific activity of β-galactosidase was measured according to Kaiser et al. (9) and expressed as nmoles of o-nitrophenol min–1 mg protein–1. The results are means from two independent measurements.
In order to see whether AtSWI3B can activate a reporter gene in yeast when tethered to DNA, a yeast expression plasmid encoding the GAL4DB-AtSWI3B fusion protein was prepared. Introduction of this plasmid into the yeast Y190 strain (Clontech), containing the GAL-dependent lacZ reporter, did not result in any activation of lacZ transcription as assayed by measurement of β-galactosidase activity (results not shown). Expression of a full GAL4 protein in the reporter strain resulted in considerable β-galactosidase activity. AtSWI3B is thus unable to activate transcription in yeast when tethered to DNA.
Direct yeast two-hybrid analysis indicates that AtSWI3B interacts with Arabidopsis homolog of SNF5 and with other Arabidopsis SWI3-type proteins
To determine whether AtSWI3B can be indeed the component of the putative plant Swi/Snf-type complex we used the yeast two-hybrid screen to seek evidence for its interaction with BSH. The latter is the only Arabidopsis SNF5-type protein and therefore a very likely component of a putative Swi/Snf-type complex. The AtSWI3B interacted with BSH in our two-hybrid analysis (Fig. 5A). AtSWI3B is also capable of forming homodimers as indicated by the activation of β-galactosidase activity in yeast expressing both AtSWI3B fused to GAL4 DNA-binding domain and AtSWI3B fused to GAL4 activation domain (Fig. 5A). We also confirmed the interactions of AtSWI3B with AtSWI3A and with AtSWI3C (Fig. 5A). Thus, AtSWI3B interacts not only with BSH but also with other members of SWI3-type family and can form homodimers (Fig. 5B).
Figure 5.
Interactions of AtSWI3B with BSH and other Arabidopsis SWI3-type proteins in yeast two-hybrid system. (A) The blue color of β-galactosidase filter assays shows two-hybrid interactions of AtSWI3B-AD with BSH-BD, AtSWI3B with AtSWI3B-BD and AtSWI3B-AD with AtSWI3A-BD as well as AtSWI3B-BD with AtSWI3C-AD. (B) Schematic representation of AtSWI3B interactions with SNF5- and SWI3-type Arabidopsis proteins revealed by yeast two-hybrid screen.
AtSWI3B interacts with FCA
To identify other proteins interacting with AtSWI3B, phage display analysis using the recombinant AtSWI3B protein was employed. Phage presenting the heptapeptide PQLRQSR were found to interact with AtSWI3B (results not shown). A search of GenBank with this peptide as the query sequence yielded the FCA_ARATH (Flowering Time Control Protein from Arabidopsis thaliana) as a candidate interacting protein. The FCA contains two RNA recognition motifs (RRM domains) and the protein-binding WW domain (16). The Arabidopsis genome encodes over 190 RRM-containing proteins, the function of most of which is unknown (17). The WW domain is found in a wide range of signaling proteins with both cytoplasmic and nuclear localization (18). The fragment of FCA with high homology to the heptapeptide interacting with AtSWI3B in phage display analysis is located close to the C-terminus and outside the putative RNA- and protein-binding domains (Fig. 6A). To see whether this region could be of any importance in FCA function the databases were searched for proteins homologous to FCA. A simple search of the SP-TrEMBL and SWISSPROT databases from September/October 2001 revealed a great number of sequences with homology to FCA. However, multiple alignment of those with the highest homology showed that none of them had the FCA-type domain arrangement. There were also no conserved elements within the C-terminal parts. To identify proteins with a similar domain arrangement to that of FCA the databases were then searched for sequences homologous to the following fragments of FCA: (i) residues 211–291, corresponding to the RNA-binding RRM2 domain; (ii) residues 292–492, which is a fragment linking the RRM2 and the GLN-rich domain; and (iii) residues 591–624, corresponding to the WW domain (all domains were according to SWISSPROT annotation). The search with the RRM2 domain produced over 1000 homologous sequences and that with a fragment linking the RMM2 and GLN-rich region, 184 homologous sequences. The search with the WW domain gave 82 homologous sequences. We then looked for the proteins which occurred in both the group identified by a search with the RRM2-GLN-rich domain linker fragment and the group identified by a search with the WW domain. Only two such proteins were found. One is FCA and the other, annotated as Q9LLX3, is the abscisic acid-binding protein (ABAP1) from barley (Fig. 6A). The alignment of these two proteins shows remarkable similarity along the entire length of Q9LLX3 including the region containing the peptide interacting with AtSWI3B in the phage display assay (Fig. 6B). Bearing in mind the evolutionary distance between barley and Arabidopsis, it may be concluded that the C-terminal most part of the FCA, located outside the WW domain, is of functional importance.
Figure 6.
Arrangement of sequence motifs in FCA and Q9LLX3, a homologous protein from barley. (A) Schematic representation of the alignment of FCA and Q9LLX3 with recognized domains and the AtSWIB-binding region marked. Open box, continuous amino acid sequence; solid line, gaps; black vertical boxes, homologous regions. (B) Alignment of the peptide interacting with AtSWI3B and homologous motifs of the FCA and Q9LLX3 proteins.
As the fusion of AtSWI3B and the GAL4 DNA-binding domain did not activate transcription in yeast, we were able to test whether AtSWI3B interacts with FCA using the yeast two-hybrid system. The cDNAs corresponding to the full-length FCA and to just the C-terminal half of this protein were subcloned into pGAD424 (encoding the GAL4 activation domain) resulting in plasmids pGADFCA and pGAD227, respectively. These plasmids and the pGBTSWI3B plasmid encoding AtSWI3B fused to the GAL4 DNA-binding domain were used in two-hybrid tests. As shown in Figure 7, both the full-length FCA and the C-terminal half alone are equally capable of activating the expression of β-galactosidase from the GAL-dependent lacZ reporter. Thus, FCA interacts with AtSWI3B, probably via its C-terminal part as was suggested by the phage display result. To verify the possibility that the domain responsible for AtSWI3B binding is located in the C-terminal most region of FCA, we performed yeast two-hybrid test with a plasmid containing the FCA cDNA from which the region encoding the putative AtSWI3B-binding domain had been removed. As shown in Figure 7, the truncated FCA does not activate the transcription of the β-galactosidase gene. We conclude that AtSWI3B interacts with FCA and that this interaction involves the C-terminal region of FCA, outside the conserved RRM2 and WW domains.
Figure 7.
Analysis by yeast two-hybrid assay of the interactions between AtSWI3B and FCA. On the left are schematic representations of the proteins used in the assay. The blue color of the replica lift shown on the right is from β-galactosidase activity and indicates interaction of the tested proteins. See Materials and Methods for details.
DISCUSSION
In yeast, mutants in genes encoding the core subunits of the canonical ySwi/Snf complex are viable, although they show defects in mating-type switching and sporulation, are unable to utilize sucrose as a carbon source and have a slow-growth phenotype (19). Yeast have another Swi/Snf-type complex called RSC which, unlike ySwi/Snf, is essential for mitotic growth (20). Like ySwi/Snf, the RSC contains a core built of homologs of SNF2 ATPase, SNF5 and SWI3 subunits. In Drosophila the genes encoding homologs of Swi/Snf-core proteins, the BRM (SNF2-type ATPase subunit), Snr1 (SNF5-type protein) and MOIRA (SWI3-type protein) belong to the trithorax group and are required to maintain the pattern of homeotic gene expression (21). In mammals there are at least two classes of Swi/Snf-type complexes containing homologs of the SNF2 ATPase, SNF5 and SWI3 proteins (22). Recent studies of the effects of targeted disruption of genes encoding these proteins in mouse have proved their critical role in embryogenesis and in brain development (23,24). Brg1 and hBRM, representing two different classes of human SNF2-type ATPase are involved in the control of cell growth and proliferation through their interactions with the Rb tumor suppressor proteins (25). In summary, the functional characteristics of proteins homologous to the core subunits of Swi/Snf complex in different organisms indicate that they play diverse regulatory roles and are critical for development. So far, no functional homolog of the complete Swi/Snf complex has been characterized in Arabidopsis. However, analysis of the Arabidopsis genome reveals homologs of all three core subunits of the canonical Swi/Snf complex (6; S.Kaczanowski and A.Jerzmanowski, unpublished results). As regards the SNF2-type ATPase and SWI3 subunit, there are a number of putative orthologs (see below). Interestingly, the previously characterized BSH protein encoded by a single copy gene has turned out to be the only Arabidopsis homolog of the SNF5 protein. BSH was shown to be capable of partial complementation of the snf5 mutation in yeast (7). Plants with targeted disruption of the BSH gene have yet to be analyzed although a decrease in BSH content achieved by an antisense strategy was correlated with some growth and developmental aberrations (7).
Based on the alignment and homology analysis, we have identified a small family of Arabidopsis SWI3-type proteins consisting of four members: AtSWI3A, AtSWI3B, AtSWI3C and AtSWI3D. In the phylogenetic tree these proteins form two clusters, one grouping the shorter and the other the longer homologs. Both yeast and human have different but highly related subtypes of SWI3 (see Fig. 1A). The human subtypes BAF170 and BAF155 coexist in two different SWI/SNF-type complexes, Brg1 and hBrm, respectively. However, it is not known whether their function is completely or only partly redundant (26). We decided to analyze first AtSWI3B, the smallest of the Arabidopsis homologs of the canonical SWI3.
We have shown that the gene encoding AtSWI3B is expressed ubiquitously in Arabidopsis. AtSWI3B has a nuclear localization and is found predominantly in chromatin and soluble protein fraction with only a small pool strongly attached to the nuclear matrix.
To determine whether AtSWI3B interacts with BSH within a putative Swi/Snf-type complex we used the anti-AtSWI3B and anti-BSH antibodies to test the large molecular weight protein complexes purified from Arabidopsis protein extracts by multi-step chromatography. However, the results of these experiments were not conclusive (data not shown). In contrast to AtSWI3B, BSH was shown to be strongly attached to nuclear matrix (K.Olczak, personal communication) and thus the disintegration of the BSH-containing complexes during extraction was highly probable. We showed that AtSWI3B interacts with BSH in the two-hybrid system, which is a strong indication that such interactions are also possible in Arabidopsis. Interestingly, AtSWI3B also interacts with other members of the Arabidopsis SWI3-type family (AtSWI3A and AtSWI3C) and is capable of forming homodimers. This may be an indication that the putative Swi/Snf complexes in Arabidopsis contain at least two SWI3-type subunits, which may form homo- or heterodimers. The Drosophila homolog of SWI3 (MOIRA) was also shown to participate in self-oligomerization (27).
The protein–protein interactions within the core of the Swi/Snf complex are not well understood. However, there are data on the interactions of the core subunits with non-core proteins including cyclins (28), HIV-1 integrase (29), SNF11 (30) and proteins with SET domain (31). These proteins, which are probably involved in mediation of different functions of Swi/Snf, may temporarily associate with the complex or may represent its tissue- or organ-specific auxiliary subunits. In this study, using the phage display screen we identified a seven amino acid peptide interacting specifically with AtSWI3B. A search of the databases for proteins with identical or highly homologous peptides revealed an Arabidopsis protein, FCA, which is known to be involved in the control of flowering time (16). The peptide maps to the C-terminal most region of FCA. Using the yeast two-hybrid system we confirmed that FCA indeed interacts with AtSWI3. Furthermore, the interaction is abolished upon removal of a small C-terminal region containing the peptide identified by phage display analysis. The sequence interacting with AtSWI3B is located outside the conserved RNA-binding (RRM) and protein-binding (WW) domains of FCA. A careful search of the databases for proteins homologous to FCA revealed that there is at least one other plant protein (the abscisic acid-binding protein from barley) with a remarkably similar domain arrangement to that of FCA, including conservation of the region containing the peptide interacting with AtSWI3B. It is thus possible that the functional role of the C-terminal sequence of FCA which is suggested by our results is also conserved in other plant homologs of this protein.
The molecular mechanisms underlying FCA function in the control of flowering time are not known. FCA is a nuclear protein (G.Simpson, personal communication). The presence of RNA-binding RRM domains in FCA strongly suggests its involvement in posttranscriptional regulation, perhaps in a complex with other protein partners interacting via the WW domain. The interaction with putative chromatin remodeling factor AtSWI3B mediated by a region outside the main conserved domain indicates a link between the FCA function and chromatin-level regulation. One possibility could be the targeting of RNA (bound via the RRM domains) to specific chromosomal sites. Whether such targeting occurs in vivo and whether it plays a role in the function of FCA in controlling flowering time remain to be elucidated. FCA has been also implicated in the regulation of FLC, a gene critical for promotion of flowering (32). On the other hand, the Arabidopsis VRN2 gene, which encodes a Polycomb-type protein, and is involved in vernalization-dependent flowering control, was shown to influence the chromatin state of the FLC locus (33). Taken together, the data strongly suggest that chromatin-based mechanisms may be used for integration of different regulatory pathways controlling flowering.
Because of the existence in Arabidopsis of four homologs of SWI3 with potentially overlapping function, the testing of the in vivo role of the SWI3-type proteins in plants will require a concomitant interference in the expression of all four Arabidopsis SWI3-type genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank John Gittins, Jan Brzeski and Piotr Zielenkiewicz for comments on the manuscript, Michał Dadlez, Jacek Sikora and Jacek Olędzki for mass spectrometric analyses and Beata Kilianczyk and Monika Banaszek for excellent technical assistance. We are grateful to Drs C. Dean, C. Petersen, K. Olczak and I. Smaczynska for donating plasmids. Plasmids p226 and p227 containing FCA cDNA were developed in Dr C. Dean’s laboratory and were provided under a licence agreement with Plant Bioscience Limited, Norwich, UK. This work was supported by a Howard Hughes Medical Institute grant No. 55000312, Polish Committee for Scientific Research grant Nos 6PO4A 00320 and PBZ-039/PO4/2001, and a Foundation for Polish Science grant No. 2/2000. The laboratory is supported by Centre of Excellence in Molecular Biotechnology.
REFERENCES
- 1.Strahl D.B. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 2.Sudarsanam P. and Winston,F. (2000) The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet., 16, 345–350. [DOI] [PubMed] [Google Scholar]
- 3.Eisen J.A., Sweder,S.K. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phelan M.L., Sif,S., Narlikar,G.J. and Kingston,R.E. (1999) Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell, 3, 247–253. [DOI] [PubMed] [Google Scholar]
- 5.Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- 6.Verbsky M.L. and Richards,E.J. (2001) Chromatin remodeling in plants. Curr. Opin. Plant Biol., 4, 494–500. [DOI] [PubMed] [Google Scholar]
- 7.Brzeski J., Podstolski,W., Olczak,K. and Jerzmanowski,A. (1999) Identification and analysis of the Arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res., 27, 2393–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng F., Cao,Y. and Laurent,B.C. (2001) Essential roles of Snf5p in Snf-Swi chromatin remodeling in vivo. Mol. Cell. Biol., 21, 4311–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaiser C., Michaelis,S. and Mitchell,A. (1994) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY, pp. 169–173.
- 10.Manzara T. and Gruissem,W. (1995) Identification of promoter sequences that interact with DNA-binding proteins. In Maliga,P., Klessig,D.F., Cashmore,A.R., Gruissem,W. and Varner,J.E. (eds), Methods in Plant Molecular Biology: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 233–260.
- 11.Reyes J.C., Muchardt,C. and Yaniv,M. (1997) Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol., 137, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wootton J.C. and Federhen,S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. (1989) PHYLIP—phylogeny inference package (Version 3.2). Cladistics, 5, 164–166. [Google Scholar]
- 14.Muse S.V. and Weir,B.S. (1992) Testing for equality of evolutionary rates. Genetics, 132, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strimmer K. and von Haeseler,A. (1996) Quartet puzzling: quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol., 13, 964–969. [Google Scholar]
- 16.Macknight R., Bancroft,I., Page,T., Lister,C., Schmidt,R., Love,K., Westphal,L., Murphy,G., Sherson,S., Cobbett,C. and Dean,C. (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell, 89, 737–745. [DOI] [PubMed] [Google Scholar]
- 17.Lorkovic Z.J. and Barta,A. (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res., 30, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sudol M., Sliwa,K. and Russo,T. (2001) Functions of WW domains in the nucleus. FEBS Lett., 490, 190–195. [DOI] [PubMed] [Google Scholar]
- 19.Peterson C.L. and Herskowitz,I. (1992) Characterization of the yeast SWI1, SWI2, SWI3 genes, which encode a global activator of transcription. Cell, 68, 573–583. [DOI] [PubMed] [Google Scholar]
- 20.Cairns B.R., Lorch,Y., Li,Y., Zhang,M., Lacomis,L., Erdjument-Bromage,H., Tempst,P., Du,J., Laurent,B. and Kornberg,R.D. (1996) RSC, an essential, abundant chromatin-remodeling complex. Cell, 87, 1249–1260. [DOI] [PubMed] [Google Scholar]
- 21.Tamkun J.W. (1995) The role of brahma and related proteins in transcription and development. Curr. Opin. Genet. Dev., 5, 473–477. [DOI] [PubMed] [Google Scholar]
- 22.Wang W., Xue,Y., Zhou,S., Kuo,A., Cairns,B.R. and Crabtree,G.R. (1996) Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev., 10, 2117–2130. [DOI] [PubMed] [Google Scholar]
- 23.Kim J.K., Huh,S.-O., Choi,H., Lee,K.-S., Shin,D., Lee,C., Nam,J.-S., Kim,H., Chung,H., Lee,H.W., Park,S.D. and Seong,R.H. (2001) Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol., 21, 7787–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guidi C.J., Sands,A.T., Zambrowicz,B.P., Turner,T.K., Demers,D.A., Webster,W., Smith,T.W., Imbalzano,A.N. and Jones,S.N. (2001) Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol., 21, 3598–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strober B.E., Dunaief,J.L., Guha,S. and Goff,S.P. (1996) Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol., 16, 1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sif S., Saurin,A.J., Imbalzano,A.N. and Kingston,R.E. (2001) Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev., 15, 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crosby M.A., Miller,C., Alon,T., Watson,K.L., Verrijzer,C.P., Goldman-Levi,R. and Zak,N.B. (1999) The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol., 19, 1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanahan F., Seghezzi,W., Parry,D., Mahony,D. and Lees,E. (1999) Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol. Cell. Biol., 19, 1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalpana G.V., Marmon,S., Wang,W., Crabtree,G.R. and Goff,S.P. (1994) Binding and stimulation of HIV-1 integrase by human homolog of yeast transcription factor SNF5. Science, 266, 2002–2006. [DOI] [PubMed] [Google Scholar]
- 30.Treich I., Cairns,B.R., Santos,T., Brewster,E. and Carlson,M. (1995) SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol. Cell. Biol., 15, 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozenblatt-Rosen O., Rozovskaia,T., Burakov,D., Sedkov,Y., Tillib,S., Blechman,J., Nakamura,T., Croce,C.M., Mazo,A. and Canaani,E. (1998) The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc. Natl Acad. Sci. USA, 95, 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macknight R., Duroux,M., Laurie,R., Dijkwel,P., Simpson,G. and Dean,C. (2002) Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell, 14, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gendall A.R., Levy,Y.Y., Wilson,A. and Dean,C. (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell, 107, 525–535. [DOI] [PubMed] [Google Scholar]