Abstract
The hairpin ribozyme can catalyze the cleavage of RNA substrates by employing its conformational flexibility. To form a catalytic complex, the two domains A and B of the hairpin-ribozyme complex must interact with one another in a folding step called docking. We have constructed hairpin ribozyme variants harboring an aptamer sequence that can be allosterically induced by flavin mononucleotide (FMN). Domains A and B are separated by distinct bridge sequences that communicate the formation of the FMN-aptamer complex to domains A and B, facilitating their docking. In the presence of a short oligonucleotide that is complementary to the aptamer, catalytic activity of the ribozyme is completely abolished, due to the formation of an extended conformer that cannot perform catalysis. However, in the presence of the small molecule effector FMN, the inhibitory effect of the oligonucleotide is competitively neutralized and the ribozyme is activated 150-fold. We thus have established a new principle for the regulation of ribozyme catalysis in which two regulatory factors (an oligonucleotide and a small molecule) that switch the ribozyme’s activity in opposite directions compete for the same binding site in the aptamer domain.
Keywords: competitive regulation, hairpin ribozyme, aptazyme, allosteric ribozyme, ribozyme engineering
INTRODUCTION
The secondary structure of the hairpin ribozyme comprises two internal loop domains, A and B, each of them flanked by two helices, that must interact with one another to permit site-specific cleavage of the substrate oligonucleotide, generating 5′-hydroxyl and 2′,3′-cyclic phosphate termini (Esteban et al. 1997; Hampel et al. 2001; Fedor 2002; Ferre-D’amare and Rupert 2002). The catalytic mechanism of the hairpin ribozyme relies on a reaction pathway in which binding of the substrate defines domain A, which is first aligned with domain B in a linear extended conformation (Earnshaw et al. 1997; Rupert and Ferre-D’Amare 2001). This conformer then folds into a docked structure in which loops A and B contact each other to form the catalytic center, stabilized by noncanonical base pairs. This docked conformation is characterized by a sharp bend at the so-called hinge region (Butcher et al. 1999; Walter et al. 1999; Pinard et al. 2001; Zhuang et al. 2002), a flexible linker that mediates the docking process (Esteban et al. 1998; Pörschke et al. 1999). Following cleavage, the docked complex unfolds back into the extended structure and the cleaved products dissociate.
Thus, the hinge region constitutes a flexible joint that mediates the docking process of domains A and B by facilitating the tertiary contacts that define the active site within the interface between the internal loops, and by stabilizing the active conformation of the ribozyme (Esteban et al. 1998; Pörschke et al. 1999). Consequently, this motif was used as a target domain for controlling the structure and folding pathway of the hairpin ribozyme by potentially binding effector oligonucleotides. In this way, hairpin ribozyme variants that could either be induced or repressed by external oligonucleotide effectors were engineered by introducing a third regulatory domain C at the junction between helix 2 and helix 3 (Najafi-Shoushtari et al. 2004). This new domain confers precise control over the formation of distinct structural motifs mediated by adaptive binding to defined effector RNA sequences such as mRNAs, miRNAs, or aptamers. In prototype approaches, we have used aptamers (Hartig et al. 2002), mRNAs (Najafi-Shoushtari et al. 2004), and different microRNAs (Hartig et al. 2004) as effector molecules. Other reporter-ribozymes that are regulated by external oligonucleotide effectors have been engineered by exploiting different regulatory mechanisms (Robertson and Ellington 1999; Komatsu et al. 2002; Wang et al. 2002; Vaish et al. 2003; Vauleon and Muller 2003).
Here, we expanded on this concept by using a domain C that directly corresponds to a functional RNA rather than a domain C that is complementary to a functional RNA, as in the previous approaches (Hartig et al. 2002, 2004; Najafi-Shoushtari et al. 2004). We describe an engineered hairpin ribozyme variant harboring the 21-nucleotide (nt)-long flavin mononucleotide (FMN)-binding RNA aptamer (Fig. 1A ▶) that permits allosteric induction of substrate cleavage by FMN. The aptamer separates domains A and B via distinct bridging sequences that, when designed properly, communicate the formation of the FMN-aptamer complex to the two domains by facilitating their docking. Oligonucleotides that can hybridize to the aptamer domain lead to complete inhibition of the allosteric ribozyme by enforcing an extended conformation that precludes stable docking of domains A and B. However, this inhibitory effect can be neutralized by FMN, presumably by shifting the equilibrium into a catalytically active conformation. The new hairpin ribozymes described here represent an example of allosterically regulated hairpin ribozymes that utilize a previously unexplored principle for the regulation of ribozyme catalysis, namely the competition of two regulatory factors, an oligonucleotide and a small molecule, for the same binding site in the aptamer domain.
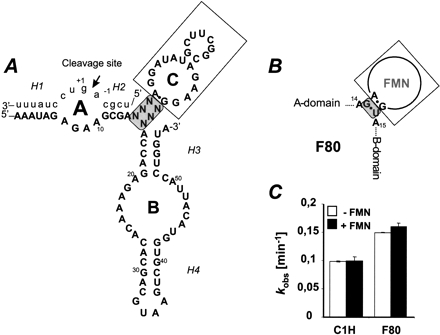
FIGURE 1.
(A) Proposed secondary structure of FMN aptamer containing hairpin ribozymes. The aptamer sequence (domain C) replaces the hinge region as an internal junction via distinct bridge sequences (gray box), which base identities are chosen in a manner to interfere with the docking process. (B) Schematic presentation of the aptamer junction in F80 ribozyme construct. (C) Catalytic rate constants of F80 and the unmodified C1H construct (Supplementary Fig.1 at http://famulok.chemie.uni-bonn.de/publications/current/index.html) in the absence and presence of 200 μM FMN.
RESULTS
As the basis for our FMN-dependent hairpin ribozyme variants, we used a kinetically well behaved version of the hairpin ribozyme that was designed according to the guidelines from the Burke laboratory (Esteban et al. 1997). Briefly, in this construct helix 4 is extended by three base pairs and a stable GUAA tetraloop to stabilize the loop B domain. In addition, the construct contains a rate-enhancing U47C mutation. Three base pairs, one in H1 and two in H2, were changed to minimize self-complementarity of the substrate (Supplementary Fig. 1 at http://famulok.chemie.uni-bonn.de/publications/current/index.html). The rest of the ribozyme is identical to the wild-type ribozyme (Chowrira and Burke 1991). This variant, which is hereafter designated as the “construct 1 hairpin (C1H),” possesses a higher cleavage activity compared to that of the wild-type hairpin ribozyme (Esteban et al. 1997).
Design of the hinge region communication modules
Based on the C1H variant, we designed a series of artificial ribozymes which harbor the 21-nt-long FMN-binding RNA aptamer (Burgstaller and Famulok 1994), inserted between helix 2 and helix 3 of domains A and B, respectively (Fig. 1A ▶). The hinge region joins domains A and B to the aptamer via distinct bridging sequences that serve as communication modules. In this design, the communication module links the aptamer via a pseudo-three-way junction at the H2 and H3 connection in the original hairpin ribozyme (Najafi-Shoushtari et al. 2004), to selectively favor the active docked conformation and to avoid unspecific interference from the aptamer domain during catalysis by stabilizing stem–loop formation.
The communication module of our initial construct F80 consisted of only a weak G·U wobble pair which we anticipated to provide the necessary flexibility at the pseudo-three-way junction formed by helices 2 and 3 and the aptamer stem A1 (Fig. 1B ▶). This design was based on structural data by Fan et al. (1996) showing that FMN binding stabilizes the noncanonical A·G-pair that elongates stem A1. We reasoned that a “minimal stem A1,” formed only by a potential G·U wobble pair, would not be stably paired in the absence of the ligand, but would form a stable helix in conjunction with the A·G-pair in the presence of FMN, thereby inducing the bent conformation that would activate the ribozyme. As shown in Figure 1C ▶, the F80 construct generally exhibited a slightly enhanced catalytic rate compared to the C1H construct without the aptamer. However, no difference in the catalytic performance of F80 was found in the presence or absence of FMN. Thus, the major fraction of the F80 construct already appears to be in a constitutively active state even in the absence of FMN and cannot be further induced by addition of the ligand.
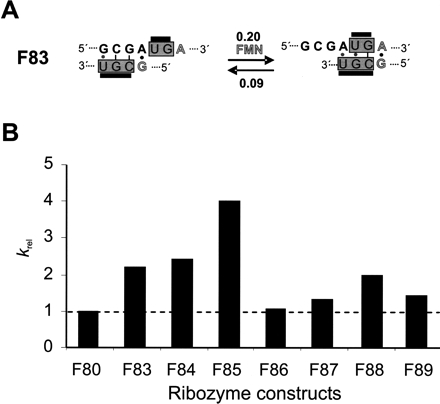
To obtain ribozymes that respond to FMN as a positive effector, seven different symmetric and asymmetric bridge elements (F83–F89) were generated and probed for allosteric regulation (Table 1 ▶), based on C1H as the parent variant. Figure 2A ▶ representatively shows the respective bridge sequence for construct F83, depicted in gray boxes. These bridge sequences, or communication modules, were designed so as to utilize a “slip-mechanism” upon binding of FMN (Fig. 2A ▶), similar to what was proposed for communication modules in allosteric hammerhead ribozymes (Soukup and Breaker 1999a). We reasoned that in the context of the hairpin ribozyme constructs used here, the communication modules would thwart the formation of the A-loop in the absence of FMN, because A-loop nucleotides would engage in pairing with complementary bases in the bridging sequence, leading to a slightly more stable helix than that formed by the communication module itself. This would result in less potent variants than the C1H or wild-type hairpin ribozymes. In the presence of FMN, the preferred formation of the terminal A·G base pair would induce a “sliding” of base pairs toward a consecutive helix starting with the terminal A·G pair, which would activate the ribozyme (Fig. 2A ▶). However, the rates obtained for the different variants show that in the absence of FMN appreciable catalysis is still achieved (Table 1 ▶). Only F85 and F88 show approximately twofold reduced catalytic performance compared to C1H; all other variants remained as active as or performed better than C1H. The catalytic performance of four of the seven variants tested could be stimulated by FMN. Constructs F83, F84, and F88 showed an approximately twofold improved krel value, whereas that of F85 improved fourfold (Fig. 2B ▶). This result is encouraging, the more so as these constructs represent the first examples of hairpin ribozymes that are allosterically responsive to a small exogenous cofactor. Only one study reported an in vitro selected ATP-dependent hairpin ribozyme (Meli et al. 2003) which, however, is very different from our hairpin ribozyme constructs in the sense that ATP does not act as an allosteric effector molecule but is rather required as a catalytic cofactor, similar to guanosine in group I introns. On the other hand, examples of allosteric aptazymes in which small cofactor-binding aptamers were linked to the hammerhead ribozyme via communication modules have shown that small molecules can lead to between 10- and 300-fold induced rate enhancements (Soukup and Breaker 1999b). We therefore sought to investigate an alternative mechanism to increase the difference between the catalytic rates of our ribozyme constructs in the absence or presence of FMN by employing inhibitory oligonucleotides that compete with the small cofactor for binding to the regulatory aptamer sequence.
TABLE 1.
Catalytic rates of different ribozyme constructs
| kobs (min−1) | |||
| Construct | Bridge sequence | −FMN | +FMN |
| C1H | — | 0.10 | 0.10 |
| F80 | G | 0.15 | 0.16 |
| U | |||
| F83 | UG | 0.09 | 0.20 |
| UGC | |||
| F84 | UGG | 0.09 | 0.22 |
| GCU | |||
| F85 | UCGG | 0.04 | 0.16 |
| GCU | |||
| F86 | GUGG | 0.15 | 0.16 |
| CGCU | |||
| F87 | UG | 0.15 | 0.20 |
| CGC | |||
| F88 | GGUG | 0.07 | 0.14 |
| UCGU | |||
| F89 | CUA | 0.14 | 0.20 |
| CGCU | |||
The concentration of the ribozymes was 200 nM; that of FMN was 200 μM.
FIGURE 2.
FMN-directed positive regulation mechanism. (A) Schematic representation of the proposed slip mechanism by FMN-responsive hairpin ribozyme constructs F83. The bridge sequence is shown in gray boxes. In the absence of FMN, this is expected to form an inhibitory base pairing with helix 2 located in the A domain. Upon FMN addition, a switch is expected in favor of an active ribozyme construct. Thereby the formation of the A·G base pair (gray letters) is essential for aptamer to bind FMN. The values at the arrows correspond to the observed rate constants (Table 1 ▶). (B) Relative rate enhancement observed in the presence of FMN. The plot shows a positive ribozyme response that is FMN-dependent.
Competitive binding by inhibitory oligonucleotide effectors
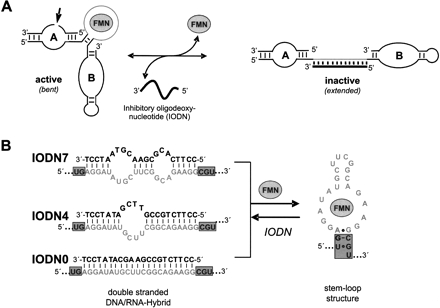
The general principle of this concept is schematically shown in Figure 3A ▶. FMN and the oligonucleotide effector molecule compete with each other for binding to the same aptamer domain, thereby shifting the equilibrium either to the active or inactive conformation of the ribozyme construct. We designed a set of three different inhibitory oligonucleotides (IODNs), each 21 nt long, that contained seven (IODN7), four (IODN4), or no mismatches (IODN0) to the FMN-aptamer domain. Annealing of IODNs to the aptamer should fully convert this domain into a double-stranded DNA-RNA hybrid (Fig. 3B ▶) which will prevent docking of domains A and B and thus inhibit catalysis. Importantly, the aptamer domain is predisposed to act as a receptor for oligonucleotide effector molecules and for the small molecule effector FMN, a prerequisite for competitive allosteric ribozyme regulation.
FIGURE 3.
(A) Principle and mechanism of a competitive allosteric regulation of hairpin ribozyme by single-stranded, 21-base DNA oligonucleotide and the FMN molecule. Whereas the oligonucleotide effector directs a negative regulation mechanism, the FMN molecule guides a positive effect. Thereby the aptamer sequence (gray) acts as a binding site for both effectors. (B) Competitive regulation mechanism conducted by different 21-nt, single-stranded inhibitory oligodeoxynucleotides (IODNs) and FMN molecule effector. IODN hybridization is expected to lead to a breakup of the bridge sequence (gray box) with subsequent inhibition of ribozyme activity. This should convert the stem–loop structure into a double-stranded helix.
We introduced the mismatches to investigate the possible balance between annealing efficiency of the oligonucleotide, and hence its inhibitory capacity, and the ability of FMN to compete with the IODN for aptamer binding by inducing the proper aptamer fold. This design was done according to our expectation that the inhibitory capacity of the oligonucleotide should increase with decreasing numbers of mismatches, whereas the activating capacity of FMN should increase with the number of mismatches. For further analysis we chose F83, despite the fact that F85 with a krel of 4 responded slightly better to FMN than F83 or F84 (Fig. 2B ▶). However, the absolute value of kobs of F85 in presence of FMN was lower than for F83 or F84 (Table 1 ▶).
Table 2 ▶ shows the competition constants at 1.0 mM FMN for the F83 construct in the presence of IODN7 and IODN4, respectively. The competition constant kcompet is the ratio of the values of kobs obtained for the IODN at the indicated concentration in presence of 1.0 mM FMN and without the ligand [kcompet = kobs+ / kobs−]. An equimolar concentration of IODN7 and ribozyme led to marginal improvement of kcompet in the case of F84 (data not shown) and F83 (Table 2 ▶), compared to the values of krel in the absence of any inhibitory oligonucleotide (Fig. 2B ▶). Similar competition constants of 3–3.5 are obtained at the same concentration of 200 nM IODN7 and IODN4. The competition constant increases slightly with increasing concentrations of IODN4 up to 4.3 at 800 nM (IODN4:F83 = 4:1), but there appears to be no clear correlation between inhibitory capacity at increasing IODN4 concentrations, either with or without FMN (Table 2 ▶). These data indicate that the inhibitory activities of the mismatched IODNs used here are suboptimal and that the response to FMN still remains weak, similar to the ribozymes without the regulatory oligonucleotides.
TABLE 2.
Inhibitory effect of various 21-nt IODN effectors in absence and presence of 1 mM FMN
| Ribozyme | F83 | |||||||||
| IODN effector | IODN7 | IODN4 | IODN0 | |||||||
| 1:1 | 1:1 | 1:2.5 | 1:4 | 1:0,5 | 1:1 | 1:2 | 1:3 | 1:4 | ||
| kobs [min−1] | −FMN | 0.06 | 0.04 | 0.013 | 0.015 | 0.06 | 0.014 | 0.007 | 0.002 | 0.0004 |
| +FMN | 0.18 | 0.13 | 0.046 | 0.065 | 0.16 | 0.15 | 0.13 | 0.09 | 0.06 | |
| kcompet | 3.0 | 3.25 | 3.5 | 4.3 | 2.7 | 10.7 | 18.5 | 45 | 150 | |
The ratios correspond to a constant ribozyme concentration of 200 nM.
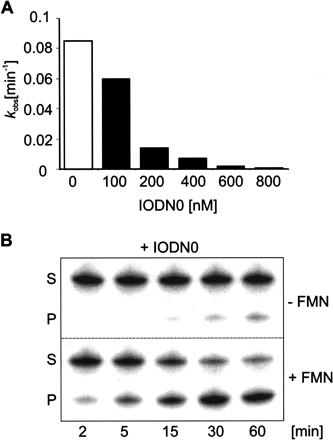
Interestingly, the fully matched oligonucleotide IODN0 (Fig. 3B ▶) shows a well correlated dose response in its inhibitory activity in the absence of FMN. Relative to the noninhibited F83 ribozyme, 1.5-fold inhibition is achieved at a 1:0.5 ratio of F83 and the IODN0 effector, sixfold inhibition at a 1:1 ratio, 13-fold inhibition at a 2:1 ratio, 45-fold inhibition at a 3:1 ratio, and 225-fold inhibition of ribozyme activity at a 4:1 ratio (Table 2 ▶; Fig. 4A ▶). However, in the presence of 1.0 mM FMN, this inhibitory effect of IODN0 can be competed efficiently even at the 800 nM concentrations of IODN0, although a continuous slight drop of the absolute values of kobs occurs with increasing concentrations of IODN0 (Table 2 ▶; see also Supplementary Fig. 2 at http://famulok.chemie.uni-bonn.de/publications/current/index.html). With a value of kobs+ of 0.06 min−1 at the 4:1 ratio, this corresponds to a competition constant of 150, or a 150-fold activation of the inhibited ribozyme. The time course of the cleavage reaction of the highest competitive effect is shown in Figure 4B ▶. A comparison of the reaction in the absence of FMN and that in the presence of 1.0 mM FMN and 800 nM IODN0 indicates that the two effector molecules compete for binding to the regulatory aptamer domain, presumably by arriving at an equilibrium of two ribozyme populations: one inactive population bound to IODN0, and an active population complexed to FMN.
FIGURE 4.
Inhibitory and competitive effects of F83 ribozyme activity by IODN0 and FMN. (A) Inactivation of F83 ribozyme construct in the presence of increasing concentrations of IODN0 as indicated. Maximal inhibition occurs at a ratio of 1:4 of IODN0 and F83. (B) Competitive allosteric effect observed by the F83 ribozyme caused by FMN and IODN0 at a ratio of 200:1. Shown are substrate (S) and 5′-cleavage product (P) resulting from ribozyme catalysis in the constant presence of 800 nM IODN0. Products were separated by denaturing PAGE following reaction in the absence or presence of 1.0 mM FMN at 37°C over a 60-min time course.
DISCUSSION
Here we describe a new class of rationally designed allosteric hairpin ribozymes that can be regulated by two different effector molecules, an oligonucleotide which acts as an inhibitor, and a small organic cofactor serving as an allosteric activator. When added together to the ribozyme, both effectors compete with each other for binding to the same allosteric domain. The design involved substitution of the hinge region of the hairpin ribozyme for various communication modules to which the FMN-binding RNA aptamer was attached to serve as the allosteric domain. In the absence of any regulatory effector molecule, these ribozymes remained constitutively active, but further twofold to fourfold activation could be achieved in the presence of FMN, which acts as a positive effector. As a negative effector we used an oligonucleotide that hybridizes to the aptamer domain, resulting in up to 225-fold down-regulation of its catalytic activity.
In the F83 hairpin ribozyme construct investigated here, a fourfold excess of IODN0 is necessary to achieve significant inhibition of the hairpin ribozyme construct F83, with kobs values of 4 × 10−4 min−1. This probably reflects the fact that the inhibitory oligonucleotide IODN0 can hybridize only to the aptamer domain itself, without including parts of the connecting communication module. This design of the IODN hybridization differs from our previously described hairpin ribozymes (Hartig et al. 2002, 2004; Najafi-Shoushtari et al. 2004), which contained the antisense sequence of functional RNAs such as aptamer, microRNAs, or mRNAs inserted as a regulatory domain via a preformed helix at the hinge region. The corresponding functional oligonucleotides exhibited their full regulatory activity already at equimolar amounts of their respective allosteric ribozyme partners. Different from the IODNs described here, they could hybridize to their anti-sense sequence including parts of the connecting helix in the hinge region, thereby disassembling it very efficiently.
The bridge elements in constructs F80–F89 exhibited marked differences in communicating the formation of the FMN-aptamer complex to the catalytic center of the ribozyme. The result that a maximum of fourfold activation by FMN could be achieved with the bridge elements used here is probably due to the fact that all ribozyme constructs are constitutively active in the absence of the ligand, exhibiting a catalytic activity that is close to that of the wild-type ribozyme, and within ± 1.5-fold of the nonregulable C1H hairpin motif. It is possible that allosteric ribozymes that can reach a much more pronounced level of activation by a small molecule regulator could be isolated by in vitro selection, as was shown for allosteric hammerhead ribozymes (Soukup and Breaker 1999a). In that study, communication modules were identified from a library of variants in which the bridging domain that connected the hammerhead ribozyme to the FMN aptamer was randomized. Likewise, Kertsburg and Soukup (2002) described a versatile communication module, also obtained by in vitro selection, that can function similarly between a variety of catalysts such as the hepatitis delta virus, hammerhead, X motif and Tetrahymena group I ribozymes, and various ligand-binding domains.
As an alternative to optimizing communication modules, we show here that a 150-fold level of activation of an allosteric ribozyme by a small molecule can be achieved by using a ribozyme in combination with a specific inhibitory oligonucleotide. The design of the IODN-aptamer pair is intuitive: each aptamer sequence inherently carries along the prescription for its IODN. Thus, this strategy should be applicable for a broad variety of aptamer-hairpin ribozyme pairs. Because this oligonucleotide binds to the same allosteric site as the regulatory small molecule effector, catalytic activity is a function of the equilibria between a population of IODN-inhibited and a population of FMN-activated hairpin ribozymes, which, in turn, depends on the relative ratios of the two effectors. In summary, we describe here a new regulatory principle that has not been applied to allosteric ribozyme regulation to date: the simultaneous regulation of a hairpin ribozyme by a small molecule and an oligonucleotide that compete for the same allosteric site. This approach of allosteric hairpin ribozyme control may be useful for applications in biosensors to detect ratios of effectors in one step, or for engineering genetic control elements that can be regulated by combinations of effector molecules.
MATERIAL AND METHODS
Oligonucleotides
Synthetic DNA templates were purchased from MWG Biotech. Ribozymes were generated by in vitro transcription of the appropriate DNA templates: The ssDNA sequences were amplified by PCR using Dap polymerase (Eurogentec) and the following primer sequences:
5′-primer: 5′-TCTAATACGACTCACTATAGGAAATAGAGAAG CGA-3′ (bold, T7 promoter sequence; italic, transcription initiating nucleotides), and
3′-primer: 5′-TACCAGGTAATGTACCACGAC-3′.
Transcription reactions (100 μL) containing 300-pmol dsDNA template were performed as described (Milligan and Uhlenbeck 1989). The transcription was carried out overnight (~16 h) at 37°C. To digest DNA template, 0.25 μL DNase I (10 U/μL) was added and the reaction incubated at 37°C for at least 30 min. The resultant transcripts were then purified by gel-electrophoresis on an 8% (v/v) denaturing polyacrylamide gel containing 8.3 M urea. Effector DNA oligonucleotides IODN7: 5′-CCTTCACGCGAACGTAATCCT-3′, IODN4: 5′-CCTTCTGCCGTTCGATATCCT-3′, and IODN0: 5′-CCTTCTGCCGAAGCATATCCT-3′ were prepared by standard solid-phase methods and were purified by denaturing (8.3 M urea) PAGE. RNA substrate (5′-UCGCAGUCC UAUUU-3′) was purchased from Eurogentec. 5′-[γ-32P]-labeled RNA substrate was generated using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]-ATP.
FMN-responsive ribozyme assay
All ribozyme reactions were carried out under single turnover conditions (STO) with excess ribozyme (200 nM) over substrate (1 nM) in a reaction buffer containing 50 mM Tris-HCl pH 7.5, 20 mM MgCl2 at 37°C. No change in either the rate or the extent of cleavage was observed at higher ribozyme concentrations, indicating that 200 nM ribozyme is saturating (data not shown). Ribozymes were pre-incubated with 200 μM FMN for 10 min at room temperature, and the cleavage reactions were initiated upon addition of the (5′-32P)-radio-labeled substrate in a total reaction volume of 50 μL. The reaction solution was aliquoted (5 μL) in a time course ranging from 0 to 60 min and added to an equal volume of LB-buffer (15 mM EDTA, 97% formamide) to stop the reaction at each time point. Cleavage products were analyzed on 20% (w/v) polyacrylamide gels containing 8.3 M urea, and the percentages of the cleaved products were quantified by measuring the radioactivity with a phosphor screen analyzer (Fuji SLA 300 Raytest PhosphorImager). The kobs values were determined by plotting the fraction of substrate versus time and fitting to the following single-exponential equation:
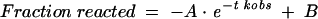 |
(1) |
where A stands for the amplitude of the exponential time course and B represents the end point of the cleavage reaction, which was typically between 0.8 and 0.9. These parameters were estimated by nonlinear regression analysis using Origin software.
Competitive allosteric ribozyme assay
Ribozyme reactions were carried out under single turnover conditions as described above. To allow competition, both effectors were added in parallel to the ribozyme containing reaction mixture at 0°C and then immediately placed into a water bath at 37°C. The competition coefficient kcompet was obtained as an equivalent to the ratio of the rate constants for the FMN-containing ribozyme reaction to that of noncontaining FMN given by the following equation:
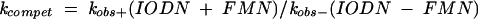 |
(2) |
Effector concentrations were as specified for each reaction setup.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft and the Volkswagen-Stiftung (to M.F.).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2840805.
REFERENCES
- Burgstaller, P. and Famulok, M. 1994. Isolation of RNA aptamers for biological cofactors by in vitro selection. Angew. Chem. Int. Ed. 33: 1084–1087. [Google Scholar]
- Butcher, S.E., Allain, F.H., and Feigon, J. 1999. Solution structure of the loop B domain from the hairpin ribozyme. Nat. Struct. Biol. 6: 212–216. [DOI] [PubMed] [Google Scholar]
- Chowrira, B.M. and Burke, J.M. 1991. Binding and cleavage of nucleic acids by the “hairpin” ribozyme. Biochemistry 30: 8518–8522. [DOI] [PubMed] [Google Scholar]
- Earnshaw, D.J., Masquida, B., Muller, S., Sigurdsson, S.T., Eckstein, F., Westhof, E., and Gait, M.J. 1997. Inter-domain cross-linking and molecular modelling of the hairpin ribozyme. J. Mol. Biol. 274: 197–212. [DOI] [PubMed] [Google Scholar]
- Esteban, J.A., Banerjee, A.R., and Burke, J.M. 1997. Kinetic mechanism of the hairpin ribozyme. Identification and characterization of two nonexchangeable conformations. J. Biol. Chem. 272: 13629–13639. [DOI] [PubMed] [Google Scholar]
- Esteban, J.A., Walter, N.G., Kotzorek, G., Heckman, J.E., and Burke, J.M. 1998. Structural basis for heterogeneous kinetics: Reengineering the hairpin ribozyme. Proc. Natl. Acad. Sci. 95: 6091–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, P., Suri, A.K., Fiala, R., Live, D., and Patel, D.J. 1996. Molecular recognition in the FMN-RNA aptamer complex. J. Mol. Biol. 258: 480–500. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. 2002. The catalytic mechanism of the hairpin ribozyme. Biochem. Soc. Trans. 30: 1109–1115. [DOI] [PubMed] [Google Scholar]
- Ferre-D’amare, A.R. and Rupert, P.B. 2002. The hairpin ribozyme: From crystal structure to function. Biochem. Soc. Trans. 30: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Hampel, K.J., Pinard, R., and Burke, J.M. 2001. Catalytic and structural assays for the hairpin ribozyme. Methods Enzymol. 341: 566–580. [DOI] [PubMed] [Google Scholar]
- Hartig, J.S., Najafi-Shoushtari, S.H., Grune, I., Yan, A., Ellington, A.D., and Famulok, M. 2002. Protein-dependent ribozymes report molecular interactions in real time. Nat. Biotechnol. 20: 717–722. [DOI] [PubMed] [Google Scholar]
- Hartig, J.S., Grune, I., Najafi-Shoushtari, S.H., and Famulok, M. 2004. Sequence-specific detection of microRNAs by signal-amplifying ribozymes. J. Am. Chem. Soc. 126: 722–723. [DOI] [PubMed] [Google Scholar]
- Kertsburg, A. and Soukup, G.A. 2002. A versatile communication module for controlling RNA folding and catalysis. Nucleic Acids Res. 30: 4599–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, Y., Nobuoka, K., Karino-Abe, N., Matsuda, A., and Ohtsuka, E. 2002. In vitro selection of hairpin ribozymes activated with short oligonucleotides. Biochemistry 41: 9090–9098. [DOI] [PubMed] [Google Scholar]
- Meli, M., Vergne, J., and Maurel, M.C. 2003. In vitro selection of adenine-dependent hairpin ribozymes. J. Biol. Chem. 278: 9835–9842. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F. and Uhlenbeck, O.C. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180: 51–62. [DOI] [PubMed] [Google Scholar]
- Najafi-Shoushtari, S.H., Mayer, G., and Famulok, M. 2004. Sensing complex regulatory networks by conformationally controlled hairpin ribozymes. Nucleic Acids Res. 32: 3212–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinard, R., Lambert, D., Heckman, J.E., Esteban, J.A., Gundlach, C.W., Hampel, K.J., Glick, G.D., Walter, N.G., Major, F., and Burke, J.M. 2001. The hairpin ribozyme substrate binding-domain: A highly constrained D-shaped conformation. J. Mol. Biol. 307: 51–65. [DOI] [PubMed] [Google Scholar]
- Pörschke, D., Burke, J.M., and Walter, N.G. 1999. Global structure and flexibility of hairpin ribozymes with extended terminal helices. J. Mol. Biol. 289: 799–813. [DOI] [PubMed] [Google Scholar]
- Robertson, M.P. and Ellington, A.D. 1999. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 17: 62–66. [DOI] [PubMed] [Google Scholar]
- Rupert, P.B. and Ferre-D’Amare, A.R. 2001. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410: 780–786. [DOI] [PubMed] [Google Scholar]
- Soukup, G.A. and Breaker, R.R. 1999a. Engineering precision RNA molecular switches. Proc. Natl. Acad. Sci. 96: 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1999b. Nucleic acid molecular switches. Trends Biotechnol. 17: 469–476. [DOI] [PubMed] [Google Scholar]
- Vaish, N.K., Jadhav, V.R., Kossen, K., Pasko, C., Andrews, L.E., McSwiggen, J.A., Polisky, B., and Seiwert, S.D. 2003. Zeptomole detection of a viral nucleic acid using a target-activated ribozyme. RNA 9: 1058–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauleon, S. and Muller, S. 2003. External regulation of hairpin ribozyme activity by an oligonucleotide effector. ChembioChem. 4: 220–224. [DOI] [PubMed] [Google Scholar]
- Walter, N.G., Burke, J.M., and Millar, D.P. 1999. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat. Struct. Biol. 6: 544–549. [DOI] [PubMed] [Google Scholar]
- Wang, D.Y., Lai, B.H., Feldman, A.R., and Sen, D. 2002. A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res. 30: 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, X., Kim, H., Pereira, M.J., Babcock, H.P., Walter, N.G., and Chu, S. 2002. Correlating structural dynamics and function in single ribozyme molecules. Science 296: 1473–1476. [DOI] [PubMed] [Google Scholar]