Abstract
Telomerase accurately synthesizes telomeric DNA by reverse transcription of a tightly defined template region in the telomerase RNA (TR). Reverse transcription past the 5′ boundary of the template can cause the incorporation of noncognate nucleotides into telomeric DNA, which can result in disruption of normal telomere function. The products synthesized by human telomerase do not contain the nucleotide cytosine, which is encoded by an hTR residue 2 nucleotides (nt) 5′ of the template boundary. We examined dCTP incorporation by a series of telomerases reconstituted with N- and C-terminally mutated human telomerase reverse transcriptases (hTERTs). We found that altering sequences in the N-terminal RNA interaction domain 1 (RID1) and C terminus caused dCTP-dependent catalytic phenotypes suggestive of reverse transcription of sequences 5′ of the template boundary. A RID1 mutant that exhibited a dCTP-dependent phenotype interacted less efficiently with a human telomerase RNA (hTR) variant in which the 5′ template boundary-defining P1b element was disrupted, whereas C-terminal mutations did not alter hTR interactions in a P1b-dependent fashion. Disruption of P1b or template linker sequences between P1b and the 5′ template boundary also impaired 5′ template usage in RID1 and C-terminal hTERT mutants. These observations identify overlapping roles for hTR sequences and structures 5′ of the template in regulating both 5′ template boundary definition and 5′ template usage, and implicate hTERT N- and C-terminal regions in 5′ template usage and suppression of noncognate nucleotide incorporation.
Keywords: telomerase, reverse transcriptase, telomere, template, processivity, fidelity
INTRODUCTION
The telomeres of linear replicons shorten with each round of DNA replication. In most eukaryotes, the enzyme that counteracts telomere shortening is telomerase. The minimal components required for reconstitution of in vitro telomerase activity are the template-bearing telomerase RNA molecule (hTR in humans) and the telomerase reverse transcriptase (hTERT in humans) (for review, see Harrington 2003).
Telomerase reverse transcribes a tightly defined template region in the telomerase RNA (TR) to synthesize the short DNA repeat that constitutes the bulk of telomeric DNA. The 5′ boundary of the TR template is strictly regulated, since reverse transcription past this boundary would result in synthesis of telomere repeats containing nontelomeric sequences. In most telomerases studied to date, 5′ template boundary definition is regulated by template-adjacent TR stem structures and intervening template linker sequences, which have been proposed to constrain movement of the template in the telomerase active site; in hTR this structure is the P1b helix (Fig. 1A; Autexier and Greider 1995; Prescott and Blackburn 1997; Tzfati et al. 2000; Lai et al. 2002; Chen and Greider 2003; Seto et al. 2003). In Saccharomyces cerevisiae and Tetrahymena telomerases, mutation of the 5′ template boundary-defining elements (H1 and Helix II, respectively) or physical disruption or mutation of template linker sequences also alters template usage, nucleotide addition processivity, and fidelity (Prescott and Blackburn 1997; Lai et al. 2002; Miller and Collins 2002; Seto et al. 2003). It is unknown whether P1b or linker sequences connecting P1b and the template regulate similar functions in human telomerase. The 5′ template boundary-defining elements of S. cerevisiae and Tetrahymena TRs interact with TERT, and TERT sequences that interact with Helix II of the Tetrahymena TR are important for 5′ template boundary definition (Miller et al. 2000; Lai et al. 2002; Seto et al. 2003; Lin et al. 2004). The hTR pseudoknot/template domain, which contains the template and P1b helix, interacts independently with hTERT, and the results of studies that have mapped potential hTERT–hTR interaction sites by mutagenesis, chemical and enzymatic footprinting, and oligonucleotide competition suggest that P1b might be one site of interaction with hTERT (Beattie et al. 2000; Mitchell and Collins 2000; Bachand and Autexier 2001; Antal et al. 2002; Keppler and Jarstfer 2004). However, the potential role of P1b in hTERT interactions has not yet been examined specifically, and no hTERT mutations that disrupt the 5′ template boundary definition or P1b interactions have been identified.
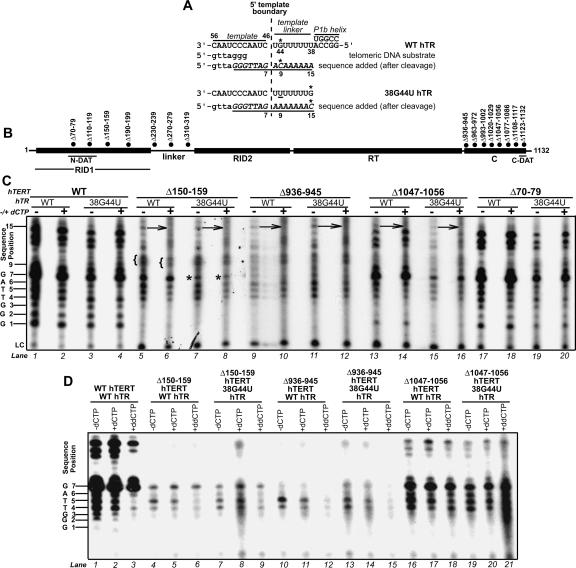
FIGURE 1.
Catalytic phenotypes of hTERT mutants in the presence of the noncognate nucleotide dCTP. (A) Schematic of the hTR template, 5′ template boundary, template linker, and P1b helix. The positions of hTR nt 38, 44, 46, and 56 are indicated. Wild-type (WT) and 38G44U hTRs are illustrated in the upper and lower panels. Alignment of the 3′ end of the DNA substrate with the template is indicated below the schematic for wild-type hTR. The products added to this primer after cleavage (underlined italics) are indicated below the schematics for wild-type and 38G44U hTRs (Huard and Autexier 2004). The positions of products resulting from addition of 7, 9, or 15 nt to the cleaved DNA substrate are indicated. The hTR positions directing dCTP incorporation and the sites of dCTP incorporation in products for wild-type and 38G44U hTRs are marked by asterisks. (B) Schematic illustrating hTERT domains and positions of mutations discussed in this study. (C ) Catalytic phenotypes of selected hTERT mutants reconstituted in RRL with wild-type or 38G44U hTRs, and assayed in the presence (+) or absence (−) of unlabeled dCTP. The dCTP-dependent catalytic phenotypes of these and other hTERT mutants not depicted in this figure are summarized in Table 1. The position numbers of each product are indicated at left. Arrows indicate products at position 15 that exhibit reduced intensity in the presence of dCTP. Opening braces ({) indicate positions at which product mobility is altered in the presence of dCTP. Asterisks indicate positions at which product intensity is reduced when telomerase is assembled with 38G44U hTR in both the presence and absence of dCTP. (LC) Loading control. A lower exposure is shown for the wild-type hTERT samples in lanes 1–4 to facilitate comparison of product pausing patterns. To enhance detection of telomerase activity, the reaction and sample volumes for all C-terminal hTERT mutants and the Δ150–159 variant were scaled up twofold with respect to wild-type hTERT and other mutants in this experiment and all others described in this study. The same amount of loading control was added to each reaction, irrespective of reaction volume. (D) Catalytic phenotypes of selected hTERT mutants reconstituted in RRL with wild-type or 38G44U hTRs and assayed in the absence of dCTP (−dCTP) or in the presence of dCTP (+dCTP) or dideoxy CTP (+ddCTP). Since the overall activity of RID1 and C-terminal mutants was very weak compared to wild-type telomerase, in some experiments such as the one shown here we could not always detect products longer than one telomere repeat. The position numbers of products are indicated at left.
hTERT, like other TERTs, contains reverse transcriptase (RT) motifs that are relatively well conserved with other RTs, and are flanked by telomerase-specific N- and C-terminal sequences (Fig. 1B; for review, see Harrington 2003). The C terminus may constitute the polymerase thumb of TERT and is important for nucleotide addition processivity (Peng et al. 2001; Huard et al. 2003). The hTERT C and N termini interact physically and functionally, may associate with DNA substrates, and also regulate the telomerase-specific property of repeat addition processivity (Beattie et al. 2001; Arai et al. 2002; Huard et al. 2003; Lee et al. 2003; Moriarty et al. 2004, 2005). The hTERT N terminus, but not the C terminus, is required for hTR interactions (Beattie et al. 2000; Bachand and Autexier 2001; Lai et al. 2001). Two regions of the N terminus are involved in hTR interactions, RNA interaction domains 1 and 2 (RID1 and RID2), which are separated by a nonconserved catalytically inessential linker (Xia et al. 2000; Armbruster et al. 2001; Moriarty et al. 2002). RID2 likely regulates telomerase assembly, via interactions with the P6.1 helix of the hTR CR4/CR5 domain (Lai et al. 2001; Moriarty et al. 2004). RID1 interacts with the hTR pseudoknot/ template domain, possibly transiently or with low affinity, is essential for repeat addition processivity, and contributes to anchor site-type catalytic functions (Moriarty et al. 2004, 2005). Similarly, yeast TERT sequences corresponding to RID1 are important for anchor site function and repeat addition processivity, and also physically contact the 5′ end of DNA substrates (Lue 2005). A sub-region of RID1, referred to as N-DAT (for dissociates activities of telomerase), is not required for wild-type levels of telomerase activity in the PCR-based TRAP (telomeric repeat amplification protocol) assay, but is important for the specificity and affinity of telomerase-DNA interactions, and is essential for telomere length maintenance in vivo (Armbruster et al. 2001; Lee et al. 2003; Moriarty et al. 2005). A short C-DAT region has also been identified at the extreme C terminus of hTERT, though C-DAT sequences contribute to human telomerase catalytic function (Banik et al. 2002; Huard et al. 2003; Moriarty et al. 2005).
In an effort to identify hTERT sequences that might mediate 5′ template boundary definition, we examined the incorporation of the noncognate nucleotide dCTP by a series of hTERT mutant telomerases. We analyzed the interaction of these mutants with the hTR P1b helix and investigated the potential roles of hTERT and the hTR P1b helix and template linker sequences in template usage. Our results implicate the hTR P1b helix and template linker sequences, and hTERT RID1 and C-terminal sequences in 5′ template usage and incorporation of noncognate nucleotides specified by hTR nucleotides 5′ of the template boundary.
RESULTS
Investigation of hTERT mutants’ catalytic phenotypes in the presence of the unlabeled noncognate nucleotide dCTP
To determine whether hTERT was implicated in 5′ template boundary regulation, we investigated the effect of the unlabeled noncognate nucleotide dCTP on the patterns of products generated by a series of rabbit reticulocyte lysate (RRL)- reconstituted N- and C-terminal hTERT mutant telomerases (Fig. 1). Human telomere repeats do not contain cytosine, and the only guanosine residue near the hTR template (nt 46–56) is located at hTR nucleotide (nt) 44 (Fig. 1A). The hTR P1b helix was previously identified as a 5′ template boundary-regulating element in human telomerase by examining product patterns generated by telomerase in the presence of unlabeled dCTP (Chen and Greider 2003). We investigated dCTP-dependent product patterns of mutant telomerases reconstituted with either wild-type hTR or an hTR substitution variant in which the guanosine residue at position 44 is moved to position 38, immediately adjacent to the P1b helix (Fig. 1A: 38G44U). This substitution is catalytically silent in the context of wild-type telomerase and has been useful in the characterization of the P1b helix as a regulator of the 5′ template boundary, as it permits unambiguous identification of dCTP-dependent products generated by read-through past the 5′ template boundary (Chen and Greider 2003). Our study focused on mutations in the hTERT N-terminal linker, RNA interaction domain 1 (RID1), and C terminus, as N-terminal hTERT RNA interaction domain 2 (RID2) variants are not catalytically active (Fig. 1B; Moriarty et al. 2004).
Mutations in the N-terminal DAT subdomain and linker region did not affect the pattern of products generated by hTERTs assembled with either wild-type hTR or the 38G44U hTR variant when these telomerases were assayed in the presence of unlabeled dCTP (Table 1, Group I mutants; for example, see Fig. 1C: WT and Δ70–79 samples, cf. lanes 1 and 2, 3 and 4, 17 and 18, and 19 and 20). In contrast, products at positions 9 and 10 generated by the Δ150–159 RID1 mutant telomerase migrated slightly faster when assays were performed in the presence of dCTP; this altered migration was observed when the Δ150–159 variant was assembled with wild-type but not 38G44U hTR (Table 1, Group II mutant; Fig. 1C: cf. products in lanes 5 and 6 marked by opening braces; note that the slightly decreased mobility of comparable products in lane 8 is due to a curve in the gel, which also affects the mobility of the loading control and other products in this lane). Different product mobilities at position 9 could be due to the incorporation of different nucleotides, as would occur if reverse transcription continued 5′ of the template (nt 44 in wild-type and 38G44U hTRs dictates incorporation of cytosine and adenosine residues, respectively). In the presence of dCTP, the intensities of products at position 15 were reduced when the activity of Δ150–159 hTERT and numerous C-terminal mutants were reconstituted with either wild-type or 38G44U hTR; products at this position were visible as distinct bands in the absence of dCTP, but not in the presence of dCTP (Table 1, Group II, III and IV mutants; for examples, see products marked by arrows in Fig. 1C: lanes 5–16). Altered product intensity at position 15 might be expected for hTERT variants assembled with 38G44U hTR, if these mutants reverse transcribed past the 5′ template boundary and incorporated a cytosine opposite position 38 in the telomerase RNA (Fig. 1A). A changed product pattern at position 15 in telomerases reconstituted with wild-type hTR (in this case dictated by hTR nt 44C) might also reflect 5′ template boundary bypass after a second round of DNA synthesis, resulting in dCTP-dependent products 15 nt in length, though this seemed less likely given the extremely weak repeat addition processivity of most of the affected hTERT mutants (Huard et al. 2003; Moriarty et al. 2004). The impaired processivity of these mutants might also contribute to the reduced intensity of products at position 15, though processivity defects cannot account for the dCTP dependence of the product pattern at this position. dCTP-dependent changes in product patterns could also be the result of dCTP misincorporation during reverse transcription of the template itself. The intensities of products shorter than 7 nt were generally reduced when wild-type, Δ150–159, Δ936–945, or Δ1047–1056 telomerase activities were assayed in the presence of dideoxy CTP (ddCTP), perhaps as a result of competition for nucleotide binding in the active site (Fig. 1D). However, inclusion of ddCTP did not result in strong pauses indicative of chain termination in the first repeat of products for either wild-type or mutant enzymes (Fig. 1D); quantification of the nucleotide addition processivities of wild-type and variant telomerases in the presence and absence of ddCTP indicated that wild-type and mutant enzyme processivities were similarly affected by inclusion of ddCTP (data not shown). These observations suggested that RID1 and C-terminal variants did not incorporate significantly more ddCTP than wild-type enzyme during reverse transcription of the template itself. We concluded that, although numerous C-terminal mutants exhibited altered patterns of product intensity in the presence of the noncognate nucleotide dCTP, this altered product pattern did not correlate well with the predicted positions of dCTP incorporation resulting from 5′ template boundary read-through in wild-type and 38G44U hTRs. In contrast, the altered mobilities of products at positions 9 and 10 generated by the Δ150–159 RID1 mutant reconstituted with wild-type hTR suggested that this mutation might disrupt 5′ template boundary definition.
TABLE 1.
Summary of dCTP-dependent phenotypes of hTERT N-terminal and C-terminal mutants
| Class of catalytic phenotypea | hTERT variant | Region of mutation | Unlabeled dCTP-dependent changes in pattern of products > +7b | Enhanced incorporation of radiolabeled dCTP at product position 9 (WT hTR)c | Enhanced overall incorporation of dCTPd |
| I | WT | − | − | − | |
| Δ70–79 | N-DAT | − | − | − | |
| Δ110–119 | N-DAT | − | − | − | |
| Δ230–239 | linker | − | − | − | |
| Δ270–279 | linker | − | − | − | |
| Δ310–319 | linker | − | − | − | |
| II | Δ150–159 | RID1 | + | − | − |
| III | Δ936–945 | C terminus | + | + | − |
| Δ963–972 | C terminus | + | + | − | |
| Δ993–1002 | C terminus | + | + | − | |
| Δ1020–1029 | C terminus | + | + | − | |
| Δ1108–1117 | C terminus | + | + | − | |
| IV | Δ1047–1056 | C terminus | + | + | + |
| Δ1077–1086 | C terminus | + | + | + | |
| Δ1123–1132 | C terminus | + | + | + |
aGroup I: No dCTP-dependent phenotype in the presence of unlabeled or radiolabeled dCTP; Group II: dCTP-dependent phenotype in the presence of unlabeled dCTP; phenotype in presence of radiolabeled dCTP unclear due to very weak overall incorporation; Group III: dCTP-dependent phenotype in the presence of unlabeled or radiolabeled dCTP; radiolabeled dCTP incorporation at positions unrelated to 5′ template boundary read-through similar to wild-type enzyme; Group IV: dCTP-dependent phenotype in the presence of unlabeled or radiolabeled dCTP; enhanced dCTP incorporation at positions unrelated to 5′ template boundary read-through compared to wild-type enzyme.
bdCTP-dependent changes in pattern of products longer than 7 nt when assembled with wild-type or 38G44U hTR (comparison of ±dCTP samples); (−) No dCTP-dependent change; (+) dCTP-dependent change. The dCTP-dependent phenotypes of each hTERT variant were examined in at least two independent experiments, with similar results.
cMutants that incorporate more radiolabeled dCTP at position 9 when assembled with wild-type versus 38G44U hTR; (−) Wild-type levels of radiolabeled dCTP incorporation; (+) enhanced radiolabeled dCTP incorporation. The dCTP-dependent phenotypes of hTERT variants were examined in two independent experiments, with similar results.
dMutants in which incorporation of radiolabeled dCTP is enhanced compared to wild-type telomerase at product positions other than position
9. (−) Wild-type levels of radiolabeled dCTP incorporation at positions other than +9; (+) Enhanced incorporation of radiolabeled dCTP at positions other than +9. The dCTP-dependent phenotypes of hTERT variants were examined in two independent experiments, with similar results.
Characterization of radiolabeled dCTP incorporation by hTR P1b mutants
To further characterize the potential role of hTERT in 5′ template boundary definition, we directly examined the ability of hTERT mutants to incorporate radiolabeled dCTP at positions dictated by the appropriate nucleotides in wild-type and 38G44U hTRs (Fig. 2). As positive controls for 5′ template boundary phenotypes, we generated several hTR constructs with mutations that are known to alter 5′ template boundary definition (P1bDown, P1bUp, P1bUp/38G44U; these mutations were previously described as 32–195-m1, 32–195m2, and 32–195-m5b, respectively) (Chen and Greider 2003), and one additional mutant whose 5′ template boundary phenotype is predicted to resemble that of P1bUp/38G44U (P1bDown/38G44U) (see Fig. 2A for a schematic). As previously reported, wild-type hTERT reconstituted with the P1bUp or P1bDown mutants displayed reduced pausing at the +8 product position when assayed in the presence of unlabeled dCTP, dATP, dTTP, and radiolabeled dGTP (Fig. 2B, lanes 3–6; Chen and Greider 2003). Unexpectedly, neither the P1bUp/38G44U nor the P1bDown/38G44U variant displayed the profoundly aberrant product pattern reported for a similar mutation in the upper strand of the P1b helix, and telomerase assembled with P1bUp/38G44U hTR was as active as wild-type enzyme (Fig. 2B, lanes 7–12) (mutant 32–195- m5b; Chen and Greider 2003). It is unclear why our data for the P1b variants differed from the results of Chen and Greider. One possible explanation is that the in vitro reconstitution experiments described by Chen and Greider (2003) employed hTRs in which all nucleotides forming the portions of the P1 helix beyond P1b were removed. In addition, these experiments were performed with mixtures of hTR fragments containing isolated pseudoknot/template and CR4/CR5 domains (Chen and Greider 2003). In contrast, all of the mutations we generated were introduced into full-length, intact hTRs, suggesting that physical separation of catalytically important hTR domains or removal of portions of the P1 helix outside P1b might affect 5′ template boundary definition or other aspects of telomerase function.
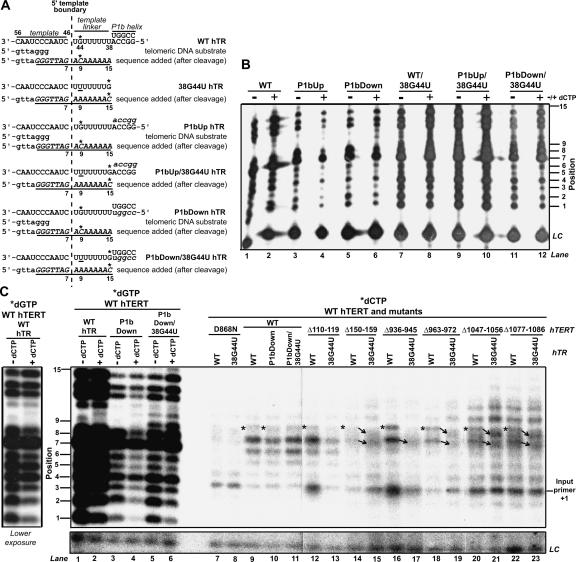
FIGURE 2.
Incorporation of radiolabeled dCTP by hTR P1b and hTERT mutants. (A) Schematic of the positions in wild-type (WT), 38G44U, P1bUp, P1bUp/38G4U, P1bDown, and P1bDown/38G44U hTRs directing incorporation of dCTP (indicated by asterisks). (B) Catalytic phenotypes of hTR P1b mutants reconstituted in RRL with wild-type hTERT. Telomerase assays were performed with radiolabeled dGTP and unlabeled dATP and dTTP in either the presence (+) or absence (−) of unlabeled dCTP. The position numbers of each product are indicated at left. (LC) Loading control. (C) Upper panel: Incorporation of radiolabeled dCTP by hTR mutants assembled with wild-type hTERT and selected hTERT mutants reconstituted with wild-type or 38G44U hTRs. The radiolabeled dCTP incorporation profiles of these and other hTERT mutants not depicted in this figure are summarized in Table 1. The position numbers of each product are indicated at left. Positions at which dCTP should be incorporated following 5′ template boundary read-through in hTERTs assembled with wild-type hTR (position 9) are marked by asterisks. Products whose mobilities were altered when hTERT mutants were reconstituted with wild-type or 38G44U hTRs are indicated by arrows. Telomerase assays performed with a catalytically inactive hTERT RT mutant (D868N) and radiolabeled dCTP indicated that some nonspecific dCTP incorporation occurred at product position 4 (lanes 7,8); nonspecific product patterns generated by D868N telomerase were similar to those observed for hTERT alone (data not shown). This might have been the result of a nonspecific terminal transferase-type activity in RRL, since a product of this size would result if a single labeled nucleotide were added to uncleaved input primer (position indicated by the label “input primer +1”). Control reactions performed with radiolabeled dGTP in the presence (+) or absence (−) of unlabeled dCTP are shown in the first six lanes. A lower exposure of wild-type hTERT samples from lanes 1 and 2 is shown at left to facilitate comparison of product pausing patterns. All reactions were performed in the same experiment and products were electrophoretically separated on the same gel. Lower panel: (LC) loading control. A longer exposure is shown because of weak LC signal in this experiment.
Radiolabeled dCTP incorporation assays were performed using P1bDown hTRs as controls (Fig. 2C). Prominent dCTP-labeled products were observed at positions 7 and 8 for wild-type hTERT telomerases reconstituted with wild-type, P1bDown, or P1bDown/38G44U hTRs (Fig. 2C, lanes 9–11), suggesting that some misincorporation of dCTP occurred during reverse transcription of hTR nucleotides 45U and/or 46C or at the first position of dGTP incorporation following enzyme translocation on the template (see Fig. 2A for a schematic of nucleotides in the template region). However, the intensities of dCTP-labeled products at position 9 were very weak relative to the product intensities at positions 7 and 8 (Fig. 2C, see asterisks in lanes 9,10). This result was unexpected, since wild-type hTERT reconstituted with P1bDown was predicted to specifically incorporate dCTP at this position (encoded by hTR nt 44C in wild-type and P1bDown hTRs), whereas wild-type hTERT assembled with P1bDown/38G44U should exhibit longer dCTP-labeled products (dCTP incorporation directed by hTR nt 38G). dCTP-labeled products were not observed at the predicted position of dCTP incorporation for P1bDown/38G44U telomerase or for any hTERT mutants reconstituted with 38G44U hTR (Fig. 2C, position 15). The earlier report of aberrant 5′ template boundary definition in P1b mutants did not analyze dCTP incorporation using the radiolabeled dCTP method described here (Chen and Greider 2003). We do not know if our failure to detect significant dCTP incorporation at the appropriate position in P1b mutant telomerase was a result of a reduced sensitivity of our assay to 5′ template boundary read-through, to reduced affinity of telomerase for radiolabeled dCTP at the concentration used in this assay, or to differences in the 5′ template boundary definition properties of the P1b mutant analyzed here and in the previous study.
Characterization of radiolabeled dCTP incorporation by hTERT mutants
In contrast to our observations for the hTR P1b variant, we found that numerous hTERT C-terminal mutants reconstituted with wild-type hTR generated a dCTP-labeled product at position 9 that was more intense than the product at the same position in the wild-type and P1bDown controls assembled with wild-type hTERT (Table 1, Group III and IV mutants; for examples, compare position 9 products in lanes 9 and 10, indicated by asterisks, to those in lanes 16,18,20,22 in Fig. 2C). Certain hTERT variants with mutations in the C-terminal half of the C terminus also incorporated more radiolabeled dCTP at positions above and below position 9, suggesting that enzyme fidelity or template usage was also affected in these variants (Table 1, Group IV mutants; for example, in Fig. 2C cf. products above and below positions 7–9 in Δ1047–1056 and Δ1077– 1086 samples to the same products in Δ936–945, Δ150–159, Δ110–119, and wild-type hTERT samples). Since all of the mutants that generated a more intense product at position 9 exhibited greatly reduced activity in radiolabeled dGTP assays compared to wild-type and P1bDown telomerases (Fig. 1C; data not shown; Huard et al. 2003), it is unlikely that the increased intensity of the position 9 product was attributable to enhanced telomerase activity. All of the C-terminal mutants that displayed a more prominent dCTP-labeled product at position 9 also generated a dCTP-dependent product pattern when telomerase activity was tested in the presence of unlabeled dCTP and radiolabeled dGTP (Table 1, Group III and IV mutants; Fig. 1C), supporting the possibility that radiolabeled dCTP incorporation at this position reflected read-through past the 5′ template boundary. The intensities of products at position 9 generated by wild-type hTERT reconstituted with wild-type or P1bDown hTR and N-DAT hTERT mutant telomerases assembled with wild-type hTR (Δ70–79 and Δ110–119) were similar (Fig. 2C, see products marked by asterisks in lanes 9,10,12; Table 1, Group I mutants). Since these N-DAT variants did not exhibit an altered pausing pattern in the presence of unlabeled dCTP (Fig. 1C; Table 1, Group I mutants), we concluded that N-DAT hTERT mutations likely did not impair 5′ template boundary definition and did not otherwise enhance the incorporation of dCTP in products. The overall levels of radiolabeled dCTP incorporated at all positions by a RID1 mutant (Δ150–159)-reconstituted with wild-type hTR were very weak compared to wild-type enzyme and other variants (Fig. 2C, lane 14). It is possible that reduced dCTP incorporation was due to the generally weak activity of this enzyme, since, like the C-terminal mutants, this RID1 variant exhibited a dCTP-dependent altered pausing pattern when its activity was assayed in the presence of unlabeled dCTP (Fig. 1C). However, many C-terminal mutants were as weakly active as the Δ150–159 variant (Fig. 1C, cf. Δ150–159 and Δ936–945), suggesting that reduced activity might not be responsible for its low levels of dCTP incorporation. dCTP-labeled products were also visible at position 9 for RID1 and C-terminal mutants reconstituted with 38G44U hTR (Fig. 2C, see products marked by upper arrows). The mobilities of these products were altered compared to the corresponding products in wild-type hTR samples, implying that the nucleotides incorporated at this position were different for telomerases reconstituted with wild-type and 38G44U hTRs (wild-type and 38G44U hTRs, respectively, encode C and A at this position) (Fig. 2C, cf. products marked by upper arrows in lanes 14–15, 18–19, 20–21, and 22–23). Collectively, these observations suggested that the Δ150–159 and C-terminal hTERT mutations might result in enhanced incorporation of dCTP at positions specified by hTR sequences 5′ of the template.
hTR P1b and template linker sequences regulate 5′ template usage in RID1 and C-terminal hTERT mutants
We also noticed that the mobilities of the dCTP-labeled products at position 8 were slightly altered for the Δ150– 159 mutant and all C-terminal variants, but not for N-DAT mutants or wild-type hTERT enzymes (for example, see products marked by lower arrows in Fig. 2C), suggesting that different nucleotides were also incorporated at this position by hTERT variants assembled with wild-type or 38G44U hTRs. Furthermore, assembly of Δ150–159 hTERT with the 38G44U variant generated an enzyme that exhibited reduced pausing in dGTP labeling assays at the position corresponding to the last G incorporated before the 5′ template boundary (Fig. 1C, cf. position 7 products marked by asterisks in lanes 7,8 and those in lanes 5,6). The altered pausing at position 7 in this hTERT mutant’s products was similar in the presence and absence of unlabeled dCTP, implying that reconstitution with the 38G44U variant affected reverse transcription at the 5′ end of the template in a fashion that was not dependent on 5′ template boundary read-through.
Since we found that the 38G44U substitution affected reverse transcription of the 5′ end of the template in the Δ150–159 variant in a dCTP-independent fashion (Fig. 1C), we decided to investigate the possible role of template linker sequences and the P1b helix in template usage by human telomerase. A subset of mutant hTERTs and wild-type hTERT were expressed in RRL in the presence of hTRs with mutations that altered the sequences of the upper and lower strands of the P1b helix (P1bUp and P1bDown, respectively), and the linker sequences between the P1b helix and the 5′ template boundary (38–45m) (Fig. 3A). A compensatory mutation that altered the sequences of the upper and lower P1b strands but restored base-pairing potential was also constructed (Fig. 3A, P1bComp). hTR variants were assembled with hTERTs bearing mutations in RID1 and at two different positions in the C terminus (Δ150–159, Δ936–945, and Δ1047–1056). These hTERT mutants were chosen because they displayed different types of catalytic defects. The Δ150–159 and Δ1047–1056 variants exhibited defects in repeat addition processivity but not nucleotide addition processivity when assembled with wild-type hTR (Fig. 3B,C; Huard et al. 2003; Moriarty et al. 2004). In contrast, nucleotide addition processivity was impaired in the Δ936–945 mutant reconstituted with wild-type hTR (Fig. 3B,C; Huard et al. 2003). Like a number of other C-terminal mutants, but not Δ936–945, the Δ1047–1056 variant also exhibited enhanced overall incorporation of radiolabeled dCTP compared to RID1 mutants and wild-type hTERT (Fig. 2C). The activities of reconstituted telomerases were examined in the absence of dCTP, to reduce the potential contributions of 5′ template boundary read-through and dCTP incorporation to catalytic phenotypes.
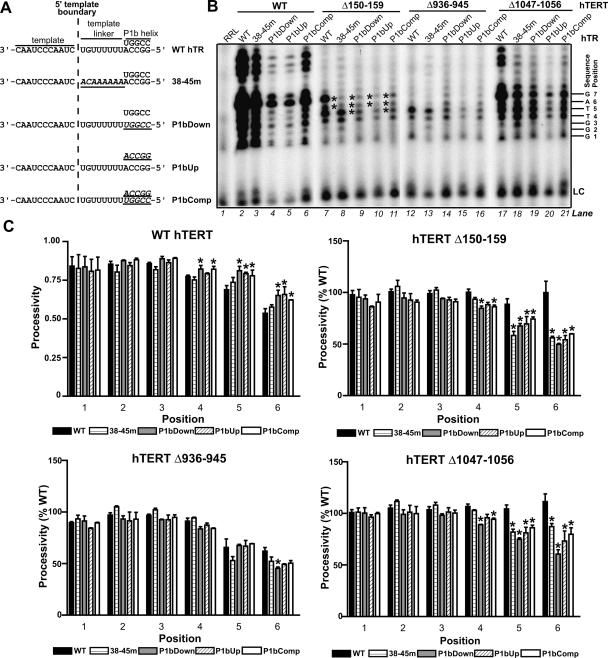
FIGURE 3.
hTR P1b and template linker sequences regulate 5′ template usage in RID1 and C-terminal hTERT mutants. (A) Schematic illustrating substitutions (underlined italicized text) introduced into the indicated hTR mutants. (B) Catalytic phenotypes of hTERT mutants reconstituted with wild-type (WT) hTR and variants and assayed in the absence of dCTP. Asterisks indicate template positions that are reverse transcribed less efficiently when Δ150–159 hTERT is assembled with hTR variants (see C ). The position numbers of each product are indicated at right of the panel. (LC) Loading control. (C ) Nucleotide addition processivities of wild-type hTERT and mutants (as indicated by graph titles) assembled with wild-type hTR and variants (identified in keys), as calculated at each product position. No processivity values are provided for position 7, because position 6 values reflect the processivity between the sixth and seventh product positions (that is, between the last 2 nt reverse transcribed before the 5′ template boundary). The processivity values of each hTERT mutant assembled with wild-type hTR and variants were expressed as a percentage of the processivity values of wild-type hTERT reconstituted with the same hTR (for example, the processivity of Δ150–159 hTERT assembled with 38–45m hTR was expressed relative to the processivity of wild-type hTERT reconstituted with 38–45m hTR). The average of these relative values from two independent experiments is graphically depicted. Asterisks indicate positions at which the mean processivity values for hTERT reconstituted with an hTR variant differed significantly (ρ < 0.05) from the mean processivity value at the same position for the same hTERT assembled with wild-type hTR.
Coexpression of wild-type hTERT with the 38–45m, P1bDown, P1bUp, or P1bComp hTR mutants generated telomerases with a reduced overall activity compared to wild-type hTR (Fig. 3B, lanes 2–6). We quantified the nucleotide addition processivities at each position of the template to permit comparison of the pausing patterns within the first repeat generated by wild-type hTR and variants that generated different levels of overall activity (Fig. 3C; Peng et al. 2001). None of the hTR mutations examined reduced the processivity of wild-type hTERT within the first repeat, though the P1bDown, P1bUp, and P1bComp mutations caused a small but significant increase in nucleotide addition processivity at the 5′ end of the template compared to wild-type hTR (ρ < 0.05) (Fig. 3C). In contrast, Δ150–159 hTERT reverse transcribed the last few nucleotides of the template very inefficiently when reconstituted with hTRs 38–45m, P1bDown, and P1bUp (Fig. 3B, lanes 7–10, see products at positions marked by asterisks; Fig. 3C). These observations implied that both the P1b helix and template linker sequences influenced 5′ template usage in this mutant. Reconstitution of Δ150–159 telomerase with the P1bComp variant only slightly enhanced reverse transcription of the last few nucleotides (Fig. 3B, lane 11; Fig. 3C), indicating that both the sequence and structure of the P1b helix affected 5′ template usage. All Δ936–945 telomerases, including those assembled with wild-type hTR, displayed impaired nucleotide addition processivity at 5′ positions in the template compared to wild-type hTERT; however, statistically significant differences between wild-type hTR and variants were observed only for the P1bDown mutant (Fig. 3C). The Δ1047–1056 telomerases reconstituted with hTR variants also reverse transcribed the last few nucleotides of the template less efficiently than the same hTERT variant reconstituted with wild-type hTR, but this defect was less pronounced than the 5′ template usage impairment of Δ150–159 telomerases (Fig. 3B,C). Together these observations indicated that sequences in the P1b helix and sequences linking the 5′ template boundary and P1b helix could influence 5′ template usage.
Examination of hTERT–hTR interactions in mutant telomerases with altered 5′ template usage phenotypes
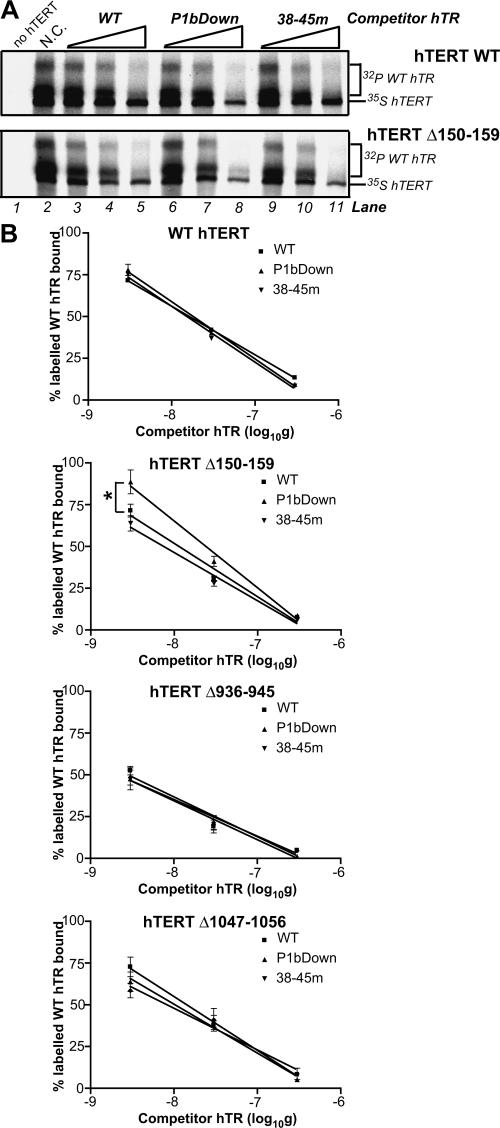
To determine whether altered 5′ template usage and incorporation of the noncognate nucleotide dCTP were attributable to altered hTERT–hTR interactions, we investigated the in vitro interactions of a subset of hTR variants with wild-type hTERT and hTERT mutants that exhibited these catalytic phenotypes (Fig. 4). Using a competitive, quantitative RNA-binding method, we found that wild-type hTERT associated equally well with wild-type hTR and the P1bDown and 38–45m variants, implying that these hTR regions are not involved in hTERT interactions or that such interactions could not be detected by this RNA-binding technique (Fig. 4B). However, all hTERT mutants exhibited altered hTR interactions compared to wild-type hTERT (Fig. 4B). The C-terminal Δ936–945 mutant that displayed reduced nucleotide addition processivity when reconstituted with either wild-type hTR or variants (Fig. 3) consistently associated more efficiently with all hTRs than wild-type hTERT (Fig. 4B, 25% greater competition efficiency at lowest concentration of competitors). As for wild-type hTERT, hTR interactions with this C-terminal hTERT variant were identical in the presence of all competitors (Fig. 4B), suggesting that hTERT residues 936–945 did not affect hTR association in a P1b-dependent fashion. hTERT mutants Δ150–159 and Δ1047–1056 exhibited a more variable association with the P1bDown and 38–45m hTRs than wild-type hTERT, especially at lower concentrations of competitor (Fig. 4B, compare WT, Δ150–159, and Δ1047– 1056 error bars and distances between data points for WT, P1bDown, and 38–45m hTRs). This suggested that their interactions with hTR variants might be less stable. However, in most cases differences in hTERT-hTR association values were not statistically significant. The only exception was the Δ150–159 mutant, which displayed significantly reduced interactions with the P1bDown hTR compared to wild-type hTR (Fig. 4B). The Δ150–159 hTERT variant exhibited more pronounced impairment of 5′ template usage and processivity when reconstituted with the P1b and 38–45m mutants compared to Δ1047–1056 (Fig. 3). This observation suggested the possibility that smaller P1b association defects that were not detectable in these assays might also be present in the less-affected Δ1047–1056 mutant. However, the Δ150–159 mutant did not display significant defects in association with hTR 38–45m (Fig. 4B). Thus, these data did not support a general correlation between hTERT–hTR interactions and 5′ template usage and processivity phenotypes in the Δ150–159 and Δ1047– 1056 mutants reconstituted with P1bDown and 38–45m variants.
FIGURE 4.
hTERT-hTR interactions in mutant telomerases with altered 5′ template usage phenotypes. (A) Representative immunoprecipitation- based competitive hTR–hTERT interaction assays performed with wild-type (WT) hTERT and the Δ150–159 hTERT mutant. The positions of hTR and hTERT are indicated at the right of each panel. Increasing concentrations of the indicated unlabeled hTRs (lanes 3–11) were added to RRL reconstitution mixtures containing identical amounts of 32P-labeled wild-type hTR. This approach ensured that each sample contained exactly the same amount of labeled input hTR and eliminated the large experimental variation that we frequently observed when different hTR variants were individually labeled and immunoprecipitated in the absence of competitors (data not shown). ′ Immunoprecipitations were performed with an antibody against hTERT. Control reactions performed in the absence of competitor (N.C.) and hTERT are shown in lanes 1 and 2. (B) Quantification of the percent labeled wild-type hTR bound after reconstitution and immunoprecipitation in the presence of increasing concentrations of unlabeled competitors (identified in keys). The wild-type hTR signal intensities for competition samples were expressed as a percentage of the wild-type hTR signal in control reactions performed in the absence of competitor (100%).
DISCUSSION
Our initial aim was to investigate hTERT sequences that might be involved in 5′ template boundary regulation in human telomerase. We identified mutations in the hTERT RID1 domain and C terminus that resulted in dCTP-dependent catalytic phenotypes (Table 1; Figs. 1, 2). A number of RID1 and C-terminal mutations also affected efficient reverse transcription of the 5′ end of the template when hTERT variants were reconstituted with hTRs containing substitutions in the P1b helix and template-linking sequences (Fig. 3). These observations are consistent with previous studies of S. cerevisiae and Tetrahymena TRs, which found that 5′ template boundary-defining elements and template-linking sequences also regulate template usage and processivity (Prescott and Blackburn 1997; Lai et al. 2002; Miller and Collins 2002; Seto et al. 2003). The Tetrahymena- specific TERT CP2 motif that regulates 5′ template- boundary definition through interactions with the 5′ template boundary-regulating element of the TR is also important for template usage, though CP2 regulates reverse transcription of the 3′ end of the template (Miller et al. 2000). Interestingly, the CP2 motif is located close to the Tetrahymena TERT sequences that correspond to hTERT RID1 (Miller et al. 2000; Xia et al. 2000; Moriarty et al. 2002). Our observation that the hTERT C terminus is important for 5′ template usage is similar to the results of previous studies indicating that the C terminus of Tetrahymena and S. cerevisiae TERTs regulates 5′ template usage and nucleotide addition processivity, respectively, though it is unknown whether the C terminus is important for suppressing incorporation of noncognate nucleotides in these enzymes (Miller et al. 2000; Peng et al. 2001; Hossain et al. 2002). Similarly, it is not known whether yeast and ciliate TERT sequences that correspond to hTERT RID1 function in template usage or 5′ template boundary definition.
P1b, hTERT RID1, and hTERT C-terminal functions in the context of the wild-type enzyme
dCTP-dependent catalytic phenotypes suggestive of 5′ template boundary bypass were observed for a P1b mutant assembled with wild-type hTERT (using unlabeled dCTP) and for RID1 and C-terminal variants reconstituted with wild-type hTR (Figs. 1, 2). However, the impaired 5′ template usage and processivity defects of certain RID1 and C-terminal mutants reconstituted with different P1b and template linker variants were not observed when wild-type hTERT was assembled with P1b and template linker mutants, nor when certain hTERT variants were reconstituted with wild-type hTR (Fig. 3). In contrast, wild-type hTERT assembled with P1b variants demonstrated increased nucleotide addition processivity at the 5′ end of the template (Fig. 3). The latter observation is consistent with the hypothesis that 5′ template boundary-regulating elements such as P1b constrain the movement of the template in the active site and with the results of a previous report indicating that disruption of the 5′ boundary-defining helix in the S. cerevisiae TR enhances processivity (Chen and Greider 2003; Seto et al. 2003). Thus, our finding that some RID1 and C-terminal hTERT mutations impair reverse transcription of the 5′ end of the template when P1b is disrupted suggests that RID1 and the C terminus may contribute to the enhanced 5′ template processivity of wild-type hTERT assembled with P1b mutants. RID1 and C-terminal sequences might promote processivity by unwinding the RNA/DNA duplex formed during reverse transcription, by regulating the movement of the P1b helix, template, or DNA substrate with respect to one another, by modulating the conformation or function of other parts of hTR, or by promoting the affinity of telomerase for DNA primer and oligonucleotide substrates.
Role of hTERT–hTR interactions in 5′ template boundary definition and 5′ template usage
We showed previously that RID1 interacts with the hTR pseudoknot/template domain (Moriarty et al. 2004), and the results presented here imply that one of the sites of RID1-pseudoknot/template domain interactions may be the P1b helix (Fig. 4). This suggests that the dCTP-dependent catalytic phenotype and pronounced 5′ template usage defect of the RID1 mutant might partly be due to altered interactions with the 5′ template boundary-regulating P1b helix. In Tetrahymena telomerase, the 5′ template boundary- regulating TR element interacts with a TERT RNA-binding domain that contains sequences corresponding to the hTERT RID2 and linker, as well as a Tetrahymena-specific motif adjacent to sequences that correspond to hTERT RID1 (Lai et al. 2001; O’Connor et al. 2005). These data suggest that TERT interactions with 5′ template boundary-defining elements in Tetrahymena and human telomerases may be quite different. The catalytic functions of the RID1 region in Tetrahymena and human telomerases may also be distinct, as deletion of hTERT RID1 eliminates repeat addition processivity, but not basic catalytic function, whereas deletion of tTERT sequences corresponding to hTERT RID1 entirely abolishes catalytic activity (Lai et al. 2001; Moriarty et al. 2004).
In contrast, our data implied that the hTERT C terminus regulates 5′ template usage and reverse transcription of hTR sequences 5′ of the template by mechanisms other than direct interaction with P1b (Fig. 4). Interestingly, the C-terminal mutant (Δ936–945) that exhibited reduced nucleotide addition processivity when assembled with either wild-type hTR or variants also associated more efficiently with all hTRs examined, suggesting the possibility that the generalized processivity defect of this mutant might be the result of enhanced affinity for hTR (Figs. 3, 4). This observation is consistent with the results of an earlier study in which hTERT C-terminal truncation mutants were found to associate more efficiently with hTR than full-length hTERT (Beattie et al. 2000). The S. cerevisiae TERT C terminus has also been implicated in interactions with the TR and RNA/DNA hybrids (Hossain et al. 2002). Since the hTERT C terminus interacts with RID1 in vitro and influences many of the same catalytic properties as RID1 (for example, repeat addition processivity, affinity for DNA substrates, and 5′ template usage) (Huard et al. 2003; Lee et al. 2003; Moriarty et al. 2004, 2005; this study), one possibility is that it indirectly influences P1b function by modulating the activity or conformation of RID1; this hypothesis is supported by previous observations that the hTERT C terminus can function in trans with respect to RID1 in complementation assays (Beattie et al. 2001; Moriarty et al. 2004). As the hTR interaction and catalytic phenotypes of the Δ936–945 and Δ1047–1056 C-terminal mutants were distinct from each other, not all regions of the C terminus might be involved in such allosteric regulation. The C terminus could also directly or indirectly alter the mobile association of hTERT with hTR by influencing the conformation or dimerization state of the hTR pseudoknot, which has been proposed as the site of a molecular switch that regulates some of the unique aspects of telomerase catalytic function (Comolli et al. 2002; Ly et al. 2003; Theimer et al. 2003, 2005; Moriarty et al. 2004).
We also found that an hTR template linker substitution that altered 5′ template usage and processivity when assembled with hTERT RID1 and C-terminal mutants did not affect interactions with wild-type or mutant hTERTs (Fig. 4). This observation implied that factors other than direct hTERT interactions with linker sequences may affect their ability to regulate these catalytic functions. Since the length of the template linker was not altered in either the 38G44U or 38–45m linker variants examined in this study, we concluded that the function of this region is likely both sequence and length dependent (Chen and Greider 2003; this study). This conclusion suggested that sequence-specific association of linker sequences with other hTR regions such as the P6.1 helix (Ueda and Roberts 2004) or other hTR molecules might affect the catalytic role of template linker sequences when the functions of the hTERT C terminus and RID1 are impaired.
Do RID1 and the C terminus regulate 5′ template boundary definition?
We found that the hTR P1bDown mutant and a RID1 hTERT variant exhibited altered product patterns at positions dictated by 5′ template boundary read-through when telomerase activity was examined in the presence of unlabeled dCTP, whereas the dCTP-dependent product patterns generated by C-terminal mutants under the same conditions were more difficult to interpret (Figs. 1, 2); it is possible that the complexity of these product patterns might have been the result of altered template usage. In contrast, incorporation of radiolabeled dCTP at positions corresponding to 5′ template boundary read-through was more readily detected for C-terminal variants than for the RID1 mutant or wild-type hTERT reconstituted with P1bDown hTR (Fig. 2; Table 1). Our observations suggest that interaction of RID1 with P1b might contribute to 5′ template boundary definition, perhaps by sterically hindering read-through past the 5′ template boundary. However, this model may not be true for the C terminus, which did not influence hTR interactions in a P1b-dependent fashion. One interesting alternative hypothesis is that the C terminus does not regulate 5′ template boundary definition per se, but could instead regulate the removal of noncognate nucleotides incorporated as a result of 5′ template boundary bypass. A template-dependent nuclease activity that removes nucleotides from DNA substrates is tightly associated with human and other telomerases, though the catalytic site of nuclease function has not yet been identified in any telomerase component (Huard and Autexier 2004; Oulton and Harrington 2004). Impairment of a potential hTERT-dependent proofreading activity might explain why the weakly active C-terminal mutants incorporated significantly more radiolabeled dCTP at positions directed by hTR sequences 5′ of the template compared to wild-type, P1bDown, and RID1 mutant telomerases. If wild-type hTERT efficiently removes noncognate nucleotides from products, then the apparent failure of P1b mutants to incorporate significant amount of radiolabeled dCTP might have resulted from removal of this nucleotide by an hTERT-dependent proofreading function. In contrast, in assays performed with radiolabeled dGTP and high concentrations of unlabeled dCTP, removal of incorporated dCTP residues may have been less efficient, resulting in the dCTP-dependent pausing phenotype observed here and previously for the P1b mutant (Chen and Greider 2003).
Conclusions
Our results implicate the hTR P1b helix and template linker sequences, and hTERT RID1 and C-terminal sequences in 5′ template usage and incorporation of noncognate nucleotides specified by hTR nucleotides 5′ of the template boundary. Although RID1 may contribute to 5′ template boundary definition and 5′ template usage through interactions with the P1b helix, the roles of the hTERT C terminus and template linker sequences in these catalytic functions remain unclear.
MATERIALS AND METHODS
Plasmid construction
All pet-hTERT constructs used in this study for in vitro reconstitution of human telomerase were described previously (Bachand and Autexier 1999; Moriarty et al. 2002; Huard et al. 2003). The construction of the phTR + 1 plasmid used to generate in vitro transcribed hTR has been described (Autexier et al. 1996). Plasmids for expression of hTR substitution mutants described in this study (38G44U, 38–45m, P1bDown, P1bUp, P1bComp, P1bDown/ 38G44U, P1bUp/38G44U) were generated by site-directed mutagenesis of phTR + 1. All hTR constructs were confirmed by sequencing.
Telomerase reconstitution by in vitro transcription/translation
Reconstitution in rabbit reticulocyte lysates (RRL) (Promega) was performed as described (Moriarty et al. 2002).
Direct primer extension telomerase assays
Direct primer extension assays were performed as described, using 2.5 μM biotinylated (TTAGGG)3 DNA primer (Huard et al. 2003). Radiolabeled dGTP assays were performed using 1 mM unlabeled dATP, dTTP, and dCTP or ddCTP (where applicable), 2.5 μM unlabeled dGTP, and 1.25 μM 800 Ci/mmol α32P dGTP (ICN/MP Biomedical). Radiolabeled dCTP assays were performed using 1mM unlabeled dATP, dGTP, and dTTP and 0.33 μM 3000 Ci/mmol α32P dCTP (ICN/MP Biomedical). All direct primer extension assays performed with Δ150–159 and C-terminal hTERT mutants were scaled up twofold with respect to wild-type hTERT, DAT mutant, and linker mutant samples to facilitate detection of signal.
Quantification of nucleotide addition processivity
Nucleotide addition processivity was quantified as described (Peng et al. 2001) and was calculated only for the first repeat of telomerase products. Statistical significance calculations were performed using a two-tailed paired Student’s t-test to detect differences between the means of wild-type and mutant samples (Excel). Standard error values and graphical plotting of values were performed using Graph- Pad Prism. Experiments in which nucleotide addition processivity values were calculated were performed independently at least twice.
In vitro RNA binding competition assays
RNA binding assays were performed as described (Moriarty et al. 2004), except that 5.4 μg/mL αhTERT antibody (Moriarty et al. 2002) were used. Unlabeled hTR competitors (3 ng, 30 ng, or 300 ng) were added to reconstitution mixtures simultaneously with 32P-labeled wild-type hTR. hTR binding efficiency was quantified as previously described (Moriarty et al. 2002), calculations of statistical significance in a two-tailed paired Student’s t-test and standard error values were performed using Excel and GraphPad Prism, respectively, and graphical plotting of values was performed using GraphPad Prism. Experiments in which hTR binding was quantified were repeated at least three times.
Acknowledgments
We thank Autexier laboratory members for critical discussion of data and A. Dulude and Y. Sun for generating hTR constructs. This work was funded by CIHR grant MOP68844 to C.A., a CIHR doctoral research scholarship and McGill Graduate Studies Fellowship award to T.J.M., and Boehringer-Ingelheim (Canada) Young Investigator and FRSQ Chercheur-Boursier awards to C.A.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2910105.
REFERENCES
- Antal, M., Boros, E., Solymosy, F., and Kiss, T. 2002. Analysis of the structure of human telomerase RNA in vivo. Nucleic Acids Res. 30: 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai, K., Masutomi, K., Khurts, S., Kaneko, S., Kobayashi, K., and Murakami, S. 2002. Two independent regions of human telomerase reverse transcriptase are important for its oligomerization and telomerase activity. J. Biol. Chem. 277: 8538–8544. [DOI] [PubMed] [Google Scholar]
- Armbruster, B., Banik, S., Guo, C., Smith, A., and Counter, C. 2001. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 21: 7775–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier, C. and Greider, C.W. 1995. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes & Dev. 15: 2227–2239. [DOI] [PubMed] [Google Scholar]
- Autexier, C., Pruzan, R., Funk, W.D., and Greider, C.W. 1996. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 15: 5928–5935. [PMC free article] [PubMed] [Google Scholar]
- Bachand, F. and Autexier, C. 1999. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem. 274: 38027–38031. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA–protein interactions. Mol. Cell. Biol. 21: 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banik, S.S.R., Guo, C., Smith, A.C., Margolis, S.S., Richardson, D.A., Tirado, C.A., and Counter, C.M. 2002. C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol. Cell. Biol. 22: 6234–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie, T., Zhou, W., Robinson, M., and Harrington, L. 2000. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11: 3329–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 21: 6151–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.-L. and Greider, C.W. 2003. Template boundary definition in mammalian telomerase. Genes & Dev. 17: 2747–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli, L.R., Smirnov, I., Xu, L., Blackburn, E.H., and James, T.L. 2002. A molecular switch underlies a human telomerase disease. Proc. Natl. Acad. Sci. 99: 16998–17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L. 2003. Biochemical aspects of telomerase function. Cancer Lett. 194: 139–154. [DOI] [PubMed] [Google Scholar]
- Hossain, S., Singh, S.M., and Lue, N.F. 2002. Functional analysis of the C-terminal extension of telomerase reverse transcriptase: a ‘putative’ thumb domain. J. Biol. Chem. 277: 36174–36180. [DOI] [PubMed] [Google Scholar]
- Huard, S. and Autexier, C. 2004. Human telomerase catalyzes nucleolytic primer cleavage. Nucleic Acids Res. 32: 2171–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard, S., Moriarty, T.J., and Autexier, C. 2003. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 31: 4059–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler, B.R. and Jarstfer, M.B. 2004. Inhibition of telomerase activity by preventing proper assemblage. Biochemistry 43: 334–343. [DOI] [PubMed] [Google Scholar]
- Lai, C., Mitchell, J., and Collins, K. 2001. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21: 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C.K., Miller, M.C., and Collins, K. 2002. Template boundary definition in Tetrahymena telomerase. Genes & Dev. 16: 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.R., Wong, M.Y., and Collins, K. 2003. Human telomerase reverse transcriptase motifs required for elongation of a telomeric substrate. J. Biol. Chem. 278: 52531–52536. [DOI] [PubMed] [Google Scholar]
- Lin, J., Ly, H., Hussain, A., Abraham, M., Pearl, S., Tzfati, Y., Parslow, T.G., and Blackburn, E.H. 2004. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl. Acad. Sci. 101: 14713–14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue, N.F. 2005. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. (in press). [DOI] [PMC free article] [PubMed]
- Ly, H., Xu, L., Rivera, M.A., Parslow, T.G., and Blackburn, E.H. 2003. A role for a novel ‘trans-pseudoknot’ RNA–RNA interaction in the functional dimerization of human telomerase. Genes&Dev. 17: 1078–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.C. and Collins, K. 2002. Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl. Acad. Sci. 99: 6585– 6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M., Liu, J., and Collins, K. 2000. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 19: 4412– 4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J. and Collins, K. 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell 6: 361–371. [DOI] [PubMed] [Google Scholar]
- Moriarty, T.J., Huard, S., Dupuis, S., and Autexier, C. 2002. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 22: 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty, T.J., Marie-Egyptienne, D.T., and Autexier, C. 2004. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 24: 3720–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty, T.J., Ward, R.J., Taboski, M.A.S., and Autexier, C. 2005. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell 16: 3152–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor, C.M., Lai, C.K., and Collins, K. 2005. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 280: 17533–17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulton, R. and Harrington, L. 2004. A human telomerase-associated nuclease. Mol. Biol. Cell 15: 3244–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y., Mian, I., and Lue, N. 2001. Analysis of telomerase processivity: Mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell 7: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Prescott, J. and Blackburn, E.H. 1997. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal non-processivity in vivo and in vitro. Genes & Dev. 11: 528–540. [DOI] [PubMed] [Google Scholar]
- Seto, A.G., Umansky, K., Tzfati, Y., Zang, A.J., Blackburn, E.H., and Cech, T.R. 2003. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 9: 1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, C.A., Finger, L.D., Trantirek, L., and Feigon, J. 2003. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. 100: 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, C.A., Blois, C.A., and Feigon, J. 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell 17: 671–682. [DOI] [PubMed] [Google Scholar]
- Tzfati, Y., Fulton, T., Roy, J., and Blackburn, E. 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288: 863–867. [DOI] [PubMed] [Google Scholar]
- Ueda, C.T. and Roberts, R.W. 2004. Analysis of a long-range interaction between conserved domains of human telomerase RNA. RNA 10: 139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, J., Peng, Y., Mian, I., and Lue, N. 2000. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 20: 5196–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]