Abstract
The signal recognition particle (SRP) from Escherichia coli consists of 4.5S RNA and protein Ffh. It is essential for targeting ribosomes that are translating integral membrane proteins to the translocation pore in the plasma membrane. Independently of Ffh, 4.5S RNA also interacts with elongation factor G (EF-G) and the 30S ribosomal subunit. Here we use a cross-linking approach to probe the conformation of 4.5S RNA in SRP and in the complex with the 30S ribosomal subunit and to map the binding site. The UV-activatable cross-linker p-azidophenacyl bromide (AzP) was attached to positions 1, 21, and 54 of wild-type or modified 4.5S RNA. In SRP, cross-links to Ffh were formed from AzP in all three positions in 4.5S RNA, indicating a strongly bent conformation in which the 5′ end (position 1) and the tetraloop region (including position 54) of the molecule are close to one another and to Ffh. In ribosomal complexes of 4.5S RNA, AzP in both positions 1 and 54 formed cross-links to the 30S ribosomal subunit, independently of the presence of Ffh. The major cross-linking target on the ribosome was protein S7; minor cross-links were formed to S2, S18, and S21. There were no cross-links from 4.5S RNA to the 50S subunit, where the primary binding site of SRP is located close to the peptide exit. The functional role of 4.5S RNA binding to the 30S subunit is unclear, as the RNA had no effect on translation or tRNA translocation on the ribosome.
Keywords: 4.5S RNA, ribosomal proteins, signal recognition particle, cross-linking
INTRODUCTION
4.5S RNA is the RNA component of the signal recognition particle (SRP) in Escherichia coli. In the complex with the protein component of the SRP, Ffh, it regulates the co-translational targeting of secretory and membrane proteins to the plasma membrane in bacteria (for reviews, see Keenan et al. 2001; Nagai et al. 2003; Doudna and Batey 2004). 4.5S RNA is a 114-nucleotide (nt) molecule containing a number of internal loops (loops A–E) connected by helical regions and a conserved GGAA tetraloop (Larsen and Zwieb 1991; Larsen et al. 1998; Figure 1). Secondary structure predictions based on thermodynamic stability and evolutionary conservation indicated a highly base-paired rodlike structure (Bourgaize et al. 1984; Poritz et al. 1988; Larsen and Zwieb 1991). However, fluorescence, melting, and chemical modification studies (Lentzen et al. 1994, 1996) suggested the possibility of an alternative, bent conformation of the full-length RNA. Ffh protects a large part of 4.5S RNA, including residues in loops A–D, from chemical modification and enzymatic cleavage (Lentzen et al. 1996). Loops A and B bind the M-domain of Ffh (Batey et al. 2000; Rosendal et al. 2003). The protections in loop C (and probably also in loop D) result from the interaction with the NG-domain of Ffh (Buskiewicz et al. 2005). In SRP bound at the peptide exit of the ribosome, 4.5S RNA appears to be present in the extended form, as indicated by the analysis of cross-links from 4.5S RNA (Rinke-Appel et al. 2002) and Ffh (Gu et al. 2003) to the ribosome.
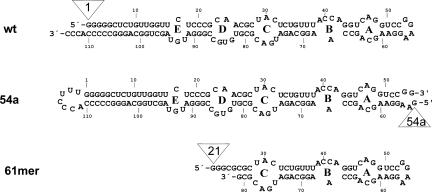
FIGURE 1.
Secondary structures of 4.5S RNA (wt), the G54a derivative (54a) with an open tetraloop, and a fragment comprising nucleotides 21–81 of 4.5S RNA (61mer). The positions where the AzP cross-linker was attached are indicated (open triangles).
4.5S RNA is essential for viability of E. coli, as the depletion of 4.5S RNA impairs protein synthesis and secretion (Jensen and Pedersen 1994; Jensen et al. 1994). The role of 4.5S RNA in SRP function is not fully understood. Binding of 4.5S RNA to Ffh stabilizes the M domain (Zheng and Gierasch 1997) and modulates the complex formation between Ffh and the SRP receptor FtsY (Peluso et al. 2000, 2001; Jagath et al. 2001). The tetraloop region of 4.5S RNA is important for SRP binding to FtsY (Jagath et al. 2001).
Several lines of evidence have suggested that in addition to its function in SRP, 4.5S RNA may be involved in other cellular functions as well. For E. coli, 4.5S RNA was reported to be present in about a fourfold excess over Ffh (Phillips and Silhavy 1992; Jensen and Pedersen 1994; Jensen et al. 1994). Mutational analysis suggested a dual role for 4.5S RNA, one involving co-translational protein secretion by a 4.5S RNA–Ffh complex, the other involving free 4.5S RNA (Brunelli et al. 2002). Point mutations in translational elongation factor G (EF-G) suppress the effect of 4.5S RNA depletion (Brown 1987). 4.5S RNA binds EF-G with moderate affinity (~2 μM) (Shibata et al. 1996; Suzuma et al. 1999; Jovine et al. 2000; Nakamura et al. 2001; Sagar et al. 2004). Furthermore, 4.5S RNA binds to the 30S ribosomal subunit (Rinke-Appel et al. 2002). The binding is independent of the presence of Ffh and the binding site must be in close proximity of the 3′ end of 16S rRNA.
In the present work, we studied the conformation of 4.5S RNA and its binding site on the ribosome by cross-linking. The UV-activatable cross-linker p-azidophenacyl bromide (AzP) was covalently attached at the 5′ end of three different 4.5S RNA constructs: at G1 in full-length wild-type 4.5S RNA, at residue G54a (“a” denoting an additional guanine added to the native nt 54) in 4.5S RNA with an opened tetraloop, and at the 5′ end (position 21 of 4.5S RNA) of a 61-nt-long 4.5S RNA fragment comprising residues 21–81. UV-induced cross-links to Ffh, vacant ribosomes, ribosome initiation complexes, or ribosome nascent chain complexes carrying an N-terminal fragment of leader peptidase (Lep-RNC) were studied. The cross-linked ribosomal components were analyzed by sucrose gradient centrifugations and immunoprecipitation (Stade et al. 1989). The effect of 4.5S RNA on the efficiency of Lep-mRNA translation and tRNA translocation was tested.
RESULTS
Attachment of the azidophenacyl group to different positions in 4.5S RNA
The experimental strategy was to introduce the UV-activatable azidophenacyl group (AzP) as a cross-linker at the 5′ end of 4.5S RNA or variants of 4.5S RNA in which the 5′ phosphate was substituted with a 35S-labeled phosphorothioate group (Burgin and Pace 1990). In full-length 4.5S RNA, two positions were chosen to attach the cross-linker: at the native 5′ end of the RNA and in the tetraloop region, i.e., the two opposite ends of the RNA in the extended two-dimensional structure (Fig. 1). To introduce the cross-linker in the tetraloop region, the tetraloop was opened between residues 54 and 55 and the original 5′ and 3′ ends of 4.5S RNA (positions 1 and 114) were connected by a UUU linker. To improve the efficiency of in vitro transcription, one more G residue (G54a) was added at the new 5′ end. AzP-modified wild-type and mutant 4.5S RNA bound to Ffh with the same affinity as the unmodified RNA (data not shown). The tetraloop region was previously shown to be nonessential for Ffh binding (Jagath et al. 2001). For cross-linking from an internal position in 4.5S RNA, truncated 4.5S RNA comprising nt 21–81, referred to as 61mer, was modified with AzP at the 5′ end (position 21) in the same way. Unmodified and AzP-modified 61mer bound to Ffh with the same affinity as full-length unmodified 4.5S RNA.
AzP-modified 4.5S RNAs were cross-linked to Ffh and to ribosomes in different functional states, and the cross-linking products were analyzed in the following way. For SDS-PAGE analysis of cross-linked proteins, i.e., Ffh or ribosomal proteins, the cross-linking products were digested with RNase T1, leaving the 35S label attached to the cross-link site on the protein, which allowed for the identification of the cross-linking products by SDS-PAGE electrophoresis and immunoprecipitation. Prior to the analysis of cross-linked ribosomal proteins, the distribution of cross-links between the two ribosomal subunits and between rRNA and ribosomal proteins, respectively, was determined by successive centrifugations through sucrose density gradients (Stade et al. 1989).
4.5S RNA cross-link to Ffh
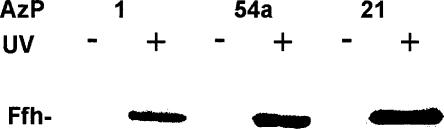
To study the cross-links from positions 1, 21, and 54a in 4.5S RNA to Ffh, samples after UV irradiation were denatured by heating, RNA was digested by RNase T1, and proteins were subjected to SDS-PAGE. Cross-links of 4.5S RNA to Ffh were observed from all three positions (Fig. 2). The efficiencies varied about fourfold, position 21 being the strongest and position 1 the weakest. No cross-linked products were observed when other proteins (FtsY, EF-Tu, or albumin) were used instead of Ffh (not shown).
FIGURE 2.
Cross-linking of AzP-modified 4.5S RNA to Ffh. SDS-PAGE lanes are labeled by the positions of AzP in 4.5S RNA (Fig. 1). After UV irradiation, RNase T1 digestion, and SDS-PAGE (see Materials and Methods), 35S-labeled Ffh was identified by autoradiography.
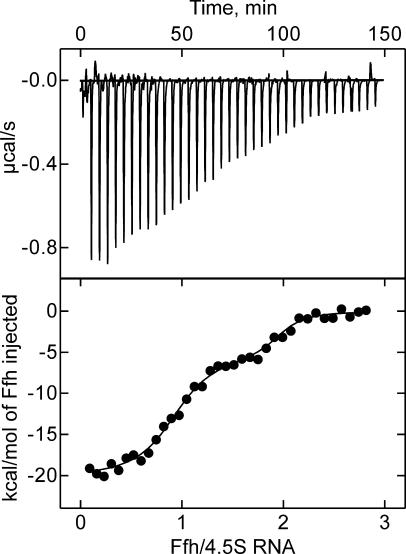
These results suggest that the 4.5S RNA molecule can assume a conformation in which not only the tetraloop region and the internal position 21 (helix e), but also the 5′ end of the RNA are within cross-linking distance to Ffh (~10 Å; Gu et al. 2003). This interpretation requires that the cross-links from both ends of 4.5S RNA are to one molecule of Ffh and do not involve two molecules of Ffh. In order to verify that a 1:1 complex of 4.5S RNA and Ffh was formed under the conditions of the cross-linking experiments, microcalorimetric titrations were performed in which Ffh was titrated to a fixed amount of 4.5S RNA (Fig. 3). The titration curve revealed a main transition at a stoichiometric ratio of 0.9, indicating the formation of a 1:1 complex. The apparent Kd of the complex was ~5 nM, close to the Kd obtained previously (Gu et al. 2003) under the buffer conditions of the present cross-linking experiments, which is higher than the Kd of 50 pM obtained under optimized buffer conditions (Batey and Doudna 2002; Buskiewicz et al. 2005). Following the saturation of the 1:1 complex, a higher-order complex was formed, probably by unspecific binding of a second molecule of Ffh to the 4.5S RNA–Ffh complex (Fig. 3). The affinity of this unspecific binding, ~0.2 μM, was ~50 times lower than the affinity of specific binding, such that the probability of unspecific binding was very low (<1% occupancy) at the conditions of the cross-linking experiments, where Ffh and 4.5S RNA were present in equimolar ratio. Another possible, though unlikely, artifact would be that cross-links were formed between two or more associated SRP complexes rather than within one SRP complex. This is most likely not the case, as the cross-links from all three AzP positions of 4.5S RNA were not affected when the concentration was lowered 10-fold below the standard concentration used for the experiments (1–0.1 μM) (data not shown), and the formation of dimers of SRP was not observed previously (Jagath et al. 2000). Thus, we conclude that the cross-links from positions 1, 21, and 54 of 4.5S RNA are formed within one Ffh–4.5S RNA complex, indicating that all three positions of 4.5S RNA must be within cross-linking distance to Ffh in the complex where Ffh is bound to the strong specific binding site in domain IV of 4.5S RNA.
FIGURE 3.
Stoichiometry of Ffh-4.5S RNA complex formation determined by microcalorimetry. Increasing amounts of Ffh were added to 4.5S RNA (10 μM) at the indicated molar ratios in a microcalorimeter, and the energy required to compensate for the binding enthalpy in order to reestablish thermal equilibrium after each addition was measured and is plotted in microcalories per second (upper panel). From the plot of the binding enthalpies (lower panel), the transitions due to saturation were obtained, which yielded stoichiometries of 0.90 ± 0.05 and 1.05 ± 0.05 for high-affinity (5 nM) and low-affinity (0.2 μM) binding of Ffh to 4.5S RNA, respectively. For details, see Materials and Methods.
4.5S RNA cross-link to the 30S subunit
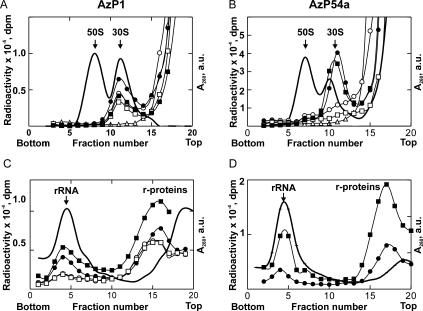
For the analysis of the distribution of cross-links between the ribosomal subunits, ribosome complexes with 35S-labeled 4.5S RNA(AzP1) or (AzP54a) were analyzed. Both derivatives bound to the ribosomes and formed UV-induced cross-links, independently of the presence of Ffh. Upon sucrose gradient centrifugation of UV-irradiated complexes, almost 10% of the input radioactivity of 4.5S RNA(AzP1) was found in the 30S subunit fractions, while no radioactivity cosedimented with the 50S subunits (Fig. 4A). At the concentrations of 4.5S RNA and Ffh used (1 μM each) and a Kd of 50 pM (Batey et al. 2001), all RNA is expected to form SRP, suggesting that both RNA alone (in the absence of Ffh) and SRP (in the presence of Ffh) were able to bind to the 30S subunit and form the cross-link. Some binding of 4.5S RNA(AzP1) or SRP was also found without UV irradiation, indicating that the respective 30S complex was stable enough to persist through sucrose gradient centrifugation without cross-linking. Analogous experiments with 4.5S RNA(AzP54a) also showed an exclusive cross-link to the 30S subunit (Fig. 4B); the apparent shift of the radioactivity peak against the absorbance peak is caused by the more extensive overlap of 30S and 50S subunits in the gradient shown in Figure 4B compared to Figure 4A. Similar results with the cross-linker at both positions of the RNA were obtained when 70S initiation complexes or Lep-RNC were used (data not shown), indicating that neither tRNA in the P site nor translation interfered with the interaction of 4.5S RNA with the 30S subunit. The 61mer, 4.5S RNA(AzP21), did not form cross-links to the ribosome.
FIGURE 4.
Identification of the cross-linking targets of 4.5S RNA(AzP1) (A,C) and 4.5S RNA (AzP54a) (B,D) on the ribosome. Ribosomal subunits (A,B) or rRNA/r-proteins (C,D) were separated by sucrose gradient centrifugation (see Materials and Methods). The positions of 30S and 50S subunits or rRNAs and r-proteins were determined by measuring optical density at A260 (solid line); cross-linked material was identified by measuring 35S radioactivity. 70S ribosomes were incubated with 35S-labeled 4.5S RNA alone (circles) or SRP (squares) and analyzed after UV irradiation (closed symbols) or without UV irradiation (open symbols).
Cross-links to the 50S subunit, due to SRP bound to the peptide exit site of the 50S subunit, as observed with AzP-modified Ffh (Gu et al. 2003), were not detected with AzP at either position of 4.5S RNA, indicating these positions of 4.5S RNA are not within cross-linking distance to the ribosome in the SRP binding site on the 50S subunit.
The material from the 30S subunit peak was subjected to a second round of sucrose density centrifugation, this time in the presence of SDS, to separate rRNA from ribosomal proteins (Fig. 4C,D). About 2%–4% of total input radioactivity was found distributed among the fractions containing 16S rRNA and 30S ribosomal proteins, when the cross-linked material from vacant ribosomes, initiation complexes, or Lep-RNC was analyzed (Table 1). The distribution of cross-links among RNA and proteins was almost the same for AzP1 and AzP54a. This result is consistent with previous studies (Rinke-Appel et al. 2002) in that both RNA and protein from the 30S subunit were cross-linked to 4.5S RNA and the cross-links were not affected by the presence of Ffh. Because the amount of cross-linked 16S rRNA was small and because cross-links from 4.5S RNA to 16S rRNA had been mapped previously (Rinke-Appel et al. 2002), the cross-links to 16S rRNA were not characterized further.
TABLE 1.
Cross-links from 4.5S RNA to ribosomes in different functional states
| Ribosomes | ||||
| Method | 4.5S RNAa | Vacant | Initiation complex | Lep-RNC |
| Sucrose gradient | AzP1 | 30S | 30S | 30S |
| AzP54a | 30S | 30S | 30S | |
| SDS-PAGE analysis | AzP1 | + | + | + |
| AzP54a | + | + | + | |
| Immunoprecipitation | AzP1 | S7, S18, S21 | S7, S18, S21 | n.d.b |
| AzP54a | S2, S7, S18, S21 | S2, S7, S18, S21 | n.d. |
aCross-links in the absence and presence of Ffh were identical.
b(n.d.) Not determined; cross-linking products were identical to those obtained with the vacant ribosomes (Fig. 5A,B) or initiation complexes as indicated by SDS-PAGE analysis.
Analysis of cross-linked 30S ribosomal proteins
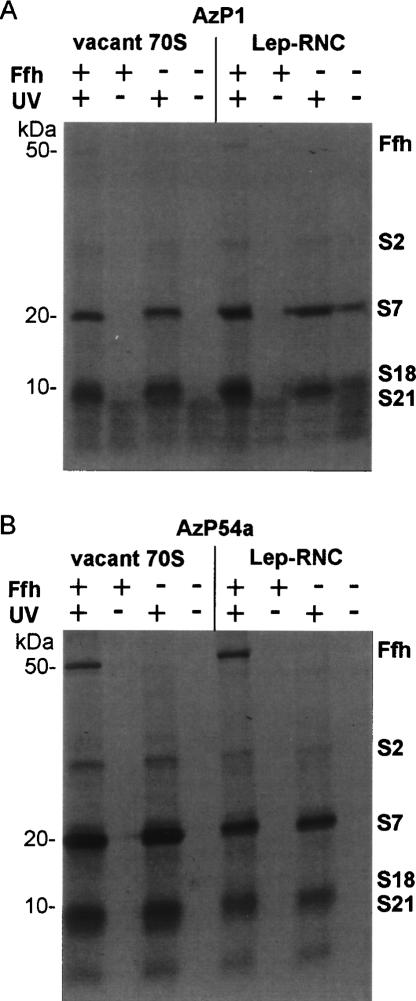
Several highly reproducible cross-links to ribosomal proteins of ~20 and 8–10 kDa were observed with 4.5S RNA(AzP1) (Fig. 5A). AzP54a gave rise to cross-links to proteins of similar size and to an additional product of ~27 kDa (Fig. 5B) The formation of the cross-links was not influenced by Ffh, and identical cross-links were observed for ribosomes in either functional state (Fig. 5; Table 1).
FIGURE 5.
Cross-linking of 4.5S RNA(AzP1) (A) and 4.5S RNA(AzP54a) (B) to vacant ribosomes (left panel) or Lep-RNC (right panel). Proteins were separated by SDS-PAGE, and 35S-labeled proteins were visualized by autoradiography. The positions of marker proteins of the indicated molecular weights (left) and ribosomal proteins (right) are indicated.
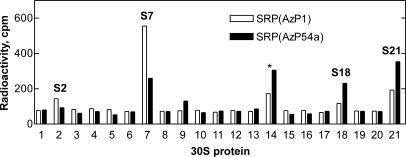
Cross-linked proteins were identified by a pull-down assay, using antibodies against individual ribosomal proteins and a secondary antibody attached to agarose beads (Gulle et al. 1988). With both AzP1 and AzP54a the predominant cross-link to vacant ribosomes was to protein S7 (19.9 kDa), whereas proteins S2 (26.6 kDa), S18 (8.9 kDa), and S21 (8.4 kDa) were cross-linked to a smaller extent (Fig. 6). The molecular weights of identified proteins were in good agreement with the size of proteins identified by the SDS-PAGE analysis (Fig. 5). Similar results were obtained with 70S initiation complexes (Table 1). The cross-links to Lep-RNC were not analyzed independently, because the pattern of the cross-linked proteins was identical to that obtained with vacant ribosomes (Fig. 5).
FIGURE 6.
Identification by immunoprecipitation of ribosomal proteins cross-linked to 4.5S RNA(AzP). Histograms show the radioactivity precipitated with agarose beads coated with antibodies against ribosomal protein of the 30S subunit. The apparent reactivity of anti-S14 (*) is due to cross-contamination with anti-S21 (Stade et al. 1989) and was not considered further. (Open bars) AzP1; (closed bars) AzP54a.
Functional role of 4.5S RNA binding to the 30S ribosomal subunit
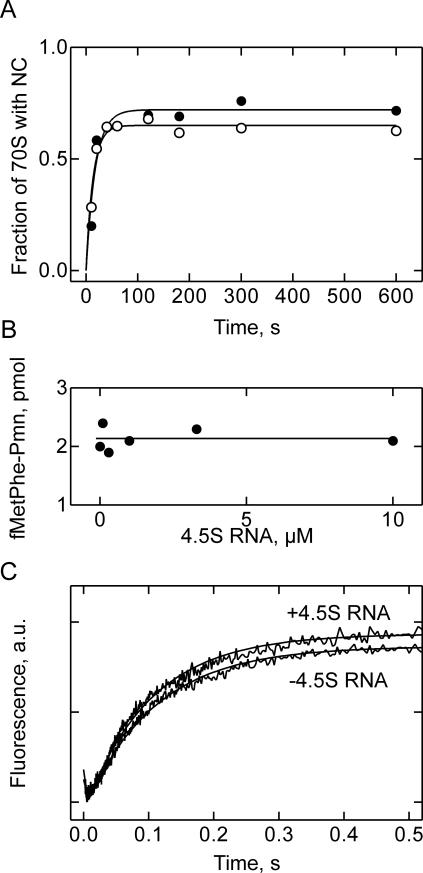
4.5S RNA may have a role in translation that is not related to its function in membrane targeting of translating ribosomes. Although bacterial 4.5S RNA lacks the Alu domain of its eukaryotic homolog, 7S RNA, that is responsible for the translational arrest (Siegel and Walter 1986; Bui and Strub 1999), a role of 4.5S RNA in translation regulation would be an attractive possibility. We tested the effect of 4.5S RNA on the translation of Lep-mRNA in a highly active translation system reconstituted from purified components (Fig. 7A). To 70S initiation complexes EF-Tu·GTP and 4.5S RNA were added, and translation was initiated by the addition of a mixture of aminoacyl-tRNAs containing [14C]Leu-tRNA and EF-G. The time course of translation was followed by determining [14C]Leu incorporation. Translation was efficient and rapid, with an average rate of amino acid incorporation of ~5 sec−1 (Fig. 7A). About 70% of ribosomes were active in translation and synthesized the Lep fragment of the expected size (75 amino acids), as indicated by the ratio of f[3H]Met in peptides to total ribosome concentration and the size of the nascent peptide as analyzed by SDS-PAGE and 14C autoradiography (not shown). Addition of 30 μM 4.5S RNA had no effect on the rate and efficiency of translation. This result argues against the possibility that 4.5S RNA alone has an influence on translation in E. coli.
FIGURE 7.
Testing the effect of 4.5S RNA on ribosome functions. (A) Translation of Lep-mRNA in the absence (open circles) or presence (closed circles) of 4.5S RNA (30 μM). (B) EF-G-dependent multiple-turnover translocation. Pretranslocation complexes (160 nM) containing f[3H]Met[14C]Phe-tRNAPhe in the A site were mixed with EF-G (0.5 nM) in the presence of GTP (1 mM) and different amounts of 4.5S RNA (0–10 μM). The extent of translocation was measured after 3 min incubation at 37°C by reaction with Pmn (1 mM) for additional 10 sec. (C ) Time course of single-round translocation. Pretranslocation complexes with fMetPhe-tRNAPhe (Prf16/17) in the A site (0.2 μM) were preincubated with 4.5S RNA (10 μM) and mixed with EF-G (0.6 μM) and GTP (1 mM) in a stopped-flow apparatus. The fluorescence of proflavin (excitated at 470 nm) was measured after passing a 500 KV cut-off filter.
Another potential effect of 4.5S RNA may be on EF-G-dependent translocation, as 4.5S RNA was shown to bind EF-G, albeit with low affinity (Shibata et al. 1996; Suzuma et al. 1999; Jovine et al. 2000; Nakamura et al. 2001; Sagar et al. 2004). Complex formation with 4.5S RNA may reduce the effective concentration of free EF-G, thereby affecting the rate of EF-G-dependent tRNA translocation on the ribosome. Furthermore, the binding site of 4.5S RNA on the 30S subunit is close to the tRNA exit (E) site (cf. Discussion), the site of transient binding for deacylated tRNA after leaving the P site during translocation, and may, therefore, interfere with tRNA movement during translocation. This prompted us to study the effect of 4.5S RNA on EF-G-dependent translocation. First, translocation was studied under conditions of multiple turnover of EF-G, using pretranslocation complexes that contained deacylated tRNAfMet in the P site and fMetPhe-tRNAPhe in the A site. Translocation, i.e., the movement of fMetPhe-tRNAPhe into the P site, was measured by the reaction with puromycin (Pmn) under initial velocity conditions where the amount of product (fMetPhe-Pmn) formed increases linearly with time. At the low concentrations of EF-G used, the factor had to turn over more than 300 times in order to accomplish translocation on all ribosomes. Under these conditions, one could expect to detect any inhibition of translocation by blocking the E site by 4.5S RNA or by reducing the concentration of free EF-G due to the formation of the 4.5S RNA·EF-G complex. However, the addition of increasing amounts of 4.5S RNA had no discernible effect on translocation (Fig. 7B).
In order to study the translocation step directly, the effect of 4.5S RNA was examined by stopped flow in a single round of translocation initiated by rapid mixing of the pretranslocation complex with excess EF-G. The fluorescence change of a reporter group (proflavin) at position 16/17 in A-site-bound fMetPhe-tRNAPhe was monitored to follow translocation (Rodnina et al. 1997). No effect of 4.5S RNA of translocation was found (Fig. 7B), as both efficiency (amplitude of the fluorescence increase) and rate (10±1 sec−1) of translocation were the same with and without 4.5S RNA. The initial small fluorescence decrease was due to EF-G binding (Rodnina et al. 1997). Thus, 4.5S RNA does not seem to have an effect on tRNA translocation under either single-round or multiple-turnover conditions.
DISCUSSION
Conformation of 4.5S RNA in the complex with Ffh
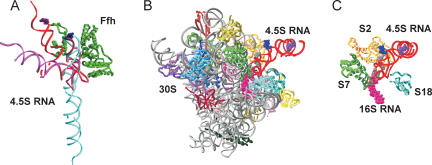
The structure of full-length 4.5S RNA in SRP is not known. Two structures of the 4.5S RNA apical fragment (nt 31–74) are available, in the complex with either full-length SRP54 from Sulfolobus solfataricus (Rosendal et al. 2003) or the M domain of E. coli Ffh (Batey et al. 2000). In either structure, the tetraloop region of 4.5S RNA is close to the M domain, which explains the efficient cross-link from AzP54a to Ffh (Fig 8A). However, another strong cross-link was observed from position 21. Assuming an extended conformation of 4.5S RNA in solution, helix E, which encompasses position 21, is 70 Å or more away from the M domain and not closer than 40–50 Å to the NG domain in the orientation suggested by Rosendal et al. (2003) or in any of the three arrangements derived from the crystal structure of free Ffh (Keenan et al. 1998). Furthermore, a significant and specific cross-link was also observed from the 5′ end of 4.5S RNA. Modeling 4.5S RNA in its extended conformation into any of the Ffh crystal structures available (Keenan et al. 1998; Rosendal et al. 2003) placed the 5′ terminus of the 4.5S RNA at a distance of at least 100 Å from the nearest residue of Ffh, independently of the arrangement of the NG and M domains of Ffh. This suggests that 4.5S RNA in SRP is present in a bent conformation where the 5′ end comes into close proximity to Ffh. Based on chemical modification results (Lentzen et al. 1996), a conformation of 4.5 S RNA bent at the internal symmetric loop C has been modeled (Gorodkin et al. 2001). In that model, position 21 of 4.5S RNA is in ~30 Å distance to the nearest residue of Ffh, indicating that additional adjustments in shape and orientation of 4.5S RNA and/or the conformation of Ffh are necessary to account for the observed cross-link. Furthermore, the model does not explain the cross-link from the 5′ end of 4.5S RNA (Fig. 8A). This strongly indicates the presence of another bend in 4.5S RNA, most likely within the asymmetric loop E, which would introduce a turn in the molecule that brings the 5′ end close to Ffh (Fig. 8A). Thus, our results suggest that 4.5S RNA in the SRP particle is folded into a globular, rather than extended, structure that is bent at both internal loops C and E. The unbound 4.5S RNA may assume the same conformation or alternate between extended and bent structures. The conformation of 4.5S RNA or SRP on the ribosome is discussed below.
FIGURE 8.
Models for 4.5S RNA interaction with Ffh and the 30S subunit. (A) Complexes of Ffh (green) with 4.5S RNA in extended (cyan), singly bent (magenta) (Gorodkin et al. 2001), or doubly bent conformation (red). The latter was derived from the singly bent conformation by introducing an additional kink in loop E. Ffh domains are depicted in an arrangement derived from the A/A arrangement of the crystal structure (Keenan et al. 1998) by moving the NG domain (right) into a position where clashes with 4.5S RNA were removed and a number of distance constraints from fluorescence resonance energy transfer and cross-linking were satisfied (Buskiewicz et al. 2005; I. Buskiewicz, unpubl. data). The position of the M domain (left) relative to the RNA is taken from the crystal structure (Batey et al. 2000). (B) Complex of the 30S subunit (1IBM; Ogle et al. 2001) with 4.5S RNA in doubly bent conformation (red) viewed from the intersubunit interface side. Cross-linkers at position 1 (purple spacefill) and 54a (blue spacefill) of 4.5S RNA are indicated. Ribosomal landmarks are color-coded as in panel C. (C ) Orientation of 4.5S RNA relative to proteins S7 (green), S2 (yellow), S18 (light blue) and the potential positions of the thiouridine cross-links from 4.5S RNA (positions 29, 32, 34, 36–38; gray spacefill) to 16S rRNA (positions 1525–1540; magenta spacefill) (Rinke-Appel et al. 2002).
The binding site of 4.5S RNA on the 30S ribosomal subunit
4.5S RNA is bound in the proximity of ribosomal protein S7 on the 30S subunit, as indicated by strong cross-links from the three positions of 4.5S RNA analyzed here, and close to S18, S21, and S2, to which weaker but significant cross-links were found. S7 is located at the head of the 30S subunit in the vicinity of the interface to the 50S subunit (Fig. 8B). S18 and S2 are found close to S7 on the solvent side of the platform. The position of S21 is not known, as it is not present in the crystal structures of Thermus thermophilus 30S subunits. The cross-links were identical on vacant, P-site-occupied and translating ribosomes, suggesting that 4.5S RNA binds to the 30S subunit in a way that does not interfere with the binding of the 50S subunit and tRNA. This places the 4.5S RNA binding site at the outer surface of the ribosome, rather than at the interface between the subunits. Previous results showed that the 3′ end of 16S rRNA (helix 45) is within zero-length cross-linking distance of 4.5S RNA (Rinke-Appel et al. 2002). Presumably, the binding site of 4.5S RNA is located between the head and the platform of the cytoplasmic surface of the 30S subunit, on the side where the ribosomal E site is located (Yusupov et al. 2001).
Conformation of 4.5S RNA on the ribosome
The analysis of cross-links from thiouracil-labeled 4.5S RNA indicated that the cross-links to helix 45 of 16S rRNA occurred from the region between U29 and U50 (Rinke-Appel et al. 2002). U45 and U50 are located in the region of 4.5S RNA that is directly involved in interactions with the M domain of Ffh (Batey et al. 2000). As the formation of the cross-links was not affected by Ffh, it is likely that one or more of the remaining thiouracil residues at U29, U32, U34, or U36–38 and nt 1525– 1540 in 16S rRNA were responsible for those cross-links (Rinke-Appel et al. 2002). Our data indicate that the 5′ end and the tetraloop region of 4.5S RNA are close to each other and both located in close proximity to proteins S7 and S18. This can be rationalized in terms of a model where 4.5S RNA binds to the 30S subunit in the double-bent conformation described above for the SRP (Fig. 8B,C,D). A kink in internal loop C (Gorodkin et al. 2001) would place the tetraloop region close to proteins S7 and S2, while the U29–U38 region is in close contact with the 3′ end of 16S rRNA. However, a second kink in 4.5S RNA would be required to bring its 5′ end into cross-linking distance of S7 and S18. By analogy to the SRP model discussed above, it appears likely that the asymmetric loop E is kinked. Thus, in SRP, SRP–30S and 4.5S RNA–30S complexes 4.5S RNA, or at least part of it, assumes a strongly bent conformation.
It should be noted that in SRP bound to the 50S binding site 4.5S RNA appears to be present in a different conformation. Cross-linking of SRP to the 50S subunit at the peptide exit channel showed that the N domain of Ffh was in close contact to ribosomal proteins L23 and L29 (Gu et al. 2003), while position 84 of 4.5S RNA was located close to nt 2828–2837 of 23S rRNA (Rinke-Appel et al. 2002; Gu et al. 2003). To span the distance between L23 and those positions of 23S rRNA, 4.5S RNA had to be modeled in extended conformation (Gu et al. 2003), rather than bent at the internal loop C (Larsen et al. 1998). The doubly bent form of 4.5S RNA suggested above for free SRP and 4.5S RNA or SRP bound to 30S subunits is inconsistent with these cross-link constraints. Recently, a cryo-EM reconstruction of eukaryotic SRP located at the peptide exit of translating ribosomes was obtained at 12 Å resolution (Halic et al. 2004). In that structure, the density attributed to protein SRP54, the eukaryotic homolog of Ffh, indicates an open conformation with the N domain facing L23, in agreement with cross-linking data from both eukaryotic (Pool et al. 2002) and prokaryotic (Gu et al. 2003; Ullers et al. 2003) SRP-ribosome complexes. The region of 7S RNA that is homologous to 4.5S RNA appeared to be present in an extended conformation and to make several contacts to 23S rRNA, one of which matches a cross-link determined for bacterial SRP (Rinke-Appel et al. 2002). Given the strong sequence conservation between bacterial SRP and its eukaryotic counterpart, these results strongly indicate that in SRP bound to the peptide exit of the ribosome, 4.5S RNA is present in an extended conformation. Thus, it appears that 4.5S RNA in SRP alternates between two conformations: In free SRP the RNA appears to be present in a globular, doubly bent conformation, which may also apply for the unbound RNA, and in SRP bound to the 50S subunit 4.5S RNA assumes an extended conformation.
No effect of 4.5S RNA on translation or tRNA translocation on E. coli ribosomes
The functional significance of 4.5S RNA binding to the 30S subunit (Rinke-Appel et al. 2002; this article) or to EF-G (Shibata et al. 1996; Suzuma et al. 1999; Jovine et al. 2000; Nakamura et al. 2001; Sagar et al. 2004) is not known. Positions of mutations that affected the in vivo requirement for 4.5S RNA, i.e., C1066U in 16S rRNA and G424A in 23S rRNA (Brunelli et al. 2002), are not involved in the binding of tRNA or translation factors (Yusupov et al. 2001) and were not reported to influence ribosome functions. Binding of 4.5S RNA in the proximity of the E site of the ribosome could affect initiation or elongation of protein synthesis. However, the present results indicate that 4.5S RNA does not compete with tRNA binding to the ribosome during initiation or translation, as the efficiency of cross-linking between 4.5S RNA and the ribosomal proteins was independent on the functional state of the ribosome. Furthermore, biochemical and kinetic measurements using a purified and highly active in vitro translation system showed that a large excess of 4.5S RNA had no effect on translation and the kinetics of single or multiple-turnover EF-G-dependent translocation (this article), and did not compete with tRNA binding to the E site (T. Bornemann, unpubl. data). The affinity of 4.5S RNA to vacant 30S subunits is in the micromolar range (I. Buskiewicz, unpubl. data). It is therefore possible that the binding of 4.5S RNA to the 30S subunit in vivo serves for storage of excessive 4.5S RNA or SRP. The low affinity of 30S binding may ensure that there is sufficient 4.5S RNA available to form SRP for membrane targeting of RNCs exposing signal peptides, whereas binding to the ribosome would allow for compartmentalization of the SRP to the ribosome. EF-G may be yet another cellular component that binds excessive free 4.5 RNA, as indicated by the affinity of 4.5S RNA to wild type EF-G and EF-G mutants (Sagar et al. 2004) that suppress the effects of 4.5S RNA depletion in vivo (Brown 1987). Another possibility is that the interaction of 4.5S RNA (or SRP) with the 30S subunit may be important under certain stress conditions, rather than at optimum translation conditions in vivo or in vitro.
MATERIALS AND METHODS
Buffer and reagents
Buffer A: 20 mM HEPES (pH 7.5), 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2. p-Azidophenacyl (AzP) bromide was from Sigma. GTP, GDPNP, pyruvate kinase, and phosphoenolpyruvate were from Roche Diagnostics. Ni-NTA agarose was from Qiagen. All other chemicals were obtained from Sigma or Merck.
Biochemical assays
Ffh was expressed and prepared as described previously (Gu et al. 2003). 70S ribosomes from E. coli MRE 600 and purified components of the translation system were prepared as described (Rodnina and Wintermeyer 1995; Matassova et al. 1999; Rodnina et al. 1999). Ribosome-nascent chain complexes (RNC) with truncated Lep-mRNA coding for the first 75 amino acids of leader peptidase (de Gier et al. 1996) were prepared and purified as described (Gu et al. 2003). The fraction of ribosomes that carried nascent chains was >70%, as determined from the number of f[3H]Met-labeled peptide chains per ribosome, and the nascent chains had the expected length, as verified by SDS-PAGE and phosphorimaging (data not shown). Pretranslocation complexes were prepared and EF-G-dependent translocation was measured as described (Rodnina et al. 1997; Savelsbergh et al. 2000).
E. coli strains and plasmids
BL21 (DE3) pLysS strain was used for expressing FtsY and Ffh from pET9-FtsY(12F128F343W)and pET24-Ffh, respectively (Jagath et al. 2000). The plasmid pT7–4.5S was used to prepare 4.5S RNA by transcription in vitro using T7RNA polymerase (Lentzen et al. 1994).
The plasmid pT7–54a coded for a 4.5S RNA(54a) variant where the 5′ and 3′ ends of RNA were introduced into the tetraloop of 4.5S RNA, while the original 3′ and 5′ ends were connected by a small loop. Plasmid pT7–54a was constructed from pT7–4.5S in the following way. The region between nt 55 and 114 and 1 and 10 of pT7–4.5S RNA was amplified by PCR using the primer pair CTATGATCATGATTACGCCAAGCTCTAATACGACTCACTA TAGAAGGAAGCAG (primer 1, BclI restriction site and T7 promotor are shown in bold; the fragment of 4.5S RNA sequence corresponding to nt 55–64 are in italics) and CAGAGCCCCCAAAGGGTGGGGGCCCTGC (primer 2; AAA, shown in bold, is the linker between the 3′ and 5′ ends of 4.5S RNA; the sequence before and after the AAA linker are complementary to nt 10–1 and 114–100 of 4.5S RNA, respectively). In the second PCR, the region between nt 110–114 and 1–64 of pT7–4.5S RNA was amplified by the primer pair GC AGGGCCCCCACCCTTTGGGGGCTCTG (primer 3, the sequence before and after AAA corresponds to positions 110–114 and 1–10 of 4.5S RNA) and CTGCTTCCTTCCGGACCTGACC (primer 4, AccIII restriction site is indicated in bold; the sequence is complementary to nt 64–43 of 4.5S RNA). The products of the two PCR reactions were used for the site overlap extension (SOE) PCR with primers 1 and 4. The SOE-PCR product was inserted into pT7/T3α-19 (Gibco-BRL) after removal of the T7 promotor of the plasmid by PvuII. Mutations were confirmed by DNA sequencing.
In vitro transcription
To prepare wild-type 4.5S RNA or 4.5S RNA(54a), the respective plasmid was linearized by Cfr42I (MBI Fermentas) or AccIII (Stratagene). The in vitro transcription reactions contained 40 mM Tris-HCl (pH 7.5), 1 mM spermidine, 10 mM DTT, 0.05% Tween-20, 8 mM MgCl2, 20 mM GMP, 2 mM of each NTP, 50 μg/mL linearized DNA template, 1600 units/mL of T7 RNA polymerase in a final volume of 1 mL. Reactions were stopped after 4 h incubation at 37°C by addition of EDTA to 10 mM. RNA was purified on Nucleobond AX cartridges (Machery and Nagel, Düren) according to the manufacturer’s protocol.
Coupling of AzP to 5′-phosphorothioate-modified RNA
To remove the 5′-phosphate, 500 pmol 4.5S RNA were incubated with 20 units of alkaline phosphatase (Roche) in 50 μL of 50 mM Tris-HCl (pH 8.5), 0.1 mM EDTA for 2 h at 50°C. Dephosphorylated RNA was labeled with 15 μL [35S]ATPγS (10 mCi/ml; ICN Biomedicals) in 50 μL of buffer 50 mM Tris-HCl (pH 8.2), 10 mM MgCl2, 0.1 mM EDTA, 5 mM DTT, 0.1 mM spermidine for 3 h at 37°C in the presence of 3 μL polynucleotide kinase (PNK; Roche). To complete phosphorothioate incorporation, nonradioactive ATPγS (4 mM) was added and the incubation continued for 3 h at 37°C. For the isolation of 35S-labeled RNA, 100 μL of the labeling mixture were applied to a NICK column (Pharmacia Biotech) equilibrated with 2 × 500 μL H2O; after washing the column, the RNA was eluted with 400 μL H2O. After phenol extraction and ethanol precipitation, the specific activity of 35S-labeled RNA typically was ~2.5 × 105 dpm/pmol.
For AzP coupling, 35S-labeled RNA was reacted with 2 mM AzP bromide in 15 mM Na2CO3 (pH 9.0) and 10% methanol for 2 h at 25°C. Unreacted cross-linker was removed by two rounds of phenol extraction followed by ethanol precipitation. The pellet was resuspended in 25 μL H2O, and the sample was frozen and stored at −80°C. Labeled RNAs were homogeneous on PAGE in 7 M urea. All steps of the AzP coupling procedure were carried out under subdued light.
UV-induced cross-linking of 4.5S RNA(AzP) to the ribosome and Ffh
Cross-linking experiments were performed as described (Gu et al. 2003) in buffer A containing additionally 0.02% Brij 35, 1 mM DTT, 0.1 mg/mL poly(U), and 10% glycerol. 4.5S RNA(AzP) or SRP(AzP) were added to vacant ribosomes or initiation complexes (1 μM each) or to RNC (0.1 μM each) and GDPNP (200 μM) to form the complex under subdued light. SRP was preformed by incubating 4.5S RNA(AzP) and Ffh for 5 min at 25°C. Mixtures were irradiated in a microtiter plate on ice for 10 min using four Philips Cleo 15-W lamps at a distance of 10 cm. To minimize photodestruction, a 305-nm cut-off filter was placed between the light source and sample.
Analysis of cross-links
Irradiated samples were denatured by boiling for 2 min, digested by RNase T1, and subjected to SDS-PAGE analysis. Proteins carrying the 35S label were visualized by phosphorimaging.
For sucrose gradient analysis of ribosome samples, irradiated samples were subjected to two rounds of sucrose gradient centrifugation without prior T1 RNase digestion. In the first run, 30S and 50S subunits were separated, and in the second, performed in the presence of SDS, rRNA and proteins were separated (Stade et al. 1989). Cross-linked RNA–ribosome complexes were precipitated with 1.1 volumes of ice-cold ethanol for 1 h at −20°C and centrifuged at 6000 rpm for 30 min. The pellet were dissociated in 200 μL buffer (50 mM Tris-HCl at pH 7.5, 500 mM ammonium chloride, 1 mM magnesium chloride) and incubated at 37°C for 5 min to facilitate resuspension. The samples were then loaded onto 11-ml sucrose gradient of 5%–20% sucrose and centrifuged for 3 h 45 min at 41,000 rpm at 4°C in a SW41 rotor (Beckman). After centrifugation, the gradient was pumped out from the bottom to the top of the tube and fractionated in 600-μL aliquots. The positions of the 50S and 30S subunits were determined spectrophotometrically at A260. Aliquots of 60 μL were taken and counted to identify the fractions containing 35S-labeled RNA. The material was pooled and precipitated with 1 volume of ice-cold ethanol and stored at −20°C for 60 min.
To separate rRNA from ribosomal proteins, a second sucrose density centrifugation was performed in the presence of SDS. Samples were dissolved in SDS loading buffer (10 mM Tris-HCl at pH 7.5, 1 mM EDTA, 5 mM DTT, 1% SDS) and loaded onto an 11-mL sucrose gradient of 5%–20% sucrose in buffer containing 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 100 mM lithium chloride, 0.1% SDS, and centrifuged for 16 h at 36,000 rpm in an SW41 rotor (Beckman) at 4°C. After centrifugation, the gradient was fractionated as described above. One-hundred-microliter aliquots were used for radioactivity counting. Protein-containing fractions were pooled and precipitated with 2 volumes of ethanol and 0.1 M sodium acetate at pH 5.3 for 2 h at −20°C and centrifuged 30 min at 6000 rpm. Pellets were washed with 80% ethanol and resuspended in dissociation buffer for further analysis.
Immunoprecipitation
Cross-linked ribosomal proteins were identified by immunoprecipitation using antibodies against 30S ribosomal proteins. The primary antibodies were conjugated to agarose beads coated with rabbit anti-goat IgG (Sigma). Five hundred microliters of 50% agarose suspension were washed with buffer AKB (0.9% NaCl, 0.05% Tween 20, 0.05% β-ME, 20 mM sodium phosphate at pH 7.7). The agarose was distributed into 21 portions of 20 μl each, corresponding to 21 antibodies against 30S ribosomal proteins. Five microliters of primary antibody solution and 180 μl of buffer AKB were added to agarose portions and incubated for 30 min under gentle mixing. Unbound primary antibodies were removed by thorough washing of agarose with AKB buffer.
Two hundred to 500 μl of cross-linked ribosomal proteins (diluted to concentrations of SDS <0.01%) were added to agarose suspensions conjugated to each individual antibody against ribosomal proteins and incubated for 2 h with gentle mixing. Unbound proteins were removed by thorough washing. Ribosomal proteins that were retained on the agarose were eluted by 1% SDS, and radioactivity was counted in LumaSafe scintillation cocktail.
Calorimetry
The interaction between 4.5S RNA and Ffh protein was quantitated by isothermal titration calorimetry in a microcalorimeter (MicroCal Inc.). Solutions of Ffh and 4.5S RNA in buffer A were dialyzed against the same reservoir of buffer A with 10% glycerol overnight. The solution of 4.5S RNA (10 μM, 1.38 mL) was thermally equilibrated against the reference cell containing buffer at 20°C. Then, 8-μl portions of a 120-μM solution of Ffh were added, and the energy required to reestablish thermal equilibrium between the two cells after each addition was measured and plotted in microcalories per second. Integration yielded the enthalpy of complex formation in kilocalories per mole. The stoichiometry of complex formation was calculated using the manufacturer’s software.
The concentration of Ffh was determined by absorbance measurements at 205 nm, using a coefficient of E(1 mg/ml; 1 cm) = 31 (Scopes 1974). The concentration of 4.5S RNA was determined from absorbance measurements at 260 nm in buffer A, using an extinction coefficient (260 nm) of 0.86 μM−1cm−1. The latter was estimated from the extinction coefficient of tRNAPhe, 0.57 μM−1cm−1, taking into account the different number of nucleotides of 4.5S RNA (114) and tRNAPhe (76).
Acknowledgments
We are indebted to Richard Brimacombe for providing a collection of antibodies against ribosomal proteins and for the protocol of the pull-down assay. We thank Christian Herrmann for help with calorimetry, Iwona Buskiewicz and Thomas Bornemann for discussion and unpublished data, Frank Peske for critical reading and comments on the manuscript, and Petra Striebeck, Astrid Böhm, Carmen Schillings, and Simone Möbitz for expert technical assistance. The work was supported by the Deutsche Forschungs-gemeinschaft (WI626/17-1), the Alfried Krupp von Bohlen und Halbach-Stiftung, and the Fonds der Chemischen Industrie.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7219805.
REFERENCES
- Batey, R.T. and Doudna, J.A. 2002. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry 41: 11703–11710. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Rambo, R.P., Lucast, L., Rha, B., and Doudna, J.A. 2000. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287: 1232–1239. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Sagar, M.B., and Doudna, J.A. 2001. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 307: 229–246. [DOI] [PubMed] [Google Scholar]
- Bourgaize, D.B., Farrell, C., Langley, K.H., and Fournier, M.J. 1984. Physical properties of the E. coli 4.5S RNA: First results suggest a hairpin helix of unusual thermal stability. Nucleic Acids Res. 12: 2019–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S. 1987. Mutations in the gene for EF-G reduce the requirement for 4.5S RNA in the growth of E. coli. Cell 49: 825–833. [DOI] [PubMed] [Google Scholar]
- Brunelli, C.A., O’Connor, M., and Dahlberg, A.E. 2002. Decreased requirement for 4.5S RNA in 16S and 23S rRNA mutants of Escherichia coli. FEBS Lett. 514: 44–48. [DOI] [PubMed] [Google Scholar]
- Bui, N. and Strub, K. 1999. New insights into signal recognition and elongation arrest activities of the signal recognition particle. Biol. Chem. 380: 135–145. [DOI] [PubMed] [Google Scholar]
- Burgin, A.B. and Pace, N.R. 1990. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 9: 4111–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz, I., Kubarenko, A., Peske, F., Rodnina, M.V., and Wintermeyer, W. 2005. Domain rearrangement of SRP protein Ffh upon binding 4.5S RNA and the SRP receptor FtsY. RNA 6: 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gier, J.W., Mansournia, P., Valent, Q.A., Phillips, G.J., Luirink, J., and von Heijne, G. 1996. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399: 307–309. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A. and Batey, R.T. 2004. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 73: 539–557. [DOI] [PubMed] [Google Scholar]
- Gorodkin, J., Knudsen, B., Zwieb, C., and Samuelsson, T. 2001. SRPDB (Signal Recognition Particle Database). Nucleic Acids Res. 29: 169–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, S.Q., Peske, F., Wieden, H.J., Rodnina, M.V., and Wintermeyer, W. 2003. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA 9: 566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulle, H., Hoppe, E., Osswald, M., Greuer, B., Brimacombe, R., and Stoffler, G. 1988. RNA–protein cross-linking in Escherichia coli 50S ribosomal subunits; determination of sites on 23S RNA that are cross-linked to proteins L2, L4, L24 and L27 by treatment with 2-iminothiolane. Nucleic Acids Res. 16: 815–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic, M., Becker, T., Pool, M.R., Spahn, C.M., Grassucci, R.A., Frank, J., and Beckmann, R. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814. [DOI] [PubMed] [Google Scholar]
- Jagath, J.R., Rodnina, M.V., and Wintermeyer, W. 2000. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. J. Mol. Biol. 295: 745–753. [DOI] [PubMed] [Google Scholar]
- Jagath, J.R., Matassova, N.B., de Leeuw, E., Warnecke, J.M., Lentzen, G., Rodnina, M.V., Luirink, J., and Wintermeyer, W. 2001. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA 7: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, C.G. and Pedersen, S. 1994. Concentrations of 4.5S RNA and Ffh protein in Escherichia coli: The stability of Ffh protein is dependent on the concentration of 4.5S RNA. J. Bacteriol. 176: 7148–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, C.G., Brown, S., and Pedersen, S. 1994. Effect of 4.5S RNA depletion on Escherichia coli protein synthesis and secretion. J. Bacteriol. 176: 2502–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovine, L., Hainzl, T., Oubridge, C., Scott, W.G., Li, J., Sixma, T.K., Wonacott, A., Skarzynski, T., and Nagai, K. 2000. Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA. Struct. Fold. Des. 8: 527–540. [DOI] [PubMed] [Google Scholar]
- Keenan, R.J., Freymann, D.M., Walter, P., and Stroud, R.M. 1998. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94: 181–191. [DOI] [PubMed] [Google Scholar]
- Keenan, R.J., Freymann, D.M., Stroud, R.M., and Walter, P. 2001. The signal recognition particle. Annu. Rev. Biochem. 70: 755–775. [DOI] [PubMed] [Google Scholar]
- Larsen, N. and Zwieb, C. 1991. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 19: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, N., Samuelsson, T., and Zwieb, C. 1998. The Signal Recognition Particle Database (SRPDB). Nucleic Acids Res. 26: 177–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentzen, G., Dobberstein, B., and Wintermeyer, W. 1994. Formation of SRP-like particle induces a conformational change in E. coli 4.5S RNA. FEBS Lett. 348: 233–238. [DOI] [PubMed] [Google Scholar]
- Lentzen, G., Moine, H., Ehresmann, C., Ehresmann, B., and Wintermeyer, W. 1996. Structure of 4.5S RNA in the signal recognition particle of Escherichia coli as studied by enzymatic and chemical probing. RNA 2: 244–253. [PMC free article] [PubMed] [Google Scholar]
- Matassova, N.B., Rodnina, M.V., Endermann, R., Kroll, H.P., Pleiss, U., Wild, H., and Wintermeyer, W. 1999. Ribosomal RNA is the target for oxazolidinones, a novel class of translational inhibitors. RNA 5: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, K., Oubridge, C., Kuglstatter, A., Menichelli, E., Isel, C., and Jovine, L. 2003. Structure, function and evolution of the signal recognition particle. EMBO J. 22: 3479–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K., Miyamoto, H., Suzuma, S., Sakamoto, T., Kawai, G., and Yamane, K. 2001. Minimal functional structure of Escherichia coli 4.5 S RNA required for binding to elongation factor G. J. Biol. Chem. 276: 22844–22849. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons, Jr., W.M., Tarry, M.J., Carter, A.P., and Ramakrishnan, V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902. [DOI] [PubMed] [Google Scholar]
- Peluso, P., Herschlag, D., Nock, S., Freymann, D.M., Johnson, A.E., and Walter, P. 2000. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science 288: 1640–1643. [DOI] [PubMed] [Google Scholar]
- Peluso, P., Shan, S.O., Nock, S., Herschlag, D., and Walter, P. 2001. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry 40: 15224–15233. [DOI] [PubMed] [Google Scholar]
- Phillips, G.J. and Silhavy, T.J. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359: 744–746. [DOI] [PubMed] [Google Scholar]
- Pool, M.R., Stumm, J., Fulga, T.A., Sinning, I., and Dobberstein, B. 2002. Distinct modes of signal recognition particle interaction with the ribosome. Science 297: 1345–1348. [DOI] [PubMed] [Google Scholar]
- Poritz, M.A., Strub, K., and Walter, P. 1988. Human SRP RNA and E. coli 4.5S RNA contain a highly homologous structural domain. Cell 55: 4–6. [DOI] [PubMed] [Google Scholar]
- Rinke-Appel, J., Osswald, M., von Knoblauch, K., Mueller, F., Brimacombe, R., Sergiev, P., Avdeeva, O., Bogdanov, A., and Dontsova, O. 2002. Cross-linking of 4.5S RNA to the Escherichia coli ribosome in the presence or absence of the protein Ffh. RNA 8: 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina, M.V. and Wintermeyer, W. 1995. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl. Acad. Sci. 92: 1945–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina, M.V., Savelsbergh, A., Katunin, V.I., and Wintermeyer, W. 1997. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385: 37–41. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Savelsbergh, A., Matassova, N.B., Katunin, V.I., Semenkov, Y.P., and Wintermeyer, W. 1999. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl. Acad. Sci. 96: 9586–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendal, K.R., Wild, K., Montoya, G., and Sinning, I. 2003. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc. Natl. Acad. Sci. 100: 14701–14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar, M.B., Lucast, L., and Doudna, J.A. 2004. Conserved but nonessential interaction of SRP RNA with translation factor EF-G. RNA 10: 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh, A., Matassova, N.B., Rodnina, M.V., and Wintermeyer, W. 2000. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J. Mol. Biol. 300: 951–961. [DOI] [PubMed] [Google Scholar]
- Scopes, R.K. 1974. Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 59: 277–282. [DOI] [PubMed] [Google Scholar]
- Shibata, T., Fujii, Y., Nakamura, Y., Nakamura, K., and Yamane, K. 1996. Identification of protein synthesis elongation factor G as a 4.5 S RNA-binding protein in Escherichia coli. J. Biol. Chem. 271: 13162–13168. [DOI] [PubMed] [Google Scholar]
- Siegel, V. and Walter, P. 1986. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature 320: 81–84. [DOI] [PubMed] [Google Scholar]
- Stade, K., Rinke-Appel, J., and Brimacombe, R. 1989. Site-directed cross-linking of mRNA analogues to the Escherichia coli ribosome; identification of 30S ribosomal components that can be cross-linked to the mRNA at various points 5′ with respect to the decoding site. Nucleic Acids Res. 17: 9889–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuma, S., Hayashi, K., Nakamura, K., and Yamane, K. 1999. Analysis of Escherichia coli 4.5S RNA binding affinity to Ffh and EF-G. FEMS Microbiol. Lett. 180: 271–277. [DOI] [PubMed] [Google Scholar]
- Ullers, R.S., Houben, E.N., Raine, A., ten Hagen-Jongman, C.M., Ehrenberg, M., Brunner, J., Oudega, B., Harms, N. and Luirink, J. 2003. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell. Biol. 161: 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]
- Zheng, N. and Gierasch, L.M. 1997. Domain interactions in E. coli SRP: Stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Mol. Cell 1: 79–87. [DOI] [PubMed] [Google Scholar]