Abstract
Although it is generally accepted that transcription and translation are spatially separated in eukaryotes, a number of recent observations have called this belief into question. In particular, several studies have shown that parts of the translation machinery, including ribosomal proteins, can be found associated with sites of active transcription in metazoans. Here we describe results of chromatin immunoprecipitation (ChIP) experiments designed to determine whether ribosomal proteins associate with nascent transcripts in Saccharomyces cerevisiae and whether this association reflects a functional engagement of the translation machinery. We find that HAT-tagged ribosomal proteins can be detected in association with nascent RNAs in budding yeast. However, our data clearly indicate that this binding is independent of transcript translatability, so is therefore not indicative of nuclear translation.
Keywords: nuclear translation, chromatin immunoprecipitation, ribosomal proteins, nascent RNA
INTRODUCTION
Whereas cotranscriptional translation is the rule in bacteria, in eukaryotes these two processes are generally believed to occur in separate cellular compartments. A major rationale for this strict spatial segregation of transcription and translation in eukaryotes is derived from the need for extensive post-transcriptional processing of nascent transcripts in these organisms. Most pertinent is the excision of introns from pre-mRNAs—for most eukaryotic messages, intron removal is required to generate the intended open reading frame.
Although the idea that transcription and translation are spatially separated in eukaryotes is now widely accepted, this was not always the case. Indeed, prior to the discovery of introns in the late 1970s, the possibility of nuclear translation was openly debated. Numerous earlier studies had been interpreted to suggest that some eukaryotic protein synthesis does occur within the nucleus (Goidl 1978). Lines of experimentation that produced the most compelling evidence at the time were (1) whole cell radioactive amino acid labeling time courses, which seemed to show that radioactive proteins appeared more rapidly in cell nuclei than one might expect from cytoplasmic synthesis alone; (2) incorporation of radioactive amino acids into protein by isolated nuclei; and (3) the existence of what seemed to be nuclear polysomes. However, because these early experiments pushed the limits of the technology available at that time, the evidence was never entirely definitive and invariably left openings for alternate interpretations (Allen 1978).
More recently the possibility of nuclear translation was rekindled. Initial evidence pointing in this direction came from mechanistic studies of nonsense-mediated mRNA decay (NMD) in mammalian cells. NMD is the process by which mRNAs containing prematurely truncated open reading frames are preferentially eliminated. Although translation of such aberrant messages is prerequisite for their destruction, biochemical separation of the nuclear and cytoplasmic compartments revealed that in many cases such mRNAs are apparently destroyed prior to their release from the nucleus (for reviews, see Maquat 1995; Li and Wilkinson 1998; Hentze and Kulozik 1999). Additionally, immunoprecipitation studies have provided evidence that in mammalian cells transcripts are subjected to NMD prior to replacement of the nuclear cap binding complex by the cytoplasmic cap binding protein, eIF4E (Ishigaki et al. 2001). Further, two different blocks to nuclear export failed to disrupt down-regulation of defective T-cell receptor messages, suggesting that NMD of these mRNAs can occur within the nucleus proper (Buhler et al. 2002). Finally, two phenomena dubbed nonsense-associated alternative splicing (NAS) and stop codon-mediated suppression of splicing (SOS), in which pre-mRNA splicing patterns are apparently influenced by ORF integrity, have been interpreted to indicate the existence of mechanisms for detecting reading frame that act contemporaneously with pre-mRNA splicing (Wang et al. 2002; Wachtel et al. 2004).
Numerous studies have also indicated that much of the translation machinery is present in the nucleus. Because they are assembled in the nucleolus and must migrate to the nuclear envelope for export to the cytoplasm, ribosomal subunits are necessarily found throughout the nucleoplasm. However, other components of the translation machinery that would not necessarily need to ever access the nucleus have also been found there. These include amino acyl tRNAs synthetases, charged tRNAs, the translation initiation factors eIF2α, eIF3, eIF4E, eIF4G, and eIF5A, the translation elongation factor eEF1, and the release factors eRF1 and eRF3 (Etchison and Etchison 1987; Lund and Dahlberg 1998; Dostie et al. 2000; Brogna et al. 2002; Studwick and Borden 2002; Dahlberg et al. 2003). In a paper that sparked considerable attention, Cook and colleagues presented in situ localization studies suggesting that discrete sites within isolated nuclei can incorporate labeled amino acids into protein. The level of this nuclear incorporation was sensitive to both translation and transcription inhibitors, suggesting to the authors that the two processes may be coupled in eukaryotes as they are in bacteria. Immunogold labeling further indicated that newly made polypeptides within the nucleus colocalized with newly made RNA, as well as with parts of the translation machinery (Iborra et al. 2001). Similar conclusions were advanced in a paper where in situ visualization studies were used to demonstrate colocalization of ribosomal proteins and translation factors with sites of RNA Pol II transcription on Drosophila polytene chromosomes (Brogna et al. 2002).
Although the authors of the papers cited above interpreted their data as indicative of nuclear translation, other interpretations have been proposed (Dahlberg et al. 2003; Dahlberg and Lund 2004). In particular, it has been noted that association of the translation machinery with active chromatin does not necessarily reflect function (Dahlberg et al. 2003). In this article, we describe the results of chromatin immunoprecipitation (ChIP) experiments designed to determine whether ribosomal proteins associate with active genes in Saccharomyces cerevisiae and, if so, whether or not this association reflects any functional engagement of the translation apparatus with nascent transcripts.
RESULTS
Choice and verification of tagged ribosomal proteins
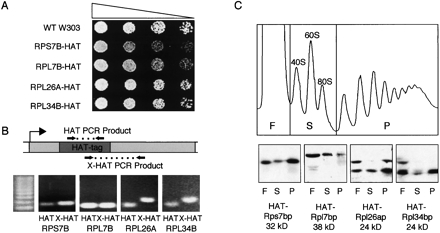
To assess the association of ribosomal proteins with sites of active transcription in S. cerevisiae, we utilized chromatin immunoprecipitation (ChIP). Strains encoding HAT-tagged ribosomal proteins were obtained from the TRIPLES library (Ross-Macdonald et al. 1999). Each of these strains, which were derived via random transposon insertion followed by Cre-mediated loxR-loxP recombination, contains the mTn-3xHA (HAT) tag within the coding region of the specified gene. For our purposes, we chose only those insertions reported to have high protein expression (Ross-Macdonald et al. 1997; Kumar et al. 2000) and demonstrating haplosufficiency (Fig. 1A). This set comprised one protein from the small ribosomal subunit, Rps7bp, and three from the large subunit, Rpl7bp, Rpl26ap, and Rpl34bp. Individual HAT-tag insertions were verified by PCR analysis with both HAT-specific and gene-specific primer pairs (Fig. 1B). Western analysis of sucrose gradient fractions verified that some fraction of all four tagged proteins cosedimented with polysomes, indicating that ribosomes containing these proteins were active for translation (Fig. 1C). However, to a varying degree, a significant fraction of each tagged protein also sedimented at the top of the gradient, presumably reflecting free protein. To ensure maximal incorporation of these tagged proteins into ribosomes, all strains were maintained in the haploid state.
FIGURE 1.
Confirmation of HAT-tagged strains. (A) Serial dilution series (5×) of haploid strains containing HAT-tagged ribosomal proteins on YPD plates. (B) HAT-tag genotype verification using PCR with primer sets as diagrammed: one set internal to the HAT-tag (HAT) and one extending from the HAT-tag into the tagged ribosomal protein gene (HAT-X). (C) Representative polysome profile and Western analysis of proteins TCA precipitated from 10% to 50% sucrose gradient fractions. Fractions were pooled into free proteins (F), 40S 60S subunits and 80S monosomes (S), and polysomes (P).
Validation of ChIP conditions for detection of transcript-associated proteins
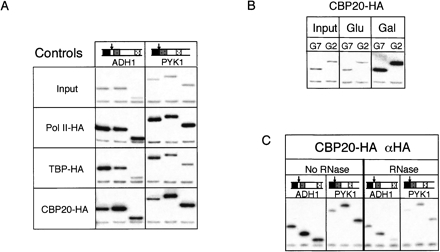
Initial ChIP experiments were performed with HA-tagged versions of TATA-binding protein (TBP-HA) and Rpb3p (Komarnitsky et al. 2000), the third largest subunit of RNA polymerase II (referred to hereafter as Pol II-HA to avoid confusion with the ribosomal proteins). Previous ChIP studies with TBP-HA and Pol II-HA had revealed distinct patterns of association across active genes (Komarnitsky et al. 2000). Whereas TBP-HA exhibits much higher association with promoters than with downstream transcribed regions, Pol II-HA yields high ChIP signals both at promoters and throughout coding regions. As previously reported, the highly transcribed ADH1 and PYK1 loci yielded reproducibly strong ChIP signals with the expected patterns in both the TBP-HA and Pol II-HA strains (Fig. 2A). Control experiments performed with this same anti-HA antibody but using chromatin prepared from a strain lacking any HA- or HAT-tag yielded no enrichment of any loci tested, confirming the specificity of the antibody (data not shown). These control experiments thus demonstrated that the ChIP method worked in our hands and validated the specificity of the anti-HA antibody (12CA5 Roche) used in all subsequent experiments.
FIGURE 2.
Chromatin immunoprecipitation (ChIP) using HA-tagged TBP, RNA Pol II, and CBP20. (A) Radiolabeled PCR products for the ADH1 and PYK1 genes from Input and ChIP samples of strains containing HA-tagged Rpb3p (Pol II-HA), TATA-binding protein (TBP-HA), and nuclear cap-binding protein (Cbp20p-HA). (B) PCR of GAL7 (G7) and GAL2 (G2) after ChIP of CBP20-HA strain grown in media containing either glucose or galactose as the carbon source. (C) ChIP of chromatin from CBP20-HA cells treated or not with RNases A and T1 prior to IP. Diagrams above gene names represent locations of individual primer sets within that gene (black, shaded, and polka dot boxes); arrows indicate 5′ end of ORF. Each PCR reaction contained two sets of primers, one for the gene of interest (upper band) and one for the extragenic control from chromosome V (lower band; Komarnitsky et al. 2000).
Both TBP and Pol-II interact directly with DNA. To determine whether our ChIP conditions could also detect proteins associated indirectly with the DNA via interaction with nascent RNA transcripts, we next examined the ability of HA-tagged Cbp20p to precipitate active genes. Cbp20p is part of the nuclear complex that binds the 7-methylguanosine cap added to all Pol II transcripts. Much like Pol II-HA, Cbp20p-HA exhibited strong association throughout the coding regions of ADH1 and PYK1 (Fig. 2A). However, Cbp20p yielded lower promoter region signals than either Pol II or TBP. This lower association with the promoter is consistent with the 7-methylguanosine cap being added to the RNA only after the start of transcription and suggests that Cbp20p does not associate with the Pol II pre-initiation complex.
That Cbp20p association is limited to active genes was verified by comparing Cbp20p-HA ChIP signals for the GAL2 and GAL7 loci from cells grown either in glucose or galactose (Fig. 2B). Whereas α-HA ChIP gave no enrichment of these loci when cells were grown in glucose, substantial enrichment was observed for cells grown in galactose. To assess the RNA dependence of this association, we also performed RNase digestions prior to immunoprecipitation (Fig. 2C). The fixation time in these latter samples was decreased from 20 to 5 min, in accordance with results from Abruzzi et al. (2004), who found that shortening the fixation time allowed for better discrimination between RNA-bound and DNA-bound factors. Our observation that the Cbp20p ChIP signals were sensitive to RNase indicates that Cbp20p association with active chromatin is due to its interaction with RNA as opposed to either direct interaction with the DNA or indirect interaction with DNA-associated proteins.
RNA-dependent association of ribosomal proteins with active genes
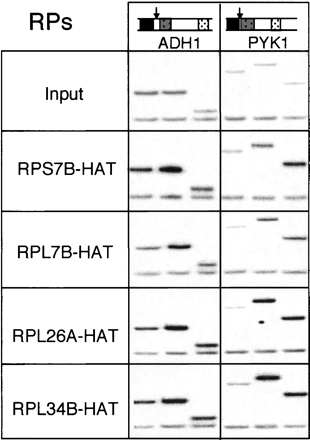
Initial ChIP experiments with the HA-tagged ribosomal proteins revealed patterns similar to those observed with Cbp20p-HA, albeit at somewhat reduced signal levels. That is, all four ribosomal proteins exhibited much stronger association with the ADH1 and PYK1 genes than with the intergenic region of chromosome V (Fig. 3). Further, within the ADH1 and PYK1 loci, all the ribosomal proteins yielded higher signals with the transcribed regions than with the promoters. Quantitation of the bands in Figure 3 showed that ChIP of ribosomal proteins enriched coding regions over the intergenic region between two- and ninefold compared to input. Although the magnitude of the ribosomal protein ChIP signals varied somewhat from experiment to experiment (e.g., cf. Fig. 3 and Fig. 4B; data not shown), they consistently showed the same patterns of association across genes and the data shown are typical.
FIGURE 3.
Radiolabeled PCR products for the ADH1 and PYK1 genes from Input and ChIP samples of strains containing indicated HAT-tagged ribosomal proteins.
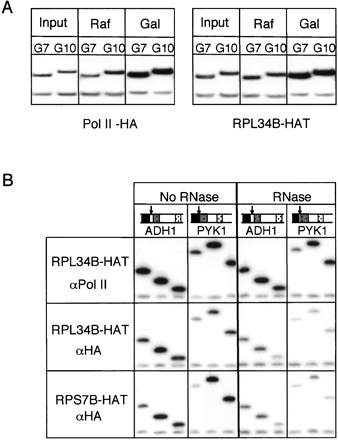
FIGURE 4.
Transcription and RNA dependence of ribosomal protein ChIP. (A) Radiolabeled PCR products for GAL7 and GAL10 from ChIP samples from indicated strains grown either in raffinose or after a 60-min shift to galactose. (B) ChIP of chromatin from RPL34B-HAT and RPS7B-HAT cells treated or not with RNases A and T1 prior to IP with α-RNA pol II (8WG16) and/or α-HA (12CA5).
Analysis of other gene loci revealed that the ChIP signal strength for ribosomal proteins roughly correlated with calculated transcription rates for those genes (Holstege et al. 1998; data not shown). To confirm that the association was indeed transcription dependent, we examined ChIP patterns for two galactose inducible loci, GAL7 and GAL10. For both Pol II-HA and Rpl34bp-HAT, cells grown in raffinose exhibited no enrichment of the GAL loci over input upon ChIP. However, ChIP signals for both proteins at these loci increased dramatically upon shifting the cells to galactose (Fig. 4A).
To determine whether the ribosomal protein ChIP signals were RNA dependent, we treated sheared chromatin with RNase prior to immunoprecipitation as performed previously for Cbp20p (see above; Fig. 2C). For this experiment we precipitated the same chromatin preparation with antibodies recognizing either endogenous Pol II (8WG16) or the HAT-tagged ribosomal proteins. Whereas pulldown with the α-Pol II antibody was largely unaffected by prior RNase treatment, αHA ChIP signals were substantially decreased (Fig. 4B). Thus, association of ribosomal proteins with active genes in S. cerevisiae is RNA dependent.
Ribosomal protein association is independent of transcript translatability
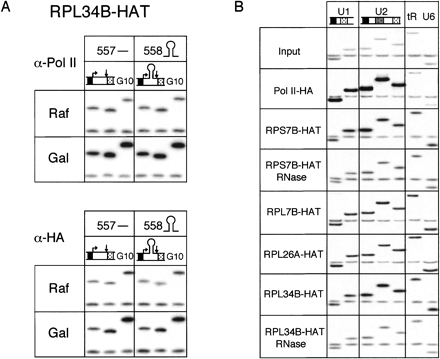
To test whether the association of ribosomal proteins with nascent transcripts represented a functional interaction analogous to mRNA translation, we next examined the effects of mRNA secondary structure on the ChIP signals. A galactose-inducible construct encoding a 26-nt hairpin in the 5′ UTR was previously shown to have a rate of transcription similar to the parental construct lacking the hairpin, but its rate of translation was reduced >5000-fold (Muhlrad et al. 1995). Presumably, the small subunit of the ribosome cannot resolve this extremely stable hairpin and becomes stalled on the 5′ UTR. For this experiment, plasmids encoding the parental (pRP557) and hairpin-containing (pRP558) PGK1/lacZ fusion constructs were transformed into the yeast strain expressing HAT-tagged Rpl34bp, as this tagged protein routinely yielded the highest ChIP signal among all the tagged ribosomal proteins. As above, the same chromatin preparations were independently precipitated with antibodies recognizing either endogenous Pol II (8WG16) or the HAT tag.
Upon galactose induction there was a noticeable increase in the association of both Pol II and Rpl34bp with the coding region of the PGK1/lacZ fusion construct. Further, compared to Pol II, Rpl34bp exhibited no difference in its association with the coding region dependent on the absence or presence of the secondary structure (Fig. 5A). That is, while the Rpl34bp coding region signal was slightly less intense for pRP558 than pRP557 (12% difference), precipitation of the same sample with α-pol II antibodies yielded almost exactly the same difference (13%). This indicated that the association of Rpl34bp more closely followed the transcriptional activity of each construct than its translatability. Thus, association of Rpl34bp with PGK1/lacZ fusion transcripts exhibited no detectable dependence on their translatability.
FIGURE 5.
ChIP of active genes by ribosomal proteins is independent of gene translatability. (A) Radiolabeled PCR products from gal-inducible PGK1/LacZ fusion gene ± 5′-UTR hairpin and chromosomal GAL10 (G10) from cells co-expressing Rpl34bp-HAT and grown either in raffinose or galactose. ChIP was performed with α-RNA pol II (8WG16) and/or α-HA (12CA5), as indicated. (B) PCR products of snRNAs U1 and U2 (pol II transcripts), snRNA U6, and tRNALysCUU (pol III transcripts) from ChIP performed using α-HA (12CA5) on chromatin from HAT-tagged ribosomal proteins, including ChIP of chromatin from RPL34B-HAT and RPS7B-HAT cells treated or not with RNases A and T1 prior to IP.
To determine whether ribosomal protein association with nascent transcripts is limited to protein-coding genes, we next analyzed their association with gene loci expressing noncoding RNAs. Loci tested included both Pol II-transcribed (U1 and U2 snRNAs) and Pol III-transcribed (U6 snRNA and tRNALysCUU) genes. As expected, Pol II-HA exhibited significant association with the U1 and U2 loci, but not the U6 or tRNALysCUU loci (Fig. 5B). In contrast, all ribosomal proteins tested exhibited comparable association with all four loci. RNase treatment of Rps7bp-HAT and Rpl34bp-HAT prior to precipitation confirmed that ribosomal protein association with these genes is RNA-dependent (Fig. 5B). Examination of the chromosomal features in the regions of these genes (http://www.yeastgenome.org/) revealed that in no case is there a predicted ORF encoded by the opposite DNA strand (data not shown). We therefore conclude that ribosomal protein association with nascent RNA is limited neither to protein-coding genes nor Pol II transcripts.
DISCUSSION
In this article, we have demonstrated that ribosomal proteins can associate with sites of active transcription in S. cerevisiae as detected by ChIP. RNase sensitivity of the ChIP signals indicates that ribosomal protein association with active gene loci occurs via nascent RNA. However, the interactions are neither limited to transcripts destined for translation nor those transcribed by RNA Pol II. We therefore conclude that the association of ribosomal proteins with nascent transcripts in S. cerevisiae does not reflect active engagement of the translation machinery in the nuclear compartment. At this juncture we cannot deduce whether the associations we observed reflect fully assembled ribosomes or merely free ribosomal proteins.
The data presented here offer an alternate explanation for the association of ribosomal proteins with sites of active transcription as observed by indirect immunofluorescence of Drosophila polytene chromosomes (Brogna et al. 2002). In that study, antibodies recognizing endogenous ribosomal proteins were found to bind polytene chromosomes with a pattern largely overlapping the sites occupied by elongating RNA Pol II. Analogous to our ChIP results in S. cerevisiae, anti-ribosomal protein antibodies in that study could be recruited to new loci upon transcriptional induction, and the resultant immunofluorescence signals were sensitive to RNase. Although these results were previously interpreted to reflect functional engagement of the translation apparatus with nascent transcripts on polytene chromosomes, at least two pieces of our data are inconsistent with that explanation: the association of ribosomal proteins with (1) an mRNA defective for translation and (2) RNAs that are not normally translated (Fig. 5). Thus, at least in budding yeast, the association of ribosomal proteins with sites of active transcription does not necessarily reflect the presence of active ribosomes.
Other data that have been offered to support the case for nuclear translation include evidence of a nuclear population of translation factors (Etchison and Etchison 1987; Lund and Dahlberg 1998; Dostie et al. 2000; Brogna et al. 2002; Studwick and Borden 2002; Dahlberg et al. 2003), perhaps associated with elongating RNA Pol II (Brogna et al. 2002), and the association of newly made proteins with newly made RNAs (Iborra et al. 2001). During the course of our studies, in addition to the tagged ribosomal proteins described here, we also created numerous strains expressing HA-tagged translation factors including eIF4A, eIF4E, eIF4G, eRF1, and eRF3. However, none of the latter strains ever yielded reproducible ChIP signals (C. Giorgi and M.J. Moore, unpubl. results). These findings are consistent with a report from Gorlich and coworkers that most translation initiation, elongation, and termination factors are strictly excluded from yeast nuclei (Bohnsack et al. 2002). We also performed a number of ChIP experiments using several different tags incorporated at the N termini of multiple polypeptides in an attempt to demonstrate association of nascent peptides with their own genes (data not shown). Again, none of these experiments ever yielded any credible signal. Thus, we conclude that there is no cotranscriptional translation in budding yeast.
This conclusion is in agreement with a recent paper reporting that NMD does not occur within the yeast nucleus (Kuperwasser et al. 2004). In that study, S. cerevisiae strains temperature sensitive for mRNA export were exploited to examine NMD of PTC-containing reporters driven by a heat shock promoter. Because no NMD was observed upon restriction of nascent transcripts to the nucleus, the authors concluded that NMD, and therefore translation, is strictly a cytoplasmic event in budding yeast. That there is no nuclear translation in budding yeast is further supported by the observation that 40S ribosomal subunits are not found in polysomes until the 18S rRNA undergoes a final maturation step (conversion of 20S to 18S) in the cytoplasm (Venema and Tollervey 1999).
At this juncture, the preponderance of the evidence overwhelmingly disfavors the existence of nuclear translation in budding yeast. What remains open for debate is the extent to which the yeast data rule out the possibility of nuclear translation in other organisms. While several papers have called into question the evidence supporting nuclear translation and/or nuclear reading frame recognition in mammalian cells (Dahlberg et al. 2003; Nathanson et al. 2003; Dahlberg and Lund 2004; Buhler and Muhlemann 2005; Mohn et al. 2005), we predict that the debate will never entirely disappear until it has been definitively proven on which side of the nuclear envelope nucleus-associated NMD occurs in mammals.
Although the interaction of ribosomal proteins with nascent RNAs observed here is not apparently due to active engagement of the translation apparatus, it is possible that these associations could reflect other functional roles for these proteins. Contrary to the expectation that ribosomal proteins might be expressed at similar relative stoichiometries, recent genome-wide protein expression level analyses have revealed that yeast ribosomal proteins vary in their abundance by over two orders of magnitude (from <103 to >105 copies per cell). This difference was suggested to reflect other roles of these very abundant cellular proteins (Beyer et al. 2004). Indeed, numerous extraribosomal functions have been previously noted for eukaryotic ribosomal proteins, several of which involve modulation of transcription or nuclear pre-mRNA splicing (Wool 1996). Yeast Rps20p participates in anti-termination by RNA Pol III (Hermann-Le Denmat et al. 1994), while the human S14 protein has been shown to bind to and regulate transcription of its own gene (Tasheva and Roufa 1995). Other ribosomal proteins bind regulatory elements in their own pre-mRNAs and inhibit splicing when the protein is in excess (Bozzoni et al. 1984; Presutti et al. 1991; Fewell and Woolford 1999; Vilardell et al. 2000; Ivanov et al. 2005). A difference between these previously reported extraribosomal activities in the nucleus and the interactions between ribosomal proteins and nascent transcripts described here is their specificity. Whereas the known auto-inhibitory feedback mechanisms tend to involve recognition of specific sequences or secondary structures in target transcripts, the interactions we observed correlated more closely with transcription rate than with particular sequences. Therefore, if the interactions of ribosomal proteins with nascent transcripts detected here do reflect some functional role, it is likely a more global one than those previously described.
METHODS
Yeast strains and plasmids
All yeast strains (Table 1) were grown at 30°C using 2% glucose as a carbon source unless otherwise noted. HAT-tagged strains were grown in YPD. Strains expressing Cbp20p-HA, Pol II-HA, and TBP-HA were grown in media lacking either leucine (Cbp20p-HA and Pol II-HA) or uracil (TBP-HA) to ensure maintenance of the tagged vector. PCR reactions for strain verification (Fig. 1) were performed as for ChIP (see below), except without radioactivity. PCR products were separated on 1% agarose gels and visualized by ethidium bromide staining.
TABLE 1.
Strains and plasmids used in this study
| Name | Strain/plasmid | Genotype and notes | Source or reference |
| Strains | |||
| W303 | MATa, ade2, his3, leu2, trp1, ura3, lys2 | M. Rosbash | |
| Pol II-HA | Z780 | MATa; ura3-52; leu2-3,112; trp1-1, his3 200; RPB3-(HA3)::LEU2 | Komarnitsky et al. (2000) |
| TBP-HA | Y611 | MATa, ade2, his3, leu2, trp1, ura3 [3XHA-TBP on YlpLac211 (URA3)] | Kuras and Struhl (1999) |
| Cbp20p-HA | Y394 | MATa, leu2-3; leu2-112; ura3-52; trp1-289; arg4; ade2; mud13::ADE2 [p366 LEU2-CEN; MUD13-(HA)] | Colot et al. (1996) |
| Y800 | Diploid parental strain : MATa/α leu2-98/leu2-98; cry1R/CRY1; ade2-101/ade2-101 HIS3/his3-200; ura3-52/ura3-52; can1R/CAN1; lys2-801/lys2-801;CHY2/chy2R;trp1-1/TRP1;/Ciro | Ross-Macdonald et al. (1997) | |
| Rpl7bp-HAT | MJM74 | Y800 MATa–RPL7B-(HAT) Haploid derived from TRIPLES Strain ID:V23C6 HAT-tag@144 of 244 amino acids in RPL7B/YPL198W | TRIPLES Database ygac.med.yale.edu/triples/ |
| Rpl26ap-HAT | MJM63 | Y800 MATα –RPL26A-(HAT) Haploid strain derived from TRIPLES Strain ID:V29H9 HAT-tag@46 of 127 amino acids in RPL26A/YLR344W | This study and TRIPLES Database |
| Rpl34bp-HAT | MJM39 | Y800 MATα –RPL34B-(HAT) Haploid derived from TRIPLES Strain ID:V75C4 HAT-tag@51 of 121 amino acids in RPL34B/YIL052C | This study and TRIPLES Database |
| Rps7bp-HAT | MJM33 | Y800 MATα –RPS7B-(HAT) Haploid derived from TRIPLES Strain ID:V141G1 HAT-tag@104 of 190 amino acids in RPS7B/YNL096C | This study and TRIPLES Database |
| Plasmids | |||
| pRP557 | GAL1-PGK1-lacZ fusion construct | Muhlrad et al. (1995) | |
| pRP558 | GAL1-SLPGK1-lacZ fusion construct (pRP557 plus 5′ UTR stem–loop) | Muhlrad et al. (1995) |
Polysome fractionation and Western analysis
Polysome fractionation of each HAT-tagged ribosomal protein strain was performed as in Arava et al. (2003) except that gradients were centrifuged at 38,000 rpm. Based on the A260 traces, fractions were pooled into three categories: (1) free proteins, (2) 40S and 60S subunits plus 80S monosomes, and (3) polysomes. TCA-precipatated proteins were separated on a 10% SDS-polyacrylamide gel. Western blots were probed with α-HA antibody (12CA5 Roche), developed using Enhanced Chemifluorescence reagent (Amersham) and scanned using a Molecular Dynamics Phosphorimager.
Chromatin immunoprecipitation
ChIP was performed using methods based on Komarnitsky et al. (2000) and Abruzzi et al. (2004) with differences in the pretreatment of cells, cell lysis, and sonication as described below. Prior to cross-linking, cells were treated with 100 μg/mL cycloheximide for 5 min. Cells were lysed for 3 min at 4°C using a Mini-Bead Beater 8 (BioSpec Products) set to “homogenize.” DNA was sheared by sonication for 2.5 min (5 sec on, 5 sec off) at 25% amplitude using a Branson Digital Sonifier 250. Immunoprecipitations were performed using the monoclonal antibodies 8WG16 (Covance) against RNA pol II (recognizes the C-terminal heptapeptide repeat present on the largest subunit of pol II with a bias for highly phosphorylated forms; Thompson et al. 1989) and 12CA5 (Roche) against the HA epitope (used for both HA and HAT tags). Protein-A Sepharose 4B (Zymed) beads were preincubated with antibody for 1 to 2 h at 4°C prior to IP overnight at 4°C.
RNase A/T1 treatment for assessing RNA-dependent associations was performed as previously described (Abruzzi et al. 2004) except that samples were diluted 1:1 with milli-Q water prior to RNase addition to lower the SDS concentration to 0.05% and then incubated for 1 h at room temperature instead of 30 min. Cross-linking time was reduced to 5 min from 20 min for cells subjected to RNase treatment (Abruzzi et al. 2004).
ChIP PCR reactions contained 0.5 U of platinum Taq DNA polymerase (Invitrogen), 0.1 mM dNTP mixture, 0.5 μM each primer (Table 2), 3.5 mM MgCl2, 0.03 μCi [α32P]-dATP, and 1× platinum Taq PCR buffer (20 mM Tris-HCl at pH 8.4, 500 mM KCl). All PCR reactions were performed using the following program: 2 min at 94°C, then 26–28 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min, followed by 72°C for 4 min.
TABLE 2.
Primer sets used in the study
| Gene | Region | Orientation/position | Sequence |
| ADH1 | Promoter | ADH1.F.−235 | TTCCTTCCTTCATTCACGCACACT |
| ADH1.R.−13 | GTTGATTGTATGCTTGGTATAGCTTG | ||
| CDS1-1 | ADH1.F.146 | ACGCTTGGCACGGTGACTG | |
| ADH1.R.372 | ACCGTCGTGGGTGTAACCAGA | ||
| CDS1-2 | ADH1.F.203 | AAGGTGCCGGTGTCGTTGTC | |
| ADH1.R.393 | GGTAGCGTATTGTTGGAAAG | ||
| CDS2 | ADH1.F.844 | TTCAACCAAGT CGTCAAGTCCATCTCTA | |
| ADH1.R.1013 | ATTTGACCCTTTTCCATCTTTTCGTAA | ||
| GAL2 | GAL2.F.32 | TTTCACAGCAACCCCAAG | |
| GAL2.R.411 | ATACCACCAAAGGCACAG | ||
| GAL7 | GAL7.F.709 | GTTGTTGTTCCATACTGGG | |
| GAL7.R.964 | GTAAAGGTGGGTAGAAATG | ||
| GAL10 | GAL10.F.823 | TGTCGTGAGTGGAACTTGGGTT | |
| GAL10.R.1084 | GCATATCTTCAGCGGAAAATCTGGC | ||
| HAT-tag | HAT-tag.F.34 | GTTAAGGCGGCCATTGAAGGTAG | |
| HAT-tag.R.222 | GAGCGTAATCTGGAACGTCATATGGATAGG | ||
| HAT-tag.F.148 | CTGACTATGCGGGCTATCC | ||
| Intergenic chromosome V | IntergenicV-1.F | GGCTGTCAGAATATGGGGCCGTAGTA | |
| IntergenicV-2.R | CACCCCGAAGCTGCTTTCACAATAC | ||
| PYK1 | Promoter | PYK1.F.−327 | GAATGCTTGTGATGTCT TCCAAGT |
| PYK1.R.−23 | TGATTGGTGTCTTGTAAATAGAAACA | ||
| CDS1 | PYK1.F.37 | GTTGCTGGTTCTGACTTGAG | |
| PYK1.R.406 | CCTTGGTGATGTTCTTGTAG | ||
| CDS2 | PYK1.F.802 | GGTATTGAAATCCCAGCCCCAGAAG | |
| PYK1.R.1059 | GACAGCGGTTTCAGCCATAGTG | ||
| RPL7B | Across HAT-tag | RPL7B.F.1093 | AACAAGATTCCACCTAAGCC |
| RPL7B.R.1286 | TGATCTTACCGAAACCTCTC | ||
| RPL26A | Across HAT-tag | RPL26A.F.558 | GGAGTTGAGAGCTCAATACGG |
| RPL26A.R.748 | GTGCAAGTTAATTGGAACGG | ||
| RPL34B | Across HAT-tag | RPL34B.F.549 | CAGGTGGTATCTTGCGTGCC |
| RPL34B.R.804 | CCTCAGTTTGTTCCTTAACGACC | ||
| RPS7B | Across HAT-tag | RPS7B.F.537 | TCCAGCTTTGTCTGCATACC |
| RPS7B.R.739 | GAAAACCATGTCTTCCAAAACC | ||
| tRNALysCUU | tRLysCUU.F.−139 | GCACTAGTTGATTCTTGTTCCAACAG | |
| tRLysCUU..R.149 | CCGTTTTTCCCCAGAGCACTTTTA | ||
| U1 | 5′ | U1.F.21 | GAGGAGATCAAGAAGTCCTACTG |
| U1.R.154 | CGCGCCGTATGTGTGTGTG | ||
| 3′ | U1.F.256 | TGTTTCTTCTGCCTGGAG | |
| U1.R.428 | GCCATACCTTCATCATAAAC | ||
| U2 | 5′ | U2.F.47 | CAGTGTAACAACTGAAATGACCTC |
| U2.R.218 | CATGAGCGAAGAAAATAGCAGC | ||
| Mid | U2.F.434 | GGAAGAAATGGTGCTATAGTAGG | |
| U2.R.670 | CAACAACTTTGGCCGATACTTGG | ||
| 3′ | U2.F.951 | ATGACTGCATCTGTTGTTTCAAGG | |
| U2.R.1153 | GAGAGAAACATCAAGCGACCAAAG | ||
| U6 | Mid | U6.F.29 | TGGTCAATTTGAAACAATACAGAG |
| U6.R.143 | AAGTCACGATACTTCACTCGATG | ||
| pPGK-lacZ Fusion | Promoter | GAL1.F.−352 | CGCGCCGCACTGCTCCGAAC |
| pRP557&pRP558 | pPGK1.R.−122 | ATTGACCAATTTATGCAAGT | |
| CDS2 | pPGK1.F.628 | CAAGGTTGCTGACAAGATTC | |
| pPGK1.R.864 | GCCGGAAACCAGGCAAAGCG |
References for sequence source: GAL10 primers, Lei et al. (2001); tRNALysCUU primers from K. Struhl STR2967/STR2968; ADH1, PYK1 and Chromosome V Intergenic primers, Komarnitsky et al. (2000), PPGK-lacZ Fusion primers from C. Giogi (unpubl.).
Acknowledgments
We are grateful to the Buratowski and Rosbash laboratories for their assistance in setting up the ChIP procedure and to S. Buratowski, R. Parker, M. Rosbash, and M. Snyder for yeast strains and plasmids. We express thanks to K. Abruzzi, C. Giorgi, and all the members of the Moore laboratory for their valuable technical assistance and S. Lacadie, C. Giorgi, and K. O’brien for helpful comments on this article. M.J.M. is a Howard Hughes Medical Institute Investigator.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2134305.
REFERENCES
- Abruzzi, K.C., Lacadie, S., and Rosbash, M. 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 23: 2620–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, W.R. 1978. Does protein synthesis occur within the nucleus? Unambiguous evidence still needed. Trends Biochem. Sci. 3: N225–N228. [Google Scholar]
- Arava, Y., Wang, Y., Storey, J.D., Liu, C.L., Brown, P.O., and Herschlag, D. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 100: 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer, A., Hollunder, J., Nasheuer, H.P., and Wilhelm, T. 2004. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell Proteomics 3: 1083–1092. [DOI] [PubMed] [Google Scholar]
- Bohnsack, M.T., Regener, K., Schwappach, B., Saffrich, R., Paraskeva, E., Hartmann, E., and Gorlich, D. 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21: 6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzoni, I., Annesi, F., Beccari, E., Fragapane, P., Pierandrei-Amaldi, P., and Amaldi, F. 1984. Splicing of Xenopus laevis ribosomal protein RNAs is inhibited in vivo by antisera to ribonucleoproteins containing U1 small nuclear RNA. J. Mol. Biol. 180: 1173–1178. [DOI] [PubMed] [Google Scholar]
- Brogna, S., Sato, T.A., and Rosbash, M. 2002. Ribosome components are associated with sites of transcription. Mol. Cell 10: 93–104. [PubMed] [Google Scholar]
- Buhler, M. and Muhlemann, O. 2005. Alternative splicing induced by nonsense mutations in the immunoglobulin mu VDJ exon is independent of truncation of the open reading frame. RNA 11: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler, M., Wilkinson, M.F., and Muhlemann, O. 2002. Intranuclear degradation of nonsense codon-containing mRNA. EMBO Rep. 3: 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot, H.V., Stutz, F., and Rosbash, M. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes & Dev. 10: 1699–1708. [DOI] [PubMed] [Google Scholar]
- Dahlberg, J.E. and Lund, E. 2004. Does protein synthesis occur in the nucleus? Curr. Opin. Cell. Biol. 16: 335–338. [DOI] [PubMed] [Google Scholar]
- Dahlberg, J.E., Lund, E., and Goodwin, E.B. 2003. Nuclear translation: What is the evidence? RNA 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie, J., Lejbkowicz, F., and Sonenberg, N. 2000. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J. Cell Biol. 148: 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison, D. and Etchison, J.R. 1987. Monoclonal antibody-aided characterization of cellular p220 in uninfected and poliovirus-infected HeLa cells: Subcellular distribution and identification of conformers. J. Virol. 61: 2702–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell, S.W. and Woolford Jr., J.L. 1999. Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol. Cell. Biol. 19: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidl, J.A. 1978. Does protein synthesis occur within the nucleus? Good evidence that it does. Trends Biochem. Sci. 3: N225–N228. [Google Scholar]
- Hentze, M.W. and Kulozik, A.E. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96: 307–310. [DOI] [PubMed] [Google Scholar]
- Hermann-Le Denmat, S., Sipiczki, M., and Thuriaux, P. 1994. Suppression of yeast RNA polymerase III mutations by the URP2 gene encoding a protein homologous to the mammalian ribosomal protein S20. J. Mol. Biol. 240: 1–7. [DOI] [PubMed] [Google Scholar]
- Holstege, F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., and Young, R.A. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., Jackson, D.A., and Cook, P.R. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293: 1139–1142. [DOI] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., and Maquat, L.E. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Ivanov, A.V., Malygin, A.A., and Karpova, G.G. 2005. Human ribosomal protein S26 suppresses the splicing of its pre-mRNA. Biochim. Biophys. Acta 1727: 134–140. [DOI] [PubMed] [Google Scholar]
- Komarnitsky, P., Cho, E.J., and Buratowski, S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 14: 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., Cheung, K.H., Ross-Macdonald, P., Coelho, P.S., Miller, P., and Snyder, M. 2000. TRIPLES: A database of gene function in Saccharomyces cerevisiae. Nucleic Acids Res. 28: 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser, N., Brogna, S., Dower, K., and Rosbash, M. 2004. Nonsense- mediated decay does not occur within the yeast nucleus. RNA 10: 1907–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, L. and Struhl, K. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399: 609–613. [DOI] [PubMed] [Google Scholar]
- Lei, E.P., Krebber, H., and Silver, P.A. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 15: 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. and Wilkinson, M.F. 1998. Nonsense surveillance in lymphocytes? Immunity 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Lund, E. and Dahlberg, J.E. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082–2085. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. 1995. When cells stop making sense: Effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1: 453–465. [PMC free article] [PubMed] [Google Scholar]
- Mohn, F., Buhler, M., and Muhlemann, O. 2005. Nonsense-associated alternative splicing of T-cell receptor β genes: No evidence for frame dependence. RNA 11: 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C.J., and Parker, R. 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15: 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson, L., Xia, T., and Deutscher, M.P. 2003. Nuclear protein synthesis: A re-evaluation. RNA 9: 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presutti, C., Ciafre, S.A., and Bozzoni, I. 1991. The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J. 10: 2215–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald, P., Sheehan, A., Roeder, G.S., and Snyder, M. 1997. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 94: 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald, P., Coelho, P.S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K.H., Sheehan, A., Symoniatis, D., Umansky, L., et al. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402: 413–418. [DOI] [PubMed] [Google Scholar]
- Studwick, S. and Borden, K.L. 2002. The emerging roles of translation factor eIF4E in the nucleus. Differentiation 70: 10–22. [DOI] [PubMed] [Google Scholar]
- Tasheva, E.S. and Roufa, D.J. 1995. Regulation of human RPS14 transcription by intronic antisense RNAs and ribosomal protein S14. Genes & Dev. 9: 304–316. [DOI] [PubMed] [Google Scholar]
- Thompson, N.E., Steinberg, T.H., Aronson, D.B., and Burgess, R.R. 1989. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J. Biol. Chem. 264: 11511–11520. [PubMed] [Google Scholar]
- Venema, J. and Tollervey, D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 261–311. [DOI] [PubMed] [Google Scholar]
- Vilardell, J., Chartrand, P., Singer, R.H., and Warner, J.R. 2000. The odyssey of a regulated transcript. RNA 6: 1773–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtel, C., Li, B., Sperling, J., and Sperling, R. 2004. Stop codon-mediated suppression of splicing is a novel nuclear scanning mechanism not affected by elements of protein synthesis and NMD. RNA 10: 1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Chang, Y.F., Hamilton, J.I., and Wilkinson, M.F. 2002. Nonsense-associated altered splicing: A frame-dependent response distinct from nonsense-mediated decay. Mol. Cell 10: 951–957. [DOI] [PubMed] [Google Scholar]
- Wool, I.G. 1996. Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 21: 164–165. [PubMed] [Google Scholar]