Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate gene expression in both plants and animals. miRNA genes have been implicated in a variety of important biological processes, including development, differentiation, apoptosis, fat metabolism, viral infection, and cancer. Similar to protein-coding messenger RNAs, miRNA expression varies between tissues and developmental states. To acquire a better understanding of global miRNA expression in tissues and cells, we have developed isolation, labeling, and array procedures to measure the relative abundance of all of the known human mature miRNAs. The method relies on rapid isolation of RNA species smaller than ~40 nucleotides (nt), direct and homogenous enzymatic labeling of the mature miRNAs with amine modified ribonucleotides, and hybridization to antisense DNA oligonucleotide probes. A thorough performance study showed that this miRNA microarray system can detect subfemtomole amounts of individual miRNAs from <1 μg of total RNA, with 98% correlation between independent replicates. The system has been applied to compare the global miRNA expression profiles in 26 different normal human tissues. This comprehensive analysis identified miRNAs that are preferentially expressed in one or a few related tissues and revealed that human adult tissues have unique miRNA profiles. This implicates miRNAs as important components of tissue development and differentiation. Taken together, these results emphasize the immense potential of microarrays for sensitive and high-throughput analysis of miRNA expression in normal and disease states.
Keywords: microRNA, miRNA, gene expression profile, oligonucleotide microarray, poly(A) polymerase
INTRODUCTION
MicroRNAs (miRNAs) are small (~22-nucleotide) RNAs that are encoded in the genomes of animals, plants, and viruses (Ambros 2004; Bartel and Chen 2004; Pfeffer et al. 2004). The short, single-stranded RNAs are generated from longer primary miRNA transcripts (pri-miRNAs) by a stepwise process (Lee et al. 2002; Murchison and Hannon 2004). The first step in miRNA biogenesis takes place in a multiprotein nuclear complex, termed the Microprocessor (Denli et al. 2004; Gregory et al. 2004). There, pri-miRNAs are cleaved by an RNase III family nuclease, Drosha, to generate precursor miRNAs (pre-miRNAs) (Lee et al. 2003; Zeng et al. 2004). The pre-miRNAs, which are predicted to form hairpins of 70–125 nt with a 2-nt 3′ overhang, are exported to the cytoplasm by a RanGTP/exportin 5-dependent mechanism (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004; Zeng and Cullen 2004). In the cytoplasm, pre-miRNAs are recognized and processed by a second RNase III-family enzyme called Dicer to generate ~22-bp RNA duplexes with 2-nt 3′ overhangs (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Zhang et al. 2004). Mature miRNAs enter RISC (RNA-induced silencing complex), where one strand of the RNA duplex will guide RISC to induce endonucleolytic cleavage or translational repression of specific target messenger RNAs (Olsen and Ambros 1999; Llave et al. 2002; Zeng et al. 2002; Doench et al. 2003; Schwarz et al. 2003; Yekta et al. 2004).
Biochemical and bioinformatics approaches have identified several hundred genes encoding miRNAs in plants, Caenorhabditis elegans, Drosophila, and mammals. As with protein-coding genes, a key to understanding how miRNAs are functioning is to determine when and where they are expressed. Northern blots are commonly used for miRNA analysis, and interesting developmental or tissue-specific miRNA expression patterns have been identified (Lagos-Quintana et al. 2002; Houbaviy et al. 2003; Lim et al. 2003; Sempere et al. 2004). A variety of improvements or adaptation to existing technologies have also been tailored to small RNA detection. These include locked nucleic-acidmodified probes for Northern blot (Valoczi et al. 2004), oligonucleotide filter macroarrays (Krichevsky et al. 2003; Sioud and Rosok 2004), RNA oligonucleotide ligation followed by RT-PCR amplification (Grad et al. 2003), fluorescence resonance energy transfer (Allawi et al. 2004), signal-amplifying ribozymes (Hartig et al. 2004), primer extension (Zeng and Cullen 2003), nuclease protection (Overhoff et al. 2004), or direct real-time PCR amplification for detection of precursor miRNAs (Schmittgen et al. 2004). In addition, a number of microarray-based methods have recently been developed (Babak et al. 2004; Barad et al. 2004; Liu et al. 2004; Miska et al. 2004; Nelson et al. 2004; Sun et al. 2004; Thomson et al. 2004; Baskerville and Bartel 2005).
Conceptually, microarrays offer an ideal method for high-throughput, multiplex miRNA gene expression analyses in tissues and cells. However, miRNAs present unique challenges that make them more difficult to analyze than mRNAs. The inherent small size of miRNAs provides very little sequence for appending label or for designing probes. Furthermore, miRNAs represent only a small fraction (~0.01%) of the mass of a total RNA sample, making it important to label them with the highest specific activity possible. Finally, miRNAs exist in three forms, short mature miRNA, hairpin pre-miRNA, and long pri-miRNA (Lee et al. 2003, 2004; Cai et al. 2004; Rodriguez et al. 2004). Since only the mature miRNA is active, it is important to eliminate array signal from the pre-miRNAs and pri-miRNAs. These factors conspire to make it difficult to detect miRNAs using standard array procedures and are not completely addressed by the procedures that have thus far been developed for miRNA array analysis. To eliminate the potential bias associated with random labeling or priming (Babak et al. 2004; Liu et al. 2004; Sun et al. 2004), detection of pre-miRNA and pri-miRNA sequences (Babak et al. 2004; Liu et al. 2004; Nelson et al. 2004; Sun et al. 2004), and PCR-based amplification or ligation strategies (Barad et al. 2004; Miska et al. 2004; Thomson et al. 2004; Baskerville and Bartel 2005), we have developed a method allowing direct labeling and hybridization of the mature and active miRNAs. The microarray system was evaluated using both artificial and real samples to determine its sensitivity, dynamic range, accuracy, and reproducibility. Finally, we used the system to analyze 26 human tissue samples, which enabled classification of normal tissues based on their miRNA expression profiles.
RESULTS
Microarray strategy, design, and optimization
Labeling a total RNA sample or fractions enriched in small RNAs results in labeled mature and precursor miRNAs. Because very little is known about how mature and precursor miRNAs accumulate in cells and tissues, we chose to purify the active, mature miRNAs from samples so that we could exclude any artifacts that might be introduced in our array experiments by including the inactive precursors of the miRNAs. Routinely, purification of nucleic acid species by size requires separating the samples on electrophoretic gels, followed by localization of the samples on the gel, excision of the region of interest, and elution of the nucleic acid from the gel pieces. To minimize variability, and the time and effort required to isolate miRNAs from a total RNA sample, we developed a gel electrophoresis device that can recover mature miRNAs in <15 min without the need to excise and extract miRNAs from the gel matrix (see Materials and Methods). Efficient and reproducible miRNA recovery was demonstrated with 5′ radiolabeled synthetic miRNA oligonucleotides spiked in total RNA samples, and by direct analysis of natural miRNAs isolated from different input amounts of total RNA (see Supplemental Fig. 1).
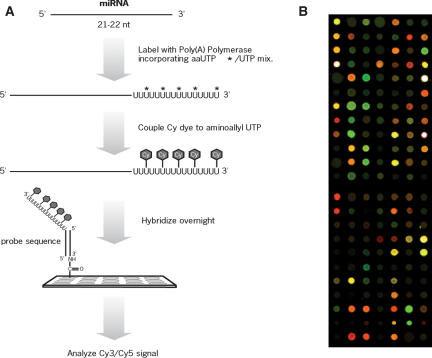
FIGURE 1.
Overview of the miRNA microarray procedure. RNA species smaller than ~40 nt are purified by a rapid electrophoretic gel fractionation method. miRNAs are 3′-end labeled with poly(A) polymerase, amine-modified nucleotides, and amine-reactive dyes. The fluorescently labeled miRNAs are hybridized to a glass slide on which miRNA-specific DNA oligonucleotide probes have been arrayed. Microarrays are processed and analyzed using standard equipment, scanner, and software. A representative result from a scanned microarray is shown in B.
To maximize the label from each miRNA without jeopardizing its capacity to hybridize to probes on a planar array, we next developed a tailing/labeling procedure. After evaluation of different template-independent polymerases and optimization of various reaction parameters, such as buffer composition, incubation time, enzyme and nucleotide type, or relative reagents concentration, we found that polynucleotide tails 20–50 nt long can reproducibly be appended to the 3′ ends of all miRNAs by the poly(A) polymerase (PAP) enzyme (data not shown). The 3′ tail is a mixture of standard and amine-modified nucleotides, and tailed miRNAs can subsequently be labeled with any mono-reactive NHS-ester dyes, such as Cy3 and Cy5 (Fig. 1). Using an optimized set of reaction conditions and ratio of modified to unmodified nucleotides improve tailing efficiency and reproducibility and reduce the fluorescence quenching that occurs when dye molecules are located on adjacent or nearby nucleotides (data not shown).
To optimize the miRNA probe design, fluorescently labeled miRNAs from human thymus, lung, and brain were hybridized to series of immobilized oligonucleotide probes with regions complementary to eight different miRNAs and various 5′ and 3′ linker lengths and sequences connecting to the attachment moiety. A matrix of hybridization and washing time, temperature, and buffer compositions was used to identify the probe design and protocol that (1) maximized signal without creating significant signal from negative control elements spotted on the same microarrays and (2) provided relative miRNA abundance data congruent to Northern blot data (data not shown). This optimal probe design and array protocol (Fig. 1; see also Materials and Methods) was used for all subsequent studies.
Microarray performances
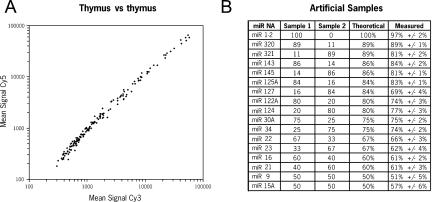
The overall performance of this new microarray system was evaluated by measuring several key parameters such as consistency, accuracy/precision, sensitivity/limit of detection, and robustness/reproducibility. First, we compared two miRNA populations independently purified from the same tissue and labeled either with Cy5 or Cy3 (self vs. self analysis). Analysis of the mean signal in each channel showed equivalent labeling between both dyes (R2 = 0.992) and consistency of the intensities (99.6% correlation) (Fig. 2A). Although microarrays are generally used to measure relative expression levels based on a given reference, we next wanted to determine how quantitative the miRNA microarray system was. Two artificial populations of 17 synthetic miRNAs mixed at different ratios were created, labeled with Cy5 or Cy3, and hybridized on the same array in triplicate (Fig. 2B). The Log2(Normalized Ratio) was measured for each miRNA feature and was used to calculate the relative representation of each miRNA in the two populations. The correlation between the theoretical and measured values was 84%. The average difference between the theoretical and measured values was 3.5%with a maximum of 15%, indicating that the microarray system can accurately report variations greater than ~20%.
FIGURE 2.
miRNA microarray accuracy and precision. (A) Self versus self analysis. A human thymus total RNA sample (20 μg) was split in half and miRNAs were independently gel purified, labeled either with Cy3 or Cy5 dyes, and hybridized to the same array carrying 162 miRNA-specific probes. The graph shows the mean signal intensities at 532 nm (Cy3) and 635 nm (Cy5) for each miRNA probe. (B) Analysis of artificial samples. Chemically synthesized RNA oligonucleotides corresponding to 17 different mature miRNA sequences were mixed as indicated (amounts are in picograms) to create two artificial miRNA populations. Each sample was labeled with Cy3 or Cy5 and hybridized to the same array. The measured percentage was calculated from the normalized Cy3/Cy5 or Cy5/Cy3 ratio from triplicate array analyses.
We next determined the sensitivity of the array system by spiking various amounts of a synthetic miRNA into a sample that lacked this miRNA. Previous reports indicate that miR-124 is specific to brain tissue (Lagos-Quintana et al. 2002). Thus, 0.14–140 fmol of synthetic miR-124 were spiked in a miRNA fraction purified from human liver (labeled with Cy5), and these samples were compared to the normal liver miRNA population (labeled with Cy3). Analysis of the mean intensities and the Log2(Normalized Ratio) of the miR-124 signal indicated that 0.42 fmol of miR-124 provides signal that is 5.8-fold above the normal liver signal, and that even 0.14 fmol of miR-124 provides signal that can be clearly distinguished from the background signal (Table 1). This level of sensitivity allows detection of miRNAs that represent as little as 0.1% of the overall miRNA population in a 10-μg total RNA sample.
TABLE 1.
miRNA microarray sensitivity
| fmol miR-124a | Mean Cy5 signal | Mean Cy5 background | Signal/background | Log2 (Cy5/Cy3)b |
| 0.14 | 213 | 55 | 3.87 | 1.90 |
| 0.42 | 403 | 65 | 6.20 | 2.53 |
| 1.41 | 796 | 53 | 15.02 | 3.61 |
| 42.4 | 2877 | 73 | 39.41 | 3.98 |
| 141 | 19245 | 55 | 349.91 | 7.94 |
aThe indicated amounts of synthetic miR-124 oligonucleotide were spiked in miRNA fractions isolated from 10 μg of human liver total RNA and subjected to the miRNA microarray process.
b(Cy3) Human liver miRNAs. (Cy5) Human liver miRNAs plus spiked miR-124 oligonucleotide.
To test the robustness and reproducibility of the system, miRNA expression profiles in human colon and prostate were compared in six independent array experiments. The Log2(Normalized Ratio) of each miRNA feature is shown in Figure 3A, and specific examples of miRNA overexpressed in prostate (let-7A, let-7C), overexpressed in colon (miR-200B) or with no significant variation (miR-16) are shown in Figure 3B. The average correlation between the six independent experiments was 98%, with a standard deviation smaller than 0.3 for individual miRNA, indicating that the miRNA microarray process is highly reproducible.
FIGURE 3.
miRNA microarray reproducibility. Total RNA was isolated from a single human prostate sample and a single human colon sample. Each total RNA sample was split into six independent samples (10 μg each). miRNAs in each sample were independently fractionated and labeled. Each of the six Cy5-labeled prostate miRNA samples was mixed with a Cy3-labeled colon sample and the six prostate/colon pairs were combined and hybridized to six arrays. (A) Distribution of average LogRatio. LogRatio and standard deviation for each of the 162 miRNAspecific probes on the array are shown. The average standard deviation of LogRatio across replicates was 0.15 and the average correlation between the replicates was 98%, ranging from 96.7% to 99.8%. (B) Examples of LogRatio and corresponding absolute fold change for let- 7A, let-7C, miR-16, and miR-200B miRNAs. Red illustrates higher expression in prostate, while green depicts higher expression in colon. LogRatio = Log2(normalized ratio prostate/colon signal).
Microarray validation
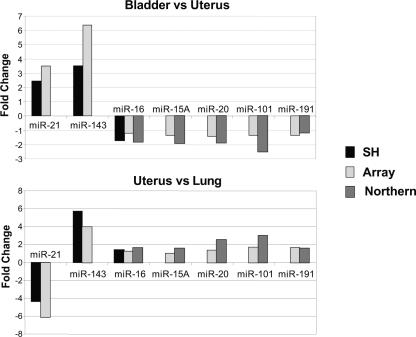
As miRNAs represent only a minuscule fraction (~0.01%) of the mass of a total RNA sample, we next asked whether the differential miRNA expression levels reported by our microarray system precisely reflected the miRNA populations present in the starting total RNA samples. To test both the gel purification and the PAP labeling steps, we first isolated total RNA from human bladder, lung, or uterus and used these samples to determine miRNA expression levels either by microarray analysis after gel purification of the miRNA fraction (162 miRNAs), or directly in the total RNA samples by two independent methods: hybridization in solution (RNase protection assay, 3 miRNAs) and hybridization on solid support (Northern blot, 5 miRNAs). Analysis of the relative differential miRNA expression by tissue pairs showed a good correlation between the three independent detection methods (Fig. 4), indicating that the gel fractionation method uniformly captures miRNAs. Further, the experiment validates the accuracy of the array system in detecting both large (e.g., miR-21 and -143) and small variations of miRNA expression (e.g., miR-15A, -16, -20, -101, and -191).
FIGURE 4.
Comparison between miRNA microarray data and other detection methods. Total RNA was isolated from human bladder, lung, or uterus. miRNA populations in 10 μg of each sample were independently fractionated, labeled, and compared by pairs on two separate arrays (162 probes). In parallel, the same total RNA samples were analyzed by solution hybridization (miR-16, -21, and -143, 1 μg total RNA) or Northern blot (miR-15A, -16, -20, -101, and -191, 5 μg total RNA) using 5′-end-labeled RNA probes. The graph shows the calculated absolute fold changes between bladder and uterus (top graph) or uterus and lung (bottom graph) deduced from microarray data analysis (Array) and Northern blot (Northern) or solution hybridization (SH) after quantitation by PhosphorImager.
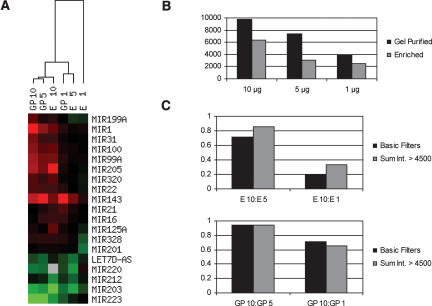
To determine whether the gel purification step provides any benefit, we compared the expression profiles obtained from gel-purified miRNA fractions and from fractions merely enriched in RNA smaller than ~200 nt. The later fractions contain mostly small structural RNAs and transfer RNAs, and miRNAs represent only ~0.1% of the samplemass. Both types of fraction were prepared from 10, 5, or 1 μg of total RNA isolated from human bladder or lung. The miRNA fraction gel purified from 10 μg total RNA, where the accuracy of miRNA expression levels was confirmed by independent detection methods (see Fig. 4; data not shown), was used as the reference sample. Hierarchical clustering analysis revealed that enriched fractions produced slightly different profiles and were less and less accurate as the sample input was reduced (Fig. 5A). These variations were accompanied by a strong reduction of the overall fluorescent signal on the array, even at the highest sample input (Fig. 5B). Further analysis showed a very poor correlation between the different inputs of enriched samples, and this loss of correlation was mainly attributed to the lowintensity signals (Fig. 5C, top). In contrast, gel-purified miRNAs from 5 and 1 μg of total RNA resulted in 94% and 70% correlation relative to the reference 10-μg sample for both high- and low-abundance miRNAs (Fig. 5C, bottom). Thus, miRNA labeling and profiling in a complex nucleic acid mixture is practical, but removing longer, unrelated RNA species by a gel purification process results in more accurate profiles and enables analysis at lower sample input.
FIGURE 5.
Comparison between enriched and gel purified miRNA fractions. One, 5, or 10 μg of total RNA from human bladder or lung were used to gel purify miRNAs (GP1, GP5, GP10) or to prepare fractions enriched in small RNA (E1, E5, E10). RNA species in the six lung fractions were labeled with Cy3 and compared with the six Cy5-labeled bladder fractions on six separate arrays (162 probes). (A) miRNA profiles were sorted based on miRNA expression levels across all sample types using hierarchical clustering. Only a few representative examples are shown. Red illustrates higher expression in bladder, green depicts higher expression in lung, and gray represents microarray data that did not pass quality filters (see Materials and Methods). (B) The sum of Cy3 and Cy5 signal intensities for each spot that passed quality filters on the six independent arrays was averaged and plotted for each sample type. The overall intensity signal for the enriched samples was 35%–60% lower than for the gel-purified samples. (C) Correlation of miRNA expression levels, i.e., Log2(normalized ratio bladder/uterus signal), in fractions prepared from 5 or 1 μg of total RNA relative to the 10 μg samples, were calculated for all the spots that passed quality filters (Basic Filters) or only for the spots that had a sum of Cy3 and Cy5 signal intensities superior to 4500 quanta (Sum Int. > 4500). The correlation between enriched fractions (top panel) was better for the high-intensity spots, showing that loss of sensitivity strongly accounts for the loss of accuracy observed with the E5 and E1 fractions.
miRNA expression in human normal tissues
Differential expression of specific miRNAs has been reported in several publications, suggesting a function for miRNA in development and differentiation of adult tissues. To gain a better understanding of the global function of miRNA in adult tissue homeostasis, we analyzed the expression profiles of 162 known human miRNAs in 26 different tissues. The miRNA population from each tissue was compared to a reference sample consisting of a pool of each total RNA sample mixed in equal amount. After gel fractionation, labeling, and hybridization to custom glass slide arrays, the Log2(Normalized Ratio) was determined for each miRNA (see Supplemental Table 1), and the tissue and miRNA profiles were sorted based on miRNA expression levels across all tissues using hierarchical clustering (Fig. 6). As expected, individual miRNAs are present at higher (red), lower (green), or similar (black) levels in different tissues relative to the pool sample, which represents the average expression level of each miRNA in the global miRNA population from the 26 tissues.
FIGURE 6.
miRNA expression profiles across 26 human normal tissues. miRNAs from 10 μg of the indicated total RNA samples were gel purified, labeled with Cy5, and compared on 26 independent microarrays against Cy3-labeled miRNAs isolated from 10 μg of the same reference pool sample. Hierarchical clustering of the tissues based on their respective miRNA profiles was performed by Average Linkage using uncentered Pearson correlation and Log2 (normalized ratio tissue/pool signal). Only miRNAs with a differential expression greater than twofold (|LogRatio| > 1) relative to the pool in at least one tissue are shown (78 out of 162 miRNAs, see Supplemental Table 1 at http://www.ambion.com/techlib/data/RNA05b.pdf for all miRNAs). Red illustrates higher expression in a given tissue, green corresponds to higher expression in the pool, and gray represents microarray data that did not pass quality filters (see Materials and Methods). A magnification of the dendrogram is presented on the right. Brain and tissues with miRNA profiles clustering together are boxed.
Interestingly, there is a subset of miRNAs that are expressed at much higher levels in one or two tissues than they are in the rest of the tissues assayed. Among these are a handful that had been previously reported to be tissue specific (e.g., miR-1, -9, -122A, and -124) as well as more than 40 other miRNAs with a strong enrichment in restricted tissues (e.g., miR-99A, -134, -181A, and -205; see also Supplemental Table 1). In contrast, only a few miRNAs, such as miR-30A, -33, -93, or -140, are expressed at roughly the same level in all adult tissues. Hierarchical clustering of the tissues based on their respective miRNA profiles also revealed that miRNA expression is similar between related tissues and distinct between unrelated tissues. For example, heart and skeletal muscle profiles are very similar, and the digestive tract tissues or the reproductive organ tissues cluster together. Strikingly, the brain miRNA profile is clearly distinct from all the other tissues analyzed.
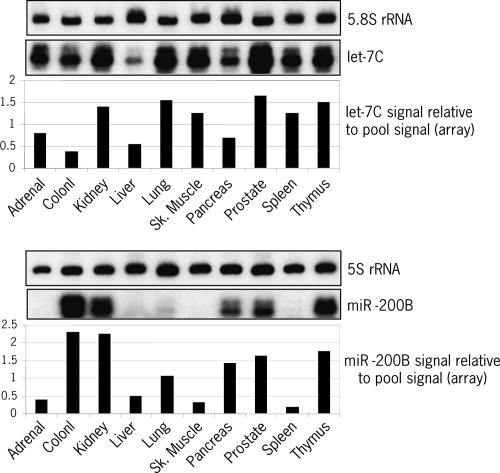
To validate the profiling data, new total RNA samples were isolated from 10 randomly selected tissues and miRNA expression levels were analyzed by independent detection methods. Both Northern blot (let-7C) and hybridization in solution (miR-200B) confirmed differential expression of individual miRNAs across tissues as well as differences in relative miRNA abundance within the same tissue (Fig. 7). miR-200B could not be detected in adrenal gland, skeletal muscle, and spleen. Tissues such as kidney, prostate, or thymus all have high expression levels of let-7C and miR- 200B, while liver poorly expresses these two miRNAs. Colon has higher expression levels of miR-200B but lower levels of let-7C relative to prostate. The same result was obtained by direct comparison between these two tissues (see Fig. 3B). Differential miRNA expression across tissues was also confirmed by pair analysis of bladder, lung, and uterus samples (see Fig. 4) or by testing other miRNAs, either broadly expressed across tissues (e.g., miR-16 and -22) or tissue specific (e.g., miR-124) (see Supplemental Fig. 2). Collectively, these data show that the miRNA microarray system described here is a powerful and robust tool for high-throughput study of miRNA expression levels and comparative analysis of miRNA expression profiles.
FIGURE 7.
Validation of miRNA expression data. Total RNA was isolated from the 10 indicated tissues, independent from the ones used in the microarray profiling study (Fig. 6). Expression levels of let-7C and miR- 200B were analyzed by Northern blot (2 μg total RNA) or solution hybridization (1 μg total RNA). As a loading control, 5S and 5.8S ribosomal RNAs were also analyzed by Northern blot. The graphs under each autoradiograph show miRNA expression levels relative to the pool signal according to the microarray data (see Supplemental Table 1 at http://www.ambion.com/techlib/data/RNA05b.pdf). The value 1 corresponds to the average expression level of the corresponding miRNA in the pool sample, i.e., Log2(normalized ratio tissue/pool signal) = 0.
DISCUSSION
Given their apparent importance in regulating gene expression, miRNAs offer a very interesting bio-molecule for life science research. A key to understanding how miRNAs are functioning in organisms is to know where and when they are expressed. To facilitate miRNA expression studies, we have adapted microarray instrumentation and protocols to analyze all of the known human miRNAs. The procedure incorporates miRNA purification, miRNA labeling through a poly-nucleotide tail appended to each miRNA, hybridization of the labeled miRNAs to a glass microarray, and microarray analysis using standard scanners and available software. The miRNA microarray system can detect subfemtomole amounts of miRNA, making it possible to detect even relatively rare miRNAs. The systemprovides high reproducibility (98% average correlation) and is reasonably quantitative, providing a great deal of confidence in the array results. Both Northern blot analysis and solution hybridization methods were used to validate relative miRNA expression levels within the same sample and across human tissues.
Most existing array-based miRNA profiling technologies use total RNA or fractions enriched in small RNA by microfiltration or reversed-phase matrices. These procedures then rely on random priming, PCR, or ligation for a labeling step prior to hybridization to immobilized probes. A key improvement in our system was to develop a rapid electrophoretic gel fractionation method that removes the large excess of unrelated RNA species and precursor miRNA sequences that can interfere with the array performance. This purification step corresponds to an enrichment by ~10,000-fold without affecting the relative miRNA abundance in individual samples (Fig. 4). The resulting fraction is ideal for efficient array labeling as well as other applications, such as cloning or enzymatic amplification (data not shown). A second improvement was to develop a method for homogenous labeling of the miRNA fraction with the highest specific activity without jeopardizing its capacity to hybridize to specific probes on a planar array. Direct enzymatic tailing with PAP and amine-modified NTP results in a homogenous tailed miRNA population that can subsequently be labeled either directly with any amine-reactive molecules, e.g., Cy or Alexa dyes, or indirectly using NHSbiotin and streptavidin coupled to fluorescent moieties (data not shown). Further, the labeled miRNA fraction is compatible with microarrays created using different immobilization chemistries, such as aldehyde, epoxy, poly-L-lysine or three-dimensional matrices glass slides (data not shown). Northern blot, solution hybridization, and microarray experiments clearly demonstrate that the combination of these two improvements does not introduce any bias in the profiling analysis, faithfully conserves miRNA representation, and allows accurate profiling at lower sample input (Figs. 4, 5, 7).
The miRNA microarray system was used to analyze a variety of different human adult tissues. We found that miRNA expression varies dramatically between tissues. Differential miRNA expression across tissues was also confirmed by direct comparison between two tissues and by independent detection methods (Figs. 3B, 4, 7). In contrast, miRNA profiles of identical tissues from multiple donors are very similar (data not shown). Furthermore, most miRNA genes likely to be processed from the same polycistronic primary transcripts have highly correlated expression patterns. As previously reported (Baskerville and Bartel 2005), correlation coefficients for pairs of miRNA genes such as miR-24 and -27a, miR-15a and -16, or miR-1 and -133 are between 0.74 and 0.91, indicating that miRNA expression is tightly regulated within individual tissues.
The miRNA expression profiles from different tissues revealed more than 50 miRNAs that are expressed primarily in one or a few related tissues (see Supplemental Table 1). Among these tissue-specific miRNAs was a set that had been previously reported in mouse and/or human: miR-1 in heart and muscle, miR-9, -124, -125, and -128 in brain, miR-122A in liver, miR-133A and -206 in muscle, miR-142 and -150 in spleen, or miR-148 in liver and stomach (Lagos-Quintana et al. 2002; Krichevsky et al. 2003; Babak et al. 2004; Sempere et al. 2004; Baskerville and Bartel 2005). In addition, miRNA expression profiles in several of the tissues assayed here had not been previously reported. The depth of our study (162 miRNAs, 26 tissues) allowed us to complete the human miRNA expression map and to identify new miRNAs with unexpected expression patterns. For example, we found that miR-125 is in fact more expressed in cervix, ovary, and uterus than in brain, miR-142 and -150 are not only expressed in spleen but are also very abundant in lymph node, ovary or, thymus, and miR-148 was detected in pancreas and ovary in addition to liver and stomach. miR- 181A was found highly expressed in brain, placenta, spleen, and thymus, while miR-205 was identified in breast, prostate, and thymus. About 20 miRNAs, such as miR-126-AS, -143, -145, -189, or -321, were found highly expressed in one or several tissues from the digestive tract cluster. Strikingly, the tissues that showed the highest restricted enrichment in individual miRNAs are brain and the cervix/uterus/ovary cluster. This observation suggests that specific miRNAs might play a critical role in brain and female reproductive system development and/or homeostasis.
Hierarchical clustering of the tissues based on their respective miRNA profiles showed that tissues are characterized by unique miRNA expression profiles. Together with the tissue specificity of several miRNAs, this observation is consistent with the concept that miRNAs are important components of tissue development, differentiation, and maintenance of differentiation in adults. Interestingly, related tissues had similar miRNA profiles, as would be predicted for global regulators of tissue differentiation. One tantalizing hypothesis is that miRNAs might directly regulate the expression levels of many of the genes that distinguish these different adult tissues or clusters of adult tissues.
Categorizing miRNAs based on their coregulation or identifying miRNAs that are differentially expressed between different tissues or cell samples will undoubtedly identify interesting miRNAs that deserve closer scrutiny. Further questions are whether these miRNA signatures are affected in disease states and, if so, at what stage. A direct comparison between a large panel of human tumors and their corresponding normal adjacent tissues revealed several miRNAs that might directly contribute to oncogenesis (K. Keiger, J. Shelton, D. Brown, E. Labourier, in prep.). This approach, combined with genetic studies in C. elegans and functional studies in human cultured cells, recently helped identify let-7 miRNA as a regulator of RAS oncogene expression and as a potential tumor suppressor disrupted in lung tumors (Johnson et al. 2005). Further, our microarray technology enables accurate miRNA expression profiling in fixed tissues (P. Powers, R. Conrad, K. Keiger, J. Shelton, D. Brown, E. Labourier, in prep.), providing access to a vast amount of information from archived clinical samples. If miRNAs are indeed involved in oncogenesis, inflammatory response, or viral infection, comparative miRNA expression studies in disease samples will certainly reveal novel therapeutic targets and diagnostic markers.
MATERIALS AND METHODS
Microarray platform
DNA oligonucleotide probes from the mirVana miRNA Probe Set (Ambion) were used to print custom microarrays. Each probe was designed using the sequence of its respective mature miRNA according to the miRNA Registry (Griffiths-Jones 2004) and was analyzed for potential secondary structure and cross hybridization. Negative control probes were designed using sequences from Escherichia coli, and each was functionally validated to have minimal cross-hybridization to a collection of chemically synthesized miRNAs as well as a series of tissue-derived miRNAs. Each probe carries a 15-nt linker at the 3′ end of the miRNA complement sequence in addition to an amine group used to couple the probes to coated glass slides. Probes were resuspended in 3× SSC at 50 μM and spotted on Nexterion Slide E (Schott) at 55% humidity with the ArrayIt SpotBot (Telechem). Each feature was spotted using SMP4 Stealth pins with a spot diameter of ~100 μm. The slides were rehydrated by incubation at room temperature in a humidity chamber for 30 min followed by a 30-min incubation at 60°C to dry. The slides were blocked in a solution containing 100 mM ethanolamine, 1 M Tris (pH 9.0), and 0.1% SDS for 15 min at 50°C, then thoroughly rinsed with water and spun dry.
RNA preparation
Total RNA isolation and small RNA enrichment procedure were performed with the mirVana miRNA Isolation Kit (Ambion) according to the manufacturer’s instructions. For miRNA expression profiling in normal human tissues, miRNA certified First- Choice Total RNA (Ambion) were used. To isolate miRNA fractions, total RNA samples were fractionated and cleaned up with the flashPAGE Fractionator and reagents (Ambion) per the manufacturer’s recommendation. Briefly, 1–20 μg of each RNA sample were loaded onto the top of a column filled with a denaturing acrylamide gel matrix and fractionated by applying an electrical current. A dye was loaded with the total RNA sample to track RNAs that are ~40 nt in size. Electrophoresis was stopped when the dye reached the bottom of the column, and miRNAs were recovered from the bottom buffer chamber using a glass fiber filter-based cleaning procedure (flashPAGE Reaction Cleanup Kit, Ambion). Approximately 1 ng of miRNA was recovered per 10 μg of total RNA.
miRNA labeling and cleanup
Chemically synthesized oligoribonucleotides (Ambion), purified miRNAs, or fractions enriched in small RNAs were labeled with the mirVana miRNA Labeling Kit (Ambion) and amine-reactive dyes as recommended by the manufacturer. Poly(A) polymerase and a mixture of unmodified and amine-modified nucleotides were first used to append a poly-nucleotide tail to the 3′ end of each miRNA. The amine-modified miRNAs were then cleaned up and coupled to NHS-ester modified Cy5 or Cy3 dyes (Amersham Bioscience). Unincorporated dyes were removed with a second glass fiber filter-based cleaning procedure.
Microarray hybridization and data analysis
A 3× miRNA Hybridization Buffer (Ambion) was added to the fluorescently labeled miRNAs and the solution was heated at 95°C for 3 min. Slides were hybridized 12–16 h at 42°C in sealed cassettes using a water bath. Following hybridization, the slides were washed and dried prior to a high-resolution scan on a GenePix 4000B Array Scanner (Axon). Each element was located and analyzed using the GenePix Pro 5.0 software package (Axon). Data were filtered for quality and significance using the Longhorn Array Database (Killion et al. 2003). Filters were based on several data quality standards, including minimum intensity and pixel consistency. All data used for analysis had a signal-to-noise ratio >5, an average sum intensity 50% higher than that of the negative control spots, and a regression ratio >0.5. Data were normalized globally per array such that the average LogRatio was 0 after normalization. Hierarchical clustering was performed by Average Linkage using uncentered Pearson correlation (Eisen et al. 1998).
Microarray data validation
miRNA expression levels in total RNA samples were measured by Northern blot (2–5 μg) or solution hybridization (1 μg) with the mirVana miRNA Detection Kit (Ambion) according to the manufacturer’s instructions. DNA or RNA probes were labeled and purified with the mirVana Probe & Marker Kit (Ambion) and [γ-32P]ATP at 6000 Ci/mmol (PerkinElmer). For Northern analyses, RNA samples were resolved on denaturing 15% polyacrylamide gels and electroblotted on BrightStar-Plus Nylon membranes (Ambion). Membranes were blocked in UltraHyb-oligo Hybridization Buffer (Ambion) for at least 1 h at 65°C, hybridized overnight at 42°C with the appropriate probe, and washed three times with NorthernMax Low Stringency Wash Buffer (Ambion) for 5 min at room temperature followed by 15 min at 42°C. Results were quantified on a PhosphorImager Storm 860 (Amersham Bioscience).
SUPPLEMENTARY MATERIAL
Supplementary material can be found at http://www.ambion.com/techlib/data/RNA05b.pdf.
Acknowledgments
We thank Eric Devroe for helpful comments on this article. This work was supported in part by National Genome Research Institute SBIR grant R43HG003430 to D.B.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2610405.
REFERENCES
- Allawi, H.T., Dahlberg, J.E., Olson, S., Lund, E., Olson, M., Ma, W.P., Takova, T., Neri, B.P., and Lyamichev, V.I. 2004. Quantitation of microRNAs using a modified Invader assay. RNA 10: 1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V. 2004. The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- Babak, T., Zhang, W., Morris, Q., Blencowe, B.J., and Hughes, T.R. 2004. Probing microRNAs with microarrays: Tissue specificity and functional inference. RNA 10: 1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad, O., Meiri, E., Avniel, A., Aharonov, R., Barzilai, A., Bentwich, I., Einav, U., Gilad, S., Hurban, P., Karov, Y., et al. 2004. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res. 14: 2486–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. and Chen, C.Z. 2004. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5: 396–400. [DOI] [PubMed] [Google Scholar]
- Baskerville, S. and Bartel, D.P. 2005. Microarray profiling of micro- RNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack, M.T., Czaplinski, K., and Gorlich, D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, X., Hagedorn, C.H., and Cullen, B.R. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli, A.M., Tops, B.B., Plasterk, R.H., Ketting, R.F., and Hannon, G.J. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235. [DOI] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad, Y., Aach, J.,Hayes, G.D., Reinhart, B.J., Church, G.M., Ruvkun, G., and Kim, J. 2003. Computational and experimental identification of C. elegans microRNAs. Mol. Cell 11: 1253–1263. [DOI] [PubMed] [Google Scholar]
- Gregory, R.I., Yan, K.P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., and Shiekhattar, R. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S. 2004. The microRNA registry. Nucleic Acids Res. 32: D109–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Hartig, J.S., Grune, I., Najafi-Shoushtari, S.H., and Famulok, M. 2004. Sequence-specific detection of MicroRNAs by signal-amplifying ribozymes. J. Am. Chem. Soc. 126: 722–723. [DOI] [PubMed] [Google Scholar]
- Houbaviy, H.B., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell-specific microRNAs. Dev. Cell 5: 351–358. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Johnson, S.M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., Labourier, E., Reinert, K.L., Brown, D., and Slack, F.J. 2005. RAS is regulated by the let-7 microRNA family. Cell 120: 635–647. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killion, P.J., Sherlock, G., and Iyer, V.R. 2003. The Longhorn Array Database (LAD): An open-source, MIAME compliant implementation of the Stanford Microarray Database (SMD). BMC Bioinform. 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., and Kosik, K.S. 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21: 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Kim, M., Han, J., Yeom, K.H., Lee, S., Baek, S.H., and Kim, V.N. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23: 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. 2003. The micro-RNAs of Caenorhabditis elegans. Genes & Dev. 17: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.G.,Calin, G.A.,Meloon, B.,Gamliel, N., Sevignani, C., Ferracin,M., Dumitru, C.D., Shimizu, M., Zupo, S., Dono, M., et al. 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. 101: 9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lund, E., Guttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. 2004. Nuclear export of microRNA precursors. Science 303: 95–98. [DOI] [PubMed] [Google Scholar]
- Miska, E.A., Alvarez-Saavedra, E., Townsend, M., Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M., and Horvitz, H.R. 2004. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 5: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison, E.P., and Hannon, G.J. 2004. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 16: 223–229. [DOI] [PubMed] [Google Scholar]
- Nelson, P.T., Baldwin, D.A., Scearce, L.M., Oberholtzer, J.C., Tobias, J.W., and Mourelatos, Z. 2004. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods 1: 155–161. [DOI] [PubMed] [Google Scholar]
- Olsen, P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671–680. [DOI] [PubMed] [Google Scholar]
- Overhoff, M.,Wunsche,W., and Sczakiel, G. 2004. Quantitative detection of siRNA and single-stranded oligonucleotides: Relationship between uptake and biological activity of siRNA. Nucleic Acids Res. 32: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, S., Zavolan, M., Grasser, F.A., Chien, M., Russo, J.J., Ju, J., John, B., Enright, A.J., Marks, D., Sander, C., et al. 2004. Identification of virus-encoded microRNAs. Science 304: 734– 736. [DOI] [PubMed] [Google Scholar]
- Rodriguez, A., Griffiths-Jones, S., Ashurst, J.L., and Bradley, A. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res. 14: 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T.D., Jiang, J., Liu, Q., and Yang, L. 2004. A highthroughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 32: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208. [DOI] [PubMed] [Google Scholar]
- Sempere, L.F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud, M. and Rosok, O. 2004. Profiling microRNA expression using sensitive cDNA probes and filter arrays. Biotechniques 37: 574–576, 578–580. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Koo, S., White, N., Peralta, E., Esau, C., Dean, N.M., and Perera, R.J. 2004. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 32: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, J.M., Parker, J., Perou, C.M., and Hammond, S.M. 2004. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods 1: 47–53. [DOI] [PubMed] [Google Scholar]
- Valoczi, A., Hornyik, C., Varga, N., Burgyan, J., Kauppinen, S., and Havelda, Z. 2004. Sensitive and specific detection of micro-RNAs by Northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 32: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta, S., Shih, I.H., and Bartel, D.P. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–596. [DOI] [PubMed] [Google Scholar]
- Yi, R., Qin, Y., Macara, I.G., and Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y. and Cullen, B.R. 2003. Sequence requirements for micro RNA processing and function in human cells. RNA 9: 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2004. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 32: 4776–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Yi, R., and Cullen, B.R. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24: 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Kolb, F.A., Jaskiewicz, L., Westhof, E., and Filipowicz, W. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68. [DOI] [PubMed] [Google Scholar]