Abstract
The helix 69 (H69) region of the large subunit (28S) rRNA of Homo sapiens contains five pseudouridine (Ψ) residues out of 19 total nucleotides (26%), three of which are universally or highly conserved. In this study, the effects of this abundant modified nucleotide on the structure and stability of H69 were compared with those of uridine. The role of a loop nucleotide substitution from A in bacteria (position 1918 in Escherichia coli 23S rRNA) to G in eukaryotes (position in 3734 in H. sapiens) was also examined. The thermodynamic parameters were obtained through UV melting studies, and differences in the modified and unmodified RNA structures were examined by 1H NMR and circular dichroism spectroscopy. In addition, a [1,3-15N]Ψ phosphoramidite was used to generate H69 analogs with site-specific 15N labels. By using this approach, different Ψ residues can be clearly distinguished from one another in 1H NMR experiments. The effects of pseudouridine on H. sapiens H69 are consistent with previous studies on tRNA, rRNA, and snRNA models in which the nucleotide offers stabilization of duplex regions through ΨN1H-mediated hydrogen bonds. The overall secondary structure and base-pairing patterns of human H69 are similar to the bacterial RNA, consistent with the idea that ribosome structure and function are highly conserved. Nonetheless, pseudouridine-containing RNAs have subtle differences in their structures and stabilities compared to the corresponding uridine-containing analogs, suggesting possible roles for Ψ such as maintaining translation fidelity.
Keywords: helix 69, 28S rRNA, pseudouridine, modified nucleotides
INTRODUCTION
One of the most important biological functions of RNA is protein synthesis, which is carried out by the ribosome machinery throughout phylogeny. The three-dimensional (3D) structures of the complete 70S ribosome and of its individual subunits have been solved at high resolution by X-ray crystallography (Cate et al. 1999; Ban et al. 2000; Carter et al. 2000; Nissen et al. 2000; Wimberly et al. 2000; Schlünzen et al. 2001; Yusupov et al. 2001). The 70S and 80S ribosome complexes have also been observed by 3D cryo-electron microscopy (EM) (Frank and Agrawal 2000; Gabashvili et al. 2000; Spahn et al. 2001; Gao et al. 2003). The details of the 70S structure demonstrate that the RNA components of the complex must play critical roles in catalyzing peptide bond formation because there are no proteins in the vicinity of the A- and P-site tRNAs (Yusupov et al. 2001). The 70S crystal structure also revealed that helix 69 (H69) of 23S rRNA is a component of the major intersubunit bridge between the small and large subunits. This bridge, referred to as B2a, connects the peptidyl-transferase center (PTC) to the decoding region. The association of the ribosomal subunits is essential for protein synthesis; therefore, the bridges must have important roles in linking structure to function.
Helix 69 is observed in the 5.5 Å resolution structure of the 70S complex (Yusupov et al. 2001) but disordered in the 2.4 Å resolution structure of the H. marismortui 50S sub-unit (Ban et al. 2000), suggesting that it has a dynamic role in structural rearrangements between the free and bound forms of the large subunit (Harms et al. 2001). It has been suggested that movement of H69 may contribute to tRNA placement and translocation (Bashan et al. 2003). Earlier cross-linking studies demonstrated that H69 of the Escherichia coli 23S rRNA contacts the decoding region at positions 1408–1411 and 1518–1520 of 16S rRNA (Mitchell et al. 1992). Related chemical probing studies showed that helix 69 of 23S RNA also interacts with the 790 loop of 16S rRNA, which is known to participate in subunit association, initiation factor binding, and tRNA binding (Joseph et al. 1997; Merryman et al. 1999). More recently, DMS modification studies have shown that methylations at A1912 and A1918 interfere with 70S ribosome function in E. coli (Maiväli and Remme 2004).
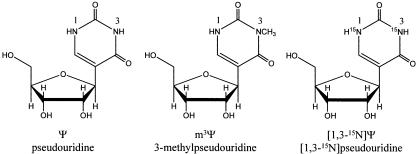
There are many known natural nucleoside modifications in RNA (Limbach et al. 1994; McCloskey and Crain 1998). The first to be discovered and one of the most common modified bases is pseudouridine (Cohn 1960; Ofengand et al. 1995; Charette and Gray 2000). Pseudouridine has its glycosidic bond between the base and sugar isomerized from a standard C1′-N1 found in uridine to a C1′-C5 bonding scheme (Fig. 1). Due to the isomerization reaction, pseudouridine has a second imino nitrogen available for hydrogen bonding. Interestingly, higher organisms have more pseudouridine residues in their large subunit rRNAs. Eukaryotic large subunit rRNAs contain 0.9%–1.4% pseudouridines compared to 0.03%–0.4% in bacteria, and the pseudouridine residues tend to be clustered in domains II, IV, and V of the large subunit rRNAs (Maden and Wakeman 1988; Charette and Gray 2000). In H. sapiens, 7.3% of uridines in the large subunit rRNA are converted to pseudouridines compared to 1.5% in E. coli (Ofengand and Bakin 1997).
FIGURE 1.
The structures of pseudouridine (Ψ), 3-methylpseudouridine (m3Ψ), and [1,3-15N]pseudouridine ([1,3-15N] Ψ).
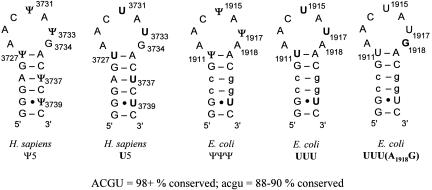
In H69 of human rRNA, which is located in domain IV of 28S rRNA, there are five pseudouridines out of 19 nucleotides and no uridine residues. Thus, the percentage of pseudouridines in H69 (26.3%) is surprisingly high relative to the percentage in the remaining eukaryotic large subunit rRNA. Conserved pseudouridines (Ψ) at positions 3727, 3731, and 3733 of H. sapiens 28S rRNA correspond to Ψ1911, Ψ1915, and Ψ1917 in E. coli 23S rRNA, of which Ψ1915 is also methylated at the N3 position (Figs. 1, 2) (Kowalak et al. 1996; Ofengand and Bakin 1997). There are two additional pseudouridines in the stem region of the H. sapiens RNA at positions 3737 and 3739.
FIGURE 2.
The helix 69 (H69) RNA hairpins from H. sapiens and E. coli. The Ψ5 and U5 RNAs are derived from positions 3722–3740 of the H. sapiens large subunit rRNA. The natural sequence (Ψ5) contains pseudouridine (Ψ) at positions 3727, 3731, 3733, 3737, and 3739, whereas the unmodified variant (U5) contains uridine (U). The corresponding ΨΨΨ and UUU RNAs are derived from positions 1906–1924 of the E. coli 23S rRNA. The UUU(A1918G) RNA contains a stem sequence corresponding to E. coli H69 and a loop of H. sapiens H69. The levels of H69 sequence conservation for bacteria and eukaryotes are indicated (ACGU indicates 98+% conservation, and acgu indicates 88%–90% conservation). A total of 431 and 115 species were compared for bacteria and eukaryotes, respectively (Cannone et al. 2002).
Pseudouridine synthase RluD modifies uridines at positions 1911, 1915, and 1917 in E. coli 23S rRNA (Wrzesinski et al. 2000). In contrast, uridines at 3731 and 3733 in the human rRNA are guided to modification by the box H/ACA snoRNA known as U19 (Bortolin and Kiss 1998). The absence of RluD is reported to alter the growth rates of E. coli, and to influence stop codon readthrough and frameshifting of tRNA (O’Connor and Dahlberg 1995; Raychaudhuri et al. 1998). Similarly, snR191, the snoRNA for the corresponding U19 in S. cerevisiae, provides a growth rate advantage to the cell (Badis et al. 2003).
The sequence of H69 of the large subunit rRNA is highly conserved among organisms (Cannone et al. 2002). Most of the stem consists of > 88% conserved Watson-Click base pairs, with the exception of one G·U mismatch (Fig. 2; Table 1). The residue on the 3′ side of the seven-nucleotide loop is different between E. coli and H. sapiens. Most (> 98%) bacteria, including E. coli, have an A residue at position 1918 (E. coli numbering). In contrast, > 98% of eukaryotes, including H. sapiens, have a G residue at the corresponding 3734 position. Highly conserved loop nucleotides (98+%) at positions 1913, 1914, and 1918 (E. coli numbering) are known to contact the minor groove of the small subunit rRNA decoding site (at residues 1408– 1410 and 1494–1495) (Yusupov et al. 2001). The first and seventh positions of the H69 loop are possible determinants for stable loop-loop interactions (Gregorian and Crothers 1995). Thus, the base difference at position 1918/3734 (A versus G) could influence ribosome stability and function between eukaryotes and prokaryotes.
TABLE 1.
Helix 69 sequence conservation
| E. coli | % Conservation in bacteria | H. sapiens | % Conservation in eukaryotes |
| G1906 | 98+ | G3722 | 98+ |
| G1907 | 98+ | G3723 | 98+ |
| C1908 | 88–90 | G3724 | 98+ |
| C1909 | 88–90 | A3725 | 98+ |
| G1910 | 98+ | G3726 | 98+ |
| U/Ψ1911 | 98+ | U/Ψ3727 | 98+ |
| A1912 | 98+ | A3728 | 98+ |
| A1913 | 98+ | A3729 | 98+ |
| C1914 | 98+ | C3730 | 98+ |
| U/Ψ1915 | 98+ | U/Ψ3731 | 98+ |
| A1916 | 98+ | A3732 | 98+ |
| U/Ψ1917 | 98+ | U/Ψ3733 | 98+ |
| A1918 | 98+ | G3734 | 98+ |
| A1919 | 98+ | A3735 | 98+ |
| C1920 | 98+ | C3736 | 98+ |
| G1921 | 88–90 | U/Ψ3737 | 98+ |
| G1922 | 88–90 | C3738 | 98+ |
| U1923 | 98+ | U/Ψ3739 | 98+ |
| C1924 | 98+ | C3740 | 98+ |
Note: 431 species of bacteria and 115 species of eukaryotes were examined using the comparative RNA Web (CRW) site (http://www.rna.icmb.utexas.edu) (Cannone et al. 2002).
Helix 69 appears to play a significant role in ribosome function; therefore, it is important to determine the structural consequences of adding modified nucleotides (Ψ) or altering its loop sequence. Since the ribosome structure has not been solved by X-ray crystallography in the unmodified form, the use of smaller model RNAs allows the comparison between modified and unmodified RNAs, and the comparison of RNAs from different organisms. In the present study, five H69 analogs (Fig. 2) based on the E. coli and H. sapiens large subunit rRNAs were examined for stability and structural differences. The U5 and UUU RNAs are the unmodified variants from human and bacteria, respectively, and contain uridines in place of pseudouridines. The Ψ5 and ΨΨΨ RNAs are the corresponding pseudouridine-modified analogs. The UUU(A1918G) RNA represents the bacterial sequence in which the loop residue at position 1918 is mutated from A to G. We employed circular dichroism (CD) spectroscopy, thermal melting experiments, and nuclear magnetic resonance (NMR) spectroscopy to analyze this set of RNAs, and we examined the effects of divalent metal ion (Mg2+) binding to each of the RNAs.
RESULTS
The effects of modification on the structure of H69 RNA analogs
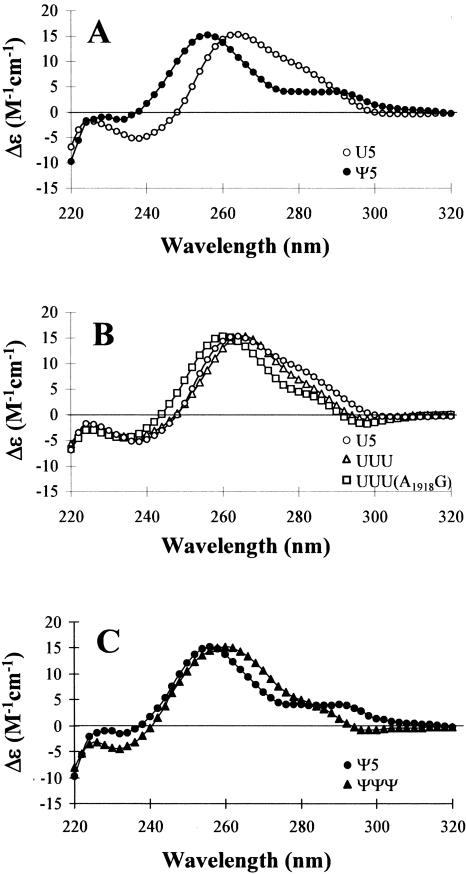
CD spectroscopy allows for indirect monitoring of conformational differences between RNAs or changes in RNA structure upon ligand binding. For example, the contributions of modified bases to RNA folding can be determined, and the conformational changes induced by metal ions can be assessed. The CD spectra of H69 RNAs were obtained in order to determine the effects of pseudouridine modification on the H. sapiens hairpin structures, and also to compare the human analogs with the previously studied E. coli RNAs (Meroueh et al. 2000; Chui et al. 2002). The spectra are shown as wave-length versusmolar ellipticity (Δɛ) in Figure 3. The CD spectra of the human-based analogs, unmodified (U5) and modified (Ψ5) H69, indicate significant differences (Fig. 3A). Both spectra are indicative of A-form RNA structures; however, differences in peak maxima, peak minima, and crossover points are observed, as well as differences in peak shapes. The U5 RNA spectrum has a peak maximum at 264 nm, a peak minimum at 238 nm, and a crossover point at 248 nm. The U5 RNA spectrum also has a broad shoulder around 280 nm. In contrast, the Ψ5 RNA spectrum has a peak maximum at 256 nm, peak minimum at 232 nm, and crossover point at 238 nm. The Ψ5 RNA spectrum also has a peak centered near 285 nm that is not observed in the U5 RNA spectrum. Similar results were observed with E. coli H69 analogs (Meroueh et al. 2000). The observed differences in the spectral features may be explained by the difference in stacking environments for uridines versus pseudouridines, due to their different arrangements of atoms and glycosidic linkages. In addition, the transition dipoles are altered for Ψ compared to U, leading to CD phase changes (Scott and Zamecnik 1969; Schweizer et al. 1971; Davis 1995). A comparison of the spectra in Figure 3B also reveals differences between the E. coli and H. sapiens RNAs, which is not surprising since they have different stem sequences. The unmodified H. sapiens H69 (U5) and E. coli H69 (UUU) have similar peak maxima, peak minima, and crossover points. The only difference is that the shoulder around 280 nm for U5 is slightly shifted to 285 nm for UUU. The spectrum of the mutant UUU(A1918G), which has a H. sapiens loop sequence and an E. coli stem sequence, is also shown in Figure 3B. This spectrum is unique and differs from both U5 and UUU spectra. The peak maximum, peak minimum, and crossover points are shifted to lower wavelengths (260 nm, 234 nm, and 244 nm, respectively) and the shoulder in the 280–285 nm region is more pronounced for UUU(A1918G). The CD spectra (Fig. 3C) of the modified RNA analogs (Ψ5 and ΨΨΨ) are also different. The extra peak in the Ψ5 spectrum at 285 nm is not due to the three pseudouridines found in both H. sapiens and E. coli (Ψ3727, Ψ3731, and Ψ3733), but rather must come from contributions of one or both of the additional pseudouridines in the stem region (Ψ3737 and Ψ3739) of the H. sapiens RNA. The positive band at 285 nm also appears in the CD spectra of a duplex RNA representing the stem region of Ψ5 and a single-stranded RNA representing residues G3722 through A3728 (5′- GGGAGΨA-3′) (data not shown); however, this band is not apparent in the CD spectrum of a single-stranded RNA representing residues A3735 though C3740 (5′- ACΨCΨC-3′). In addition, the peak at 285 nm is diminished when the Ψ5 or duplex RNA spectra are obtained at 85° C. These results suggest that the CD spectral differences between Ψ5 and U5 are due to sequence (Ψ versus U) and first-neighbor differences rather than any conformational differences caused by the modified nucleotides.
FIGURE 3.
CD spectra of unmodified U5 (○) and modified Ψ5 (•) H. sapiens H69 RNAs (A), U5 RNA overlaid with unmodified UUU E. coli RNA (▵) and UUU(A1918G) RNA (□) (B), and Ψ5 RNA and modified ΨΨΨ E. coli RNA (▴) (C) are shown. Each spectrum is an average of four scans, and the RNA concentrations were 2.6 μM.
Circular dichroism spectroscopy is typically more useful for examining changes in RNA structures upon ligand binding or changes in solution conditions. This method was used to examine the over-all structures of the U5 and Ψ5 RNAs in the presence of magnesium. Magnesium chloride was added to the RNA samples at total concentrations of 1, 3, and 5 mM. The CD spectra of the two human H69 RNAs did not show any significant differences, such as overall shape, crossover points, and peak intensities, upon addition of divalent metal ions (data not shown). For both RNAs, only a slight shift in the peak maxima and crossover points to longer wavelengths (1–2 nm) was observed with a slight increase in peak intensities at the maxima (~0.5 ellipticity units) upon addition of 5 mM magnesium. Thus, these data indicate that MgCl2 does not significantly affect the structures of U5 and Ψ5.
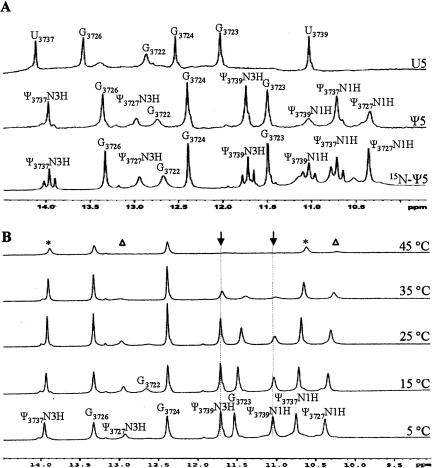
NMR spectroscopy was employed to determine the hydrogen-bonding patterns for the human H69 RNAs, U5 and Ψ5. The imino proton regions of the 1D 1H NMR spectra of U5 and Ψ5 are shown in Figure 4. The assignments of the peaks in Figure 4 were made using 1D nuclear Overhauser effect (NOE) difference spectroscopy (data not shown). The U5 spectrum (Fig. 4A, upper panel) shows five sharp imino proton resonances and a sixth broad peak between 10 and 14 ppm. In contrast, the Ψ5 spectrum (Fig. 4A, middle panel) shows 10 imino proton resonances in this region. The assignments are consistent with the formation of five base pairs for the unmodified RNA (U5) (three G-C pairs, one A-U pair, and a G·U mismatch), and six base pairs for the modified RNA (Ψ5) (three G-C pairs, two A-Ψ pairs, and a G·Ψ mismatch). The closing G-C base pair (G3722-C3740) is observed for both of the H. sapiens H69 RNAs. A loop-closing base pair of the unmodified RNA (U5), U3727-A3735, is not observed. In contrast, the Ψ3727-A3735 pair is observed in the modified RNA (Ψ5), consistent with earlier studies on the E. coli H69 variants (Meroueh et al. 2000). The fact that the U3727-A3735 (loop-closing) base pair is not observed suggests that these residues undergo rapid exchange with solvent due to possible conformational differences between U5 and Ψ5.
FIGURE 4.
The 1D imino proton (uridine H3, guanine H1, and pseudouridine N1H and N3H) NMR spectra of H. sapiens H69 RNAs, U5, Ψ5, and 15N-Ψ5. The spectra in (A) were obtained at 15° C. The spectra in (B) were obtained at 5° C, 15° C, 25° C, 35° C, and 45° C. The buffer employed was 30 mM NaCl, 10 mM sodium phosphate, and 0.5 mM Na2EDTA in 90% H2O and 10% D2O, pH 6.5. Nucleotides are assigned based on 1D NOE spectroscopy (data not shown). In B, the symbols are used to indicate resonances derived from the same nucleotide (asterisks for Ψ3737, triangles for Ψ3727, and arrows for Ψ3739).
In the Ψ5 1H NMR spectrum (Fig. 4A, middle panel), the resonance at 14.0 ppm is assigned as the Ψ3737N3 proton because of a strong NOE to the resonance assigned as G3726 at 13.3 ppm and A3725H2 at 7.6 ppm. The resonance assignment of the Ψ3737N3 proton is further confirmed by employing an 15N-labeled sample of Ψ5 (15N-Ψ5). The 15N-Ψ5 RNA contains 15N at positions N1 and N3 of the stem pseudouridines, Ψ3737 and Ψ3739 (Fig. 1). The level of 15N enrichment is 50%; therefore, for each 15N (N1 and N3 of Ψ), a combination of a doublet (J ≈ 85–95 Hz) and singlet peaks is observed. For Ψ3737N3H, a doublet and singlet centered at 14.0 ppm are observed (Fig. 4A, lower panel, 15N-Ψ5). The identity of Ψ3727N3H is confirmed by an NOE to A3735H2. The intensity of the Ψ3727N3H peak is less than that of the peaks for Ψ3737N3H and Ψ3739N3H, likely due to its location at the end of the helix and greater exposure to solvent.
There are significant differences in the chemical shifts between the U5 and Ψ5 RNAs which were not previously observed with the E. coli modified and unmodified RNAs (Meroueh et al. 2000). For example, G3723 makes a mismatch pair with Ψ3739 in Ψ5, and its imino proton peak is shifted upfield by ~0.5 ppm relative to G3723 in the unmodified U5 RNA (Fig. 4A). Similarly, the chemical shifts for the U3739 and Ψ3739 N3 protons differ by ~0.7 ppm (Ψ3739 N3H is downfield) in the U5 and Ψ5 spectra, respectively. As with the previously mentioned CD spectral changes, the chemical shift changes are likely a result of sequence differences (Ψ versus U) and are not necessarily a result of conformational differences between the modified and unmodified RNAs.
The three stem ΨN1 imino protons (Ψ3739, Ψ3737, and Ψ3727N1H) are protected from chemical exchange with solvent and are therefore observed in the 1H NMR spectrum of Ψ5 (Fig. 4A, middle panel). These protons exchange at increased temperatures (Fig. 4B), and in two cases (Ψ3739N1H and Ψ3727N1H) can no longer be observed at 45°C, except for a slight broadened peak for Ψ3727N1H. For the two A-Ψ pairs, the loss of the ΨN1 and ΨN3 proton resonance intensities (Ψ3727 andΨ3737) does not occur simultaneously (at the same temperature), in contrast to previous studies (Hall and McLaughlin 1991). Similarly, in the case of the G-Ψ pair, the Ψ3739N3 proton resonance is much more intense than the Ψ3739N1H resonance at elevated temperature.
The assignments of the ΨN1H resonances are based on the fact that they exhibit strong NOEs to upfield peaks between 7.0 and 7.4 ppm assigned as ΨH6 (Hall and McLaughlin 1992; Meroueh et al. 2000; Newby and Greenbaum 2001). For example, the identity of Ψ3727N1H is verified by an NOE to Ψ3727H6. Cross-peaks are also observed using two-dimensional (2D) nuclear Overhauser effect spectroscopy (NOESY) (data not shown) between the ΨN1 proton and ΨH6 for each stem pseudouridine, consistent with previous NMR reports on pseudouridine-containing RNAs (Meroueh et al. 2000). Both Ψ3739N1H and Ψ3737N1H can also be clearly assigned in the 15N-Ψ5 spectrum, because they appear as a combination of doublet and singlet peaks centered around 11.0 and 10.7 ppm, respectively (Fig. 4A, lower panel). The Ψ3739N1 proton can be distinguished from Ψ3737N1H because it has strong NOEs to the resonances assigned as G3723 and G3724. Similarly, Ψ3737N1H has NOEs to the neighboring G3726 and G3724 (data not shown). The Ψ3727N1 proton, which does not contain 15N, appears upfield as a singlet at 10.3 ppm.
NOEs observed at 7.6 ppm for U3737N3H or Ψ3737N3H to A3725H2 in the U5 and Ψ5 samples, respectively, are typical for duplex RNA structures. Similarly, a strong NOE is observed between Ψ3727N3H and A3735H2 at 7.4 ppm in the Ψ5 RNA, whereas no peak is observed for the corresponding U3727N3H in U5. These results indicate that the modified and unmodified human H69 RNAs are both forming standard duplex structures in their stem regions, with no evidence of unusual secondary structures. Overall, NMR studies do not reveal any unusual features of the Ψ5 structure in comparison to U5.
The CD and NMR results presented here suggest that there are only subtle differences between the structures of the modified and unmodified H69 RNAs. In addition, failure to observe imino proton resonances from the RNA loop residues suggests that they are not protected from exchange by a specific RNA conformation. In order to further assess any possible differences between the Ψ5 and U5 RNAs, chemical probing studies were carried out (Peattie and Gilbert 1980). The patterns of diethylpyrocarbonate (DEPC) modification are nearly identical for both RNAs at 37°C and 90°C, suggesting that the loop structures are similar (data not shown).
The effects of modification on stability of the helix 69 RNA analogs
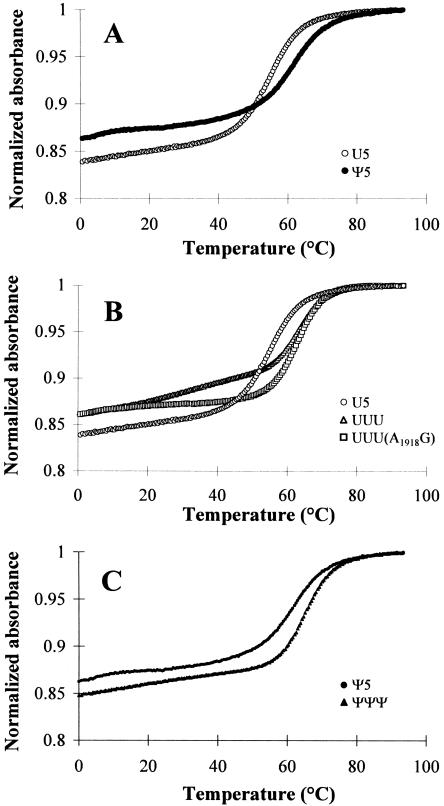
The melting curves of the H69 RNA analogs were obtained to compare the effects of modification on hairpin stability. For the three RNAs, U5, Ψ5, and UUU(A1918G), absorbance versus temperature profiles were obtained at pH 7.0 in low salt conditions (35 mM Na+) and compared to previously studied E. coli RNAs, UUU and ΨΨΨ (Meroueh et al. 2000). The curves were analyzed in terms of melting temperature (Tm), ΔH°ΔS°, and ΔG°37. Representative normalized absorbance plots at single RNA concentrations are shown in Figure 5. All melting curves (except that of UUU) follow a two-state model with unfolded and folded conformations of the H69 analogs. The helix to random coil transitions occurred between 40°C and 80°C and were independent of RNA concentration in the range of 5–100 μM, suggesting unimolecular unfolding of the hairpin structures. The curve for UUU indicates that the hairpin melts in approximately two stages. The lower temperature transition (< 40°C) likely corresponds to loop destacking because it is concentration-independent. ESI and MALDITOF mass spectrometry data and nondenaturing polyacrylamide gel analysis do not reveal any evidence for duplex formation for any of the RNAs (data not shown). The slightly greater hypochromicity observed with U5 compared to Ψ5 might suggest that the loop is more stacked, but such small differences are difficult to assess within the experimental error of the melting data.
FIGURE 5.
The melting curves of unmodified U5 (○) and modified Ψ5 (•) H69 H. sapiens RNAs (A), U5 RNA and unmodified E. coli H69 RNA (▵) and the UUU(A1918G) RNA (□) (B), and Ψ5 overlaid with modified ΨΨΨ E. coli RNA (▴) (C). All melting curves are normalized at 95°C. Buffer: 15 mM NaCl, 20 mM sodium cacodylate, 0.5 mM Na2EDTA, pH 7.0.
The most significant difference between the U5 and Ψ5 RNAs is found by comparing their thermodynamic stabilities. The thermodynamic parameters for all five RNAs are listed in Table 2. The ΔG°37 values of the unmodified and modified H. sapiens RNAs are −2.7 (U5) and −3.8 (Ψ5) kcal/mol, respectively. Thus, they are 1–2 kcal/mole less stable than the E. coli RNAs (ΔG°37 values are −4.8 kcal/mole for UUU and −5.0 kcal/mole for ΨΨΨ). The difference in stability between the E. coli and H. sapiens RNAs is in part due to the stem region sequences of the hairpins. The H. sapiens H69 has an A3725-U3737/Ψ3737 base pair in place of the E. coli C1909-G1921 pair. Previous studies demonstrated that pseudouridines in the closing base-pair position stabilize hairpins (Hall and McLaughlin 1991; Durant and Davis 1999; Yarian et al. 1999; Meroueh et al. 2000). In contrast, pseudouridines in loop positions 1915 and 1917 of the E. coli sequence slightly destabilize the hairpin (Meroueh et al. 2000). Thus, the overall differences in ΔG°37 values are negligible between unmodified (UUU) and modified (ΨΨΨ) E. coli-based RNAs (0.2 kcal/mol), whereas a much larger difference in ΔG°37 values of 1.1 kcal/mole is observed between H. sapiens unmodified (U5) and modified (Ψ5) RNAs. The major sequence differences between E. coli and H. sapiens H69 RNAs are the seventh base of the loop (3′ side, positions 1918 and 3734, respectively) and two additional pseudouridines (positions 3737 and 3739) for H. sapiens. The UUU(A1918G) H69 variant has a H. sapiens loop and E. coli stem sequence. The ΔG°37 values of UUU and UUU(A1918G) are similar (−4.8 versus −4.7 kcal/mol); therefore, the difference in ΔG°37 between U5 and UUU is more likely due to variations in the stem sequences rather than loop sequences. The two additional pseudouridines also contribute ~1.2 kcal/mole to the stability of the H. sapiens H69. Similarly, previous studies showed that incorporation of a single pseudouridine in the eukaryotic U2 snRNA can stabilize the duplex by −0.7 kcal/mole (ΔG°37) (Newby and Greenbaum 2001).
TABLE 2.
Thermodynamics of helix 69 RNAs
| RNA | ΔG°37 (kcal/mole) | ΔΔG°37 (kcal/mole) | ΔH° (kcal/mole) | ΔS° (e.u.) | Tm (°C) |
| Without Mg2+ a | |||||
| UUU | −4.8 ± 0.1 | −61.1 ± 0.5 | −181.4 ± 1.6 | 63.5 | |
| UUU(A1918G) | −4.7 ± 0.1 | −60.1 ± 0.8 | −178.5 ± 2.3 | 63.5 | |
| Ψ Ψ Ψ | −5.0 ± 0.1 | −59.2 ± 0.4 | −174.6 ± 1.2 | 65.7 | |
| U5 | −2.7 ± 0.1 | −50.2 ± 1.5 | −153.2 ± 4.6 | 54.5 | |
| Ψ5 | −3.8 ± 0.2 | −49.5 ± 1.1 | −147.2 ± 3.2 | 62.9 | |
| With 1.0 mM Mg2+ b | |||||
| UUU | −6.4 ± 0.1 | −1.6c | −63.7 ± 0.5 | −184.6 ± 1.2 | 71.8 |
| UUU(A1918G) | −6.3 ± 0.1 | −1.6 | −63.2 ± 0.8 | −183.4 ± 2.3 | 71.6 |
| Ψ Ψ Ψ | −6.6 ± 0.1 | −1.6 | −61.9 ± 0.7 | −178.6 ± 2.0 | 73.7 |
| U5 | −3.9 ± 0.1 | −1.2 | −53.9 ± 2.3 | −161.2 ± 6.8 | 61.0 |
| Ψ5 | −5.1 ± 0.1 | −1.3 | −54.3 ± 0.8 | −158.3 ± 2.4 | 69.5 |
aThe buffer conditions were 15 mM NaCl, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA, pH 7.0.
bThe buffer conditions were 15 mM NaCl, 20 mM sodium cacodylate, 0.5 mM Na2EDTA, and 1.0 mM MgCl2, pH 7.0.
cΔΔG°37 = Δ G°37 (1.0 mM Mg2+) − Δ G°37 (0 mM Mg2+).
In the present study, it was also demonstrated that Mg2+ has a stabilizing effect on both E. coli- and H. sapiens-based H69 analogs. Such a result is not surprising given that the magnesium ions can be counter ions for the negatively charged RNA phosphate backbone. The ΔΔG°37 value for both unmodified (UUU) and modified (ΨΨΨ) E. coli RNAs in the absence and presence of 1.0 mM Mg2+ is −1.6 kcal/mole (Table 2). In contrast, the ΔΔG°37 values of unmodified (U5) and modified (Ψ5) H. sapiens RNA (−1.2 and −1.3 kcal/mol, respectively) are lower than the E. coli values. The ΔΔG°37 value of UUU(A1918G) in the presence and absence of 1.0 mM Mg2+ is the same as that of the E. coli sequences (−1.6 kcal/mol); therefore, the trend of the ΔΔG°37 values shows that the UUU(A1918G) variant with the H. sapiens loop sequence and E. coli stem sequence behaves more like the bacterial RNA. The differences in magnesium effects on hairpin stability must therefore arise from differences in the stem sequences, and not from the presence or absence of pseudouridine residues.
DISCUSSION
It has been noted that the number of nucleotide modifications in rRNA increases with evolution (Ofengand et al. 1995). Helix 69 of H. sapiens 28S rRNA contains five pseudouridines compared to three in the corresponding hairpin in E. coli 23S rRNA. Furthermore, modifications tend to occur in functionally important regions of the ribosome, such as the peptidyl transferase center. Several pseudouridines (Ψ1911 and Ψ1917 in E. coli 23S rRNA) in H69 are universally conserved, and provide a growth advantage to cells (Ofengand and Bakin 1997; Raychaudhuri et al. 1998). Similarly, the A residue at position 1918 of E. coli 23S rRNA is highly conserved among bacteria. The corresponding position (3734 of 28S rRNA) in H. sapiens and other eukaryotes is a > 98% conserved G residue.
Helix 69 of the large subunit rRNA occurs in the bridge region of the 70S and 80S ribosomes referred to as B2a. Cryo-EM and X-ray crystallography studies have shown that the conformation of H69 in the large subunit (unbound, 50S) and complete ribosome (bound, 70S) is different (Harms et al. 2001; Yusupov et al. 2001). Thus, H69 is flexible and has a role extending beyond just a structural support for the intersub-unit bridge. This flexibility may allow H69 to participate directly in tRNA translocation. For this reason, H69 has been referred to as a ‘molecular crane’ (Bashan et al. 2003). A recent study to determine the location of the ribosome release factor RRF on the 70S ribosome revealed another important role for H69 (Agrawal et al. 2004). Contacts with the highly conserved regions of RRF and the loop region of H69 are attributed to specific pseudouridine residues. Direct contact of RRF with these residues may be important for conformation changes in this region and ultimately subunit dissociation.
The UV melting (Tm) data presented in these studies establish that the three pseudouridine residues in the stem region of H. sapiens H69 provide stabilization to the RNA. In contrast, substitution at position 1918 with a G residue in eukaryotes in place of an A found in bacteria does not affect the RNA stability. The UUU(A1918G) has an E. coli stem region and H. sapiens loop region, but the ΔG°37 value is similar to those of the unmodified or modified E. coli H69 RNAs. Previous studies showed that individually, Ψ1911, Ψ1915, and Ψ1917 have different effects on the stability of E. coli H69; however, when combined, the three Ψs have little effect on the overall stability of the hairpin (Meroueh et al. 2000). The ΔG°37 value of the unmodified human H69 is less than that of the E. coli H69 (U5 versus UUU) by > 2 kcal/mol. Therefore, additional pseudouridines may be necessary in order to compensate for sequence changes that destabilize the stem of the human analog (the eukaryotic sequence has an A-U pair in place of a C-G pair found in the bacterial RNA). Fahlman et al. (2004) proposed such a strategy in aminoacyl transfer RNAs binding to the ribosome. They suggest that different patterns of contact between the tRNA and ribosome sites have evolved through changes in tRNA sequence and levels of modification such that binding is uniform.
The addition of divalent metal ions such as Mg2+ leads to stabilization of the modified RNAs, but at slightly different magnitudes (the ΔΔG°37 values, defined as ΔG°37 in the presence of Mg2+ minusΔG°37 in the absence of Mg2+, differ by 0.3 to 0.4 kcal/mol) for the H. sapiens- and E. coli-derived H69 RNAs. These results indicate that the electrostatic environments of the E. coli and H. sapiens H69 RNAs are slightly different. However, these charge differences are clearly derived from sequence changes, not from modified nucleotides. These conclusions are based on the fact that the same ΔΔG°37 values are found for unmodified (U) and modified (Ψ) RNAs in both E. coli and H. sapiens (−1.6 kcal/mole for E. coli and −1.2 to −1.3 for human RNAs; Table 2). The E. coli sequence has a G-C base pair in the stem that is replaced with an A-Ψ pair in the human sequence. Previous structural and computational studies revealed the presence of certain magnesium-guanine interactions (Juneau et al. 2001; Petrov et al. 2002).
The CD and NMR data both reveal that U5 and Ψ5 form canonical A-form helical structures, and substitution of pseudouridine with uridine has subtle effects on the secondary structure of H69. More specifically, the closing base pair Ψ/U3727-A3735 is observed in theΨ5 RNA, but not in the U5 RNA. Also, the NMR data clearly show evidence of additional hydrogen- bonding interactions involving the N1H positions of the Ψ stem residues (Fig. 4). The use of 15N-labeled Ψ residues was useful for assigning these resonances in the imino proton NMR spectrum. The presence of these resonances is consistent with the formation of water-mediated hydrogen bonds from pseudouridine to the RNA backbone, as has been observed in other Ψ-containing RNAs (Arnez and Steitz 1994; Davis 1995; Newby and Greenbaum 2002).
Another unique feature of the H. sapiens H69 stem is the presence of the G·Ψ mismatch. The related G·U mismatch has been identified and characterized in a number of locations within rRNAs (Gautheret et al. 1995), and can provide specific recognition signals in RNA because of its unique structure (Hou and Schimmel 1988; McClain and Foss 1988). A G·U mismatch is present in the E. coli H69 stem, and a G·Ψ mismatch occurs in the human analog in the identical position. Our NMR data indicate that a G·Ψ mismatch does indeed form in the stem of human H69. The hydrogen-bonding pattern appears to be similar to that of the wobble G·U mismatch involving the GN1H and ΨN3H; however, the 3D structure and base orientation remain to be determined.
The roles of the first and seventh nucleotides of the H69 loop also remain to be determined. Even though G3734 is highly conserved in eukaryotic rRNAs and A1918 is similarly conserved in bacterial rRNAs (Fig. 2; Table 1), this sequence difference does not appear to affect the hairpin structure or stability in a significant manner. This nucleotide may play a role in mediating the interactions with other RNA domains within the ribosome, or in altering the dynamics of the loop in the presence and absence of modified nucleotides. Thus, the 15N-labeled RNAs that we have generated will be quite useful for further studies.
The general structures of the E. coli and H. sapiens H69 RNAs appear to be similar in that they are A-form helices with a loop-closing base pair stabilized by an A-Ψ pair. Thus, flexibility of H69 is likely to be localized in the loop regions for both eukaryotic and bacterial RNAs. Further investigation into the structural differences and similarities between bacterial and human rRNAs is important for the future development of ribosome-binding drugs that can target the intersubunit domains and act as potential antibiotics. 15NNMR relaxation studies will also lead to a better understanding of the dynamics of regions such as H69 in mediating ribosome function.
MATERIALS AND METHODS
Preparation of RNAs
The 19-nucleotide (19-nt) RNAs used in this study were obtained from Dharmacon Research. These RNAs were chemically synthesized on a 1.0 μmol scale and deprotected as described (Meroueh et al. 2000). The sequences of the five RNAs are as follows:
5′-G1906GCCGUAACUAUAACGGUC1924-3′ (UUU),
5′-G1906GCCGΨAACΨAΨAACGGUC1924-3′ (ΨΨΨ),
5′-G1906GCCGUAACUAUG1918ACGGUC1924-3′ (UUU (A1918G)),
5′-G3722GGAGUAACUAUGACUCUC3740-3′ (U5), and
5′-G3722GGAGΨAACΨAΨGACΨCΨC3740-3′ (Ψ5).
The numbering for UUU, ΨΨΨ, and UUU(A1918G) is based on full-length E. coli 23S rRNA. The numbering for U5 and Ψ5 is based on full-length H. sapiens 28S rRNA. TheΨ5 variant was also synthesized with site-specific 15N labels at the N1 and N3 positions of Ψ3737 and Ψ3739 (5′-G3722GGAGΨAACΨAΨGAC(15N-Ψ)3737C(15N-Ψ)3739C3740-3′; referred to as 15N-Ψ5). The 15N-containing pseudouridine and its corresponding phosphoramidite were synthesized as described (Grohar and Chow 1999; Meroueh et al. 2000; Hanessian and Machaalani 2003), except that [1,3-15N] uracil (SantaLucia et al. 1995) (from[15N]urea) was employed as a starting material.
The deprotected RNAs were purified on 20% denaturing poly-acrylamide gels (typically 350 V for 4 h). Prior to loading on the gel, the RNA sample contained urea loading buffer [7 M urea, 50% (w/v) glycerol, and 0.1 % (w/v) each of xylene cyanol and bromo-phenol blue]. The RNA bands were visualized on the gel by UV shadowing with a fluorescent TLC plate. The bands corresponding to the 19-nt RNAs were excised from the gel, and the RNAs were eluted with 0.5X TBE buffer (45 mM Tris, 45 mM boric acid, 0.5 mM EDTA, pH 8.2) at 200 V for 2.5 h on an Amicon Centrilutor device. The resulting RNAs were desalted over C18 Sep-Pak columns (Waters), and then dialyzed for 2–3 d against RNase-free, double-deionized water. The 15N-containing RNA was purified by HPLC on an XTerra MS C18 column (2.5 μm, 10 × 50 mm, Waters) in which the eluent was 0.1 M TEAA buffer, pH 7.0 with a 7%–11% linear gradient of acetonitrile over 17 min at a flow rate of 4.0 mL/min. After HPLC purification, the RNA was desalted by ethanol precipitation and dialysis for 2–3 d against RNase-free, double-deionized water.
RNA concentrations were calculated using Beer’s law and single-stranded extinction coefficients (ɛ) of 188,860 M−1cm−1 for UUU and ΨΨΨ, 186,500 M−1 cm−1 for UUU(A1918G), and 191,300 M−1 cm−1 for U5 and Ψ5. The same extinction coefficient was used for uridine and pseudouridine (1.0 × 104 cm−1 M−1 at pH 7.0) (Richards 1975). The RNAs were renatured in 10 mM Tris-HCl, pH 7.5 by heating to 90°C for 2 min followed by slowly cooling to room temperature.
Circular dichroism (CD) spectroscopy
CD spectra were obtained on a Jasco J600 spectropolarimeter (220–320 nm) at room temperature in 15 mM NaCl, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA at pH 7.0. The RNA concentrations were 2.6 μM for all CD experiments. The ΔA values were converted to molar ellipticity (Δɛ) expressed in moles of RNA molecules (Cantor and Schimmel 1980). The RNA concentrations of each CD sample were determined from the UV absorbance values (260 nm) at 95°C. Magnesium chloride (0–5 mM MgCl2 ·6H2O) was also titrated into the CD samples. Small volumes (< 5 μL) were added such that concentration changes were negligible.
Thermal melting
The absorbance versus temperature changes were monitored on an Aviv 14DS UV-visible spectrophotometer with a five-cuvette thermoelectric controller. Microcuvettes with two different path lengths (0.1 and 0.2 cm; volumes of 60 and 120 μL, respectively) were employed. The buffer used for this experiment contained 15 mM NaCl, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA, pH 7.0. The RNA concentrations of each sample were determined from the UV absorbance values (260 nm) at 95°C. The melting curves were obtained from 0°C to 90°C at 280 nm. The thermo-dynamic parameters were calculated by using Meltwin 3.5 and assuming a two-state model (McDowell and Turner 1996). The thermal melting experiments just described were also repeated in the presence of magnesium chloride (1, 3, and 5 mM).
NMR spectroscopy
The NMR spectra for the U5, Ψ5, and 15N-Ψ5 were obtained on a Bruker AVANCE-AQS 700 MHz spectrometer equipped with a 5-mm triple-resonance cryoprobe. The RNA were dissolved in buffer containing 10 mM Na2HPO4, 0.5 mM Na2EDTA, 30 mM NaCl, and 20 μM TSP in 90% H2O and 10% D2O, pH 6.5. The final RNA concentrations were ~750 μM. All NMR spectra were acquired at 15°C, except for variable temperature experiments that ranged from 5°C to 45°C. 1D proton NMR spectra were acquired for all samples using Digital Quadrature Detection for at least 16,000 data points. Samples in 9:1 H2O/D2O required additional Watergate solvent suppression pulse schemes to be included in the pulse program. Pure D2O samples were acquired with added 1-sec presaturation pulses for HDO signal suppression. 1D NOE difference spectra included a 500-msec presaturation pulse centered on imino peaks of interest. Resulting selective saturation spectra were subtracted from a reference spectrum to determine resonances involved in NOEs. To improve the signal to noise ratio, at least 128 scans were acquired for each experiment. NMR data were processed with zero-filling one time, 1–3 Hz of line broadening, and solvent signal filters and base-line correction.
Acknowledgments
This work is supported by the National Institutes of Health (grant GM54632). We thank Prof. J. SantaLucia for the use of the Aviv spectrometer and many helpful discussions. B. Ksebati, M. Meroueh, and R. Aduri provided technical assistance and useful discussions with the NMR experiments.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2320605.
REFERENCES
- Agrawal, R.K., Sharma, M.R., Kiel, M.C., Hirokawa, G., Booth, T.M., Spahn, C.M.T., Grassucci, R.A., Kaji, A., and Frank, J. 2004. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc. Natl. Acad. Sci. 101: 8900–8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnez, J.G. and Steitz, T.A. 1994. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33: 7560–7567. [DOI] [PubMed] [Google Scholar]
- Badis, G., Fromont-Racine, M., and Jacquier, A. 2003. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA 9: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Bashan, A., Agmon, I., Zarivach, R., Schlünzen, F., Harms, J., Berisio, R., Bartels, H., Franceschi, F., Auerbach, T., Hansen, H.A.S., et al. 2003. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell 11: 91–102. [DOI] [PubMed] [Google Scholar]
- Bortolin, M.-L. and Kiss, T. 1998. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA 4: 445–454. [PMC free article] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor, C.R. and Schimmel, P.R. 1980. Biophysical chemistry. Part II: Techniques for the study of biological structure and function. W. H. Freeman, San Francisco.
- Carter, A.P., Clemons, W.M., Broderson, D.E., Morgan-Warren, R.J., Wimberly, B.T., and Ramakrishnan, V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Yusupov, M.M., Yusupova, G.Zh., Earnest, T.N., and Noller, H.F. 1999. X-ray crystal structure of 70S ribosome functional complexes. Science 285: 2095–2104. [DOI] [PubMed] [Google Scholar]
- Charette, M. and Gray, M.W. 2000. Pseudouridine in RNA: What, where, how, and why. IUBMB Life. 49: 341–351. [DOI] [PubMed] [Google Scholar]
- Chui, H.M.-P., Desaulniers, J.-P., Scaringe, S.A., and Chow, C.S. 2002. Synthesis of helix 69 of Escherichia coli 23S rRNA containing its natural modified nucleosides, m3Ψ and Ψ. J. Org. Chem. 67: 8847–8854. [DOI] [PubMed] [Google Scholar]
- Cohn, W.E. 1960. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: Isolation, structure, and chemical characteristics. J. Biol. Chem. 235: 1488–1498. [PubMed] [Google Scholar]
- Davis, D.R. 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 23: 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant, P.C. and Davis, D.R. 1999. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 285: 115–131. [DOI] [PubMed] [Google Scholar]
- Fahlman, R.P., Dale, T., and Uhlenbeck, O.C. 2004. Uniform binding of aminoacyl transfer RNAs to the ribosomal A and P sites. Mol. Cell 16: 799–805. [DOI] [PubMed] [Google Scholar]
- Frank, J. and Agrawal, R.K. 2000. A ratchet-like inter-subunit reorganization of the ribosome during translation. Nature 406: 318–322. [DOI] [PubMed] [Google Scholar]
- Gabashvili, I.S., Agrawal, R.K., Spahn, C.M.T., Grassucci, R.A., Svergun, D.I., Frank, J., and Penczek, P. 2000. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100: 537–549. [DOI] [PubMed] [Google Scholar]
- Gao, H., Sengupta, J., Valle, M., Korostelev, A., Eswar, N., Stagg, S.M., Van Roey, P., Agrawal, R.K., Harvey, S.C., Sali, A., et al. 2003. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113: 789–801. [DOI] [PubMed] [Google Scholar]
- Gautheret, D., Konings, D., and Gutell, R.R. 1995. G·U base pairing motifs in ribosomal RNAs. RNA 1: 807–814. [PMC free article] [PubMed] [Google Scholar]
- Gregorian Jr., R.S. and Crothers, D.M. 1995. Determinants of RNA hairpin loop-loop complex stability. J. Mol. Biol. 248: 968–984. [DOI] [PubMed] [Google Scholar]
- Grohar, P.J. and Chow, C.S. 1999. A practical synthesis of the modified RNA nucleoside pseudouridine. Tet. Lett. 40: 2049–2052. [Google Scholar]
- Hall, K.B. and McLaughlin, L.W. 1991. Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry 30: 1795–1801. [DOI] [PubMed] [Google Scholar]
- ———. 1992. Properties of pseudouridine N1 imino protons located in the major groove of an A-form RNA duplex. Nucleic Acids Res. 20: 1883–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanessian, S. and Machaalani, R. 2003. A highly stereocontrolled and efficient synthesis of α- and β-pseudouridines. Tet. Lett. 44: 8321–8323. [Google Scholar]
- Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688. [DOI] [PubMed] [Google Scholar]
- Hou, Y.-M. and Schimmel, P. 1988. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333: 140–145. [DOI] [PubMed] [Google Scholar]
- Joseph, S., Weiser, B., and Noller, H.F. 1997. Mapping the inside of the ribosome with an RNA helical ruler. Science 278: 1093–1098. [DOI] [PubMed] [Google Scholar]
- Juneau, K., Podell, E., Harrington, D.J., and Cech, T.R. 2001. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA-solvent interactions. Structure 9: 221–231. [DOI] [PubMed] [Google Scholar]
- Kowalak, J.A., Bruenger, E., Hashizume, T., Peltier, J.M., Ofengand, J., and McCloskey, J.A. 1996. Structural characterization of U*-1915 in domain IV from Escherichia coli 23S ribosomal RNA as 3-methylpseudouridine. Nucleic Acids Res. 24: 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbach, P.A., Crain, P.F., and McCloskey, J.A. 1994. Summary: The modified nucleosides of RNA. Nucleic Acids Res. 22: 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden, B.E.H. and Wakeman, J.A. 1988. Pseudouridine distribution in mammalian 18 S ribosomal RNA. Biochem. J. 249: 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiväli, Ü . and Remme, J. 2004. Definition of bases in 23S rRNA essential for ribosomal subunit association. RNA 10: 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain, W.H. and Foss, K. 1988. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science 240: 793–796. [DOI] [PubMed] [Google Scholar]
- McCloskey, J.A. and Crain, P.F. 1998. The RNA modification data-base –1998.Nucleic Acids Res. 26: 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.A. and Turner, D.H. 1996. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry 35: 14077–14089. [DOI] [PubMed] [Google Scholar]
- Meroueh, M., Grohar, P.J., Qiu, J., SantaLucia Jr., J., Scaringe, S.A., and Chow, C.S. 2000. Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 Region of Escherichia coli 23S rRNA. Nucleic Acids Res. 28: 2075–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merryman, C., Moazed, D., Daubresse, G., and Noller, H.F. 1999. Nucleotides in 23 S rRNA protected by the association of 30 S and 50 S ribosomal subunits. J. Mol. Biol. 285: 107–113. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Osswald, M., and Brimacombe, R. 1992. Identification of intermolecular RNA cross-links at the subunit interface of the Escherichia coli ribosome. Biochemistry 31: 3004–3011. [DOI] [PubMed] [Google Scholar]
- Newby, M.I. and Greenbaum, N.L. 2001. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branchsite architecture. RNA 7: 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. 99: 12697–12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- O’Connor, M. and Dahlberg, A.E. 1995. The involvement of two distinct regions of 23S ribosomal RNA in tRNA selection. J. Mol. Biol. 254: 838–847. [DOI] [PubMed] [Google Scholar]
- Ofengand, J. and Bakin, A. 1997. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266: 246–268. [DOI] [PubMed] [Google Scholar]
- Ofengand, J., Bakin, A., Wrzesinski, J., Nurse, K., and Lane, B.G. 1995. The pseudouridine residues of ribosomal RNA. Biochem. Cell. Biol. 73: 915–924. [DOI] [PubMed] [Google Scholar]
- Peattie, D.A. and Gilbert, W. 1980. Chemical probes for higher-order structure in RNA. Proc. Natl. Acad. Sci. 77: 4679–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, A.S., Lamm, G., and Pack, G.R. 2002. Water-mediated magnesium-guanine interactions. J. Phys. Chem. B 106: 3294–3300. [Google Scholar]
- Raychaudhuri, S., Conrad, J., Hall, B.G., and Ofengand, J. 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E.G. 1975. Use of tables in calculation of absorption, optical rotary dispersion, and circular dichroism of polyribonucleotides. In Handbook of biochemistry and molecular biology, pp. 596–599. Nucleic acids. CRC Press, Cleveland, OH.
- SantaLucia Jr., J., Shen, L.X., Cai, Z., Lewis, H., and Tinoco Jr., I. 1995. Synthesis and NMR of RNA with selective isotopic enrichment in the bases. Nucleic Acids Res. 23: 4913–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlünzen, F., Zarivach, R., Harms, J., Bashan, A., Tocilj, A., Albrecht, R., Yonath, A., and Franceschi, F. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413: 814–821. [DOI] [PubMed] [Google Scholar]
- Schweizer, M.P., Thedford, R., and Slama, J. 1971. Synthesis and conformational properties of diribonucleoside monophosphates containing modified nucleosides as found in transfer RNA. Biochim. Biophys. Acta 232: 217–226. [DOI] [PubMed] [Google Scholar]
- Scott, J.F. and Zamecnik, P.C. 1969. Some optical properties of diadenosine- 5′-phosphate. Proc. Natl. Acad. Sci. 64: 1308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, C.M.T., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., and Frank, J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae – tRNA-ribosome and subunit-sub-unit interactions. Cell 107: 373–386. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Broderson, D.E., Clemons Jr., W.M., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Wrzesinski, J., Bakin, A., Ofengand, J., and Lane, B.G. 2000. Isolation and properties of Escherichia coli 23S-RNA pseudouridine 1911, 1915, 1917 synthase (RluD). IUBMB Life. 50: 33–37. [DOI] [PubMed] [Google Scholar]
- Yarian, C.S., Basti, M.M., Cain, R.J., Ansari, G., Guenther, R.H., Sochacka, E., Czerwinska, G., Malkiewicz, A., and Agris, P.F. 1999. Structural and functional roles of the N1- and N3-protons of Ψ at tRNA’s position 39. Nucleic Acids Res. 27: 3543–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H.D., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]