Abstract
We have used a compartmentalized in vitro selection method to directly select for ligase ribozymes that are capable of acting on and turning over separable oligonucleotide substrates. Starting from a degenerate pool, we selected a trans-acting variant of the Bartel class I ligase which statistically may have been the only active variant in the starting pool. The isolation of this sequence from the population suggests that this selection method is extremely robust at selecting optimal ribozymes and should, therefore, prove useful for the selection and optimization of other trans-acting nucleic acid catalysts capable of multiple turnover catalysis.
Keywords: trans-acting ribozyme, in vitro compartmentalization, in vitro selection
INTRODUCTION
In vitro selection methods have proven to be quite powerful for generating and optimizing numerous nucleic acid catalysts (Jaeger 1997; Pan 1997). However, all methods used to date select for single turnover catalysts. For example, ribozymes that can ligate primer-binding sites to themselves have been selected (Bartel and Szostak 1993; Jaeger et al. 1999; Robertson and Ellington 1999; Rogers and Joyce 2001), as have ribozymes that can self-cleave (Pan and Uhlenbeck 1992; Jayasena and Gold 1997; Tang and Breaker 2000).
In general, it has proven possible to re-engineer ribozymes to perform reactions in trans. This procedure is often as simple as breaking a ribozyme into arbitrarily defined “substrate” and “catalyst” domains. While the single turnover selections may yield catalysts that are capable of multiple turnovers, there is no selection per se for multiple turnover.
In particular, a fast and complex ligase selected in trans, the Bartel Class I ligase (Ekland et al. 1995) has been engineered to carry out multiple turnover catalysis (Ekland and Bartel 1995). However, the turnover is in general product-limited, since ligation increases the number of base pairs between the product and the catalytic core of the ribozyme. Indeed, this problem (also known as the “parabolic replicator” problem) has been the bane of those who attempt to understand the origins of self-replicating nucleic acids (von Kiedrowski 1986, 1993). Thus, it would be of great utility to be able to select directly for ribozymes capable of performing multiple turnover catalysis in trans.
The major impediment to performing nucleic acid selections in “trans” is linking genotype with phenotype. To this end, cells have previously been used as compartments for the selection of functional nucleic acids (Ferber and Maher 1998; Soukup and Maher 1998; Zimmerman and Maher 2002; Buskirk et al. 2003; Wadhwa et al. 2004). However, cell-based selections afford little control over the components and conditions of ribozyme reactions.
Recent advances in the directed evolution of protein catalysts may prove useful for better controlling selection conditions and for the selection and optimization of multiple turnover ribozymes. More specifically, a novel technique called in vitro compartmentalization (IVC) coupled with microbead display allows molecular genotypes and phenotypes to be linked within individual droplets in a water-in-oil emulsion. IVC has previously been successfully used for the selection of peptide ligands and improved protein catalysts (Griffiths and Tawfik 2003; Ghadessy et al. 2004). In addition, because this method is not cell-based, the components of each compartment can be more precisely controlled.
We have now adapted the IVC microbead display system to the selection of ligase ribozymes that function in trans. To proof this selection method we generated a doped ribozyme library based on the Bartel Class I ligase. Trans-ligation activity was detected after only two rounds of selection. The dominant clone identified after four rounds of selection was only five mutations away from the parental ribozyme and displayed comparable kinetics. Statistically, an active variant was expected to occur roughly once in the starting population. These results suggest that IVC selections are extremely robust in identifying optimal ribozymes and in scanning large tracts of sequence space.
RESULTS
Design of the selection scheme
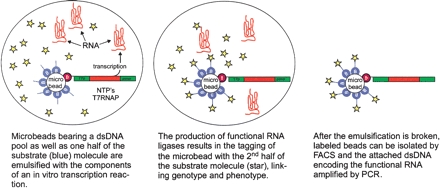
The scheme for the direct, in vitro selection of trans-acting ribozyme ligases is shown in Figure 1. In this scheme, a double-stranded DNA pool is immobilized on a microbead at a ratio of approximately one gene per bead. In addition, the beads are labeled with an RNA oligonucleotide that can serve as a ligase substrate (Fig. 1, blue). The beads are then emulsified with the components necessary for in vitro transcription (T7 RNA polymerase and NTPs), as well as the other RNA oligonucleotide substrate, which also contains an appropriate handle for the attachment of a fluorophore (Fig. 1, star). After incubation, functional RNA ligases produced via transcription will act within the emulsion to ligate the two substrates together. Thus, ligation products will become immobilized on the same beads that carry the template for the ribozyme that has led to the ligation. Because ligation also leads to immobilization of a fluorescent label, when the emulsification is broken any labeled beads can be isolated by FACS. Following sorting, the attached double-stranded DNA (dsDNA) templates encoding functional ribozyme ligases can be amplified by PCR. Multiple cycles of emulsification, selection, and amplification should lead to the isolation of those catalysts best able to work in trans and perhaps to multiple turnover catalysts.
FIGURE 1.
Selection scheme for the isolation of trans-acting ribozyme ligases.
In order to proof this novel ribozyme selection method, we initially performed control reactions using a trans-acting variant of the Bartel Class I ligase (b1-207t) which is capable of rapidly (kobspH 8 = 16 min−1) (Bergman et al. 2000) joining ligase substrates to form 3′–5′ phosphodiester bonds. The trans ligase also turns over substrates efficiently, although at pH >6.5, product release appears to be the rate-limiting step in the reaction (kcpH 8 = 375 min−1) (Bergman et al. 2000).
Initial proofs focused on simple solution-based assays rather than emulsified reactions to ensure that both the ribozyme ligation reaction and our detection scheme worked. Genes encoding the trans-acting Class I ligase were immobilized onto microbeads at a ratio of approximately one gene per bead along with the 3′-hydroxyl-bearing RNA oligonucleotide substrate (SOH) at a ratio of ~75,000 per bead. After washing to remove any unbound species, ~108 beads were suspended in a 20 μL transcription reaction containing a 5′-triphosphate-bearing RNA oligonucleotide substrate (PPPS). The ultimate ratio of PPPS to bead was ~1.2 × 106 to 1. After a 20-h reaction at room temperature, the beads were washed thoroughly and labeled for analysis by FACS using a BD FACScalibur flow cytometer.
Sensitivity studies with this instrument indicated that when using a single fluorophore such as fluorescein or AlexaFluor488, the minimum number of fluorescence events that could be observed over background was ~1000 per bead (data not shown). However, under our reaction conditions, this led to only an approximately twofold increase in the mean fluorescence of the population. In order to increase the observed signal, we utilized an antibody-based amplification scheme. The 5′-fluorescein on PPPS was bound by a fluorescently labeled antifluorescein antibody, which was in turn bound by a secondary fluorescently labeled antibody.
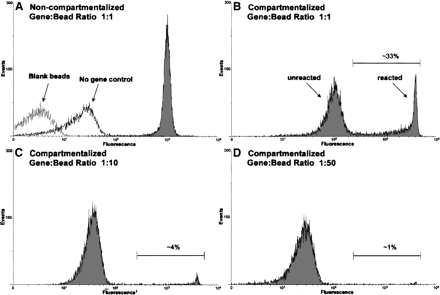
Antibody-mediated changes in fluorescence were again scanned using the BD FACScalibur. As shown in Figure 2A, a strong fluorescent signal was observed from reactions carried out with beads bearing a single dsDNA molecule encoding the Class I ribozyme (Fig. 2A, gray). In the absence of this gene (Fig. 2A, no gene control) or in the presence of a non-ribozyme encoding template (data not shown), there was only a small (<10-fold) increase in fluorescence intensity when compared to blank beads (Fig. 2A, blank beads). This increase in fluorescence was most likely due to nonspecific interactions that occurred during antibody labeling.
FIGURE 2.
FACS analysis of compartmentalized and noncompartmentalized ligation reactions. Reactions were conducted as described in the text using the b1-207t ribozyme. In the absence of compartmentalization, only a single fluorescent population was observed (A, gray). Compartmentalization resulted in the formation of two distinct populations, a highly fluorescent “reacted” population and a less fluorescent “unreacted” population (B). Decreasing the gene-to-bead ratio (B–D) resulted in a proportional decrease in size of the “reacted” population. The fraction of events that have a fluorescent signal greater than that of the “unreacted” population has been indicated as a percentage. The number of fluorescent events is indicated on the Y-axis and the relative level of fluorescence on the X-axis.
Compartmentalized reactions were then prepared, and the gene-to-bead ratio was varied. Transcription reactions were assembled as before, but the reaction volume was increased to 200 μL. Compartmentalization was achieved by adding the reaction mixture to 500 μL of oil mixture (Ghadessy et al. 2001), and the resulting emulsion was again incubated for 20 h at room temperature. Following incubation, the emulsion was broken by extraction with mineral oil followed by extraction with hexane, and the beads were washed to remove any unbound species. The beads were again labeled and analyzed by FACS.
As shown in Figure 2B–D, emulsification of the micro-beads resulted in the formation of two distinct populations: a “reacted” population that displayed a high level of fluorescence and an “unreacted” population that displayed a lower level of fluorescence. These results were markedly different than those observed with reactions carried out in solution (Fig. 2A, non-emulsified), where beads were uniformly labeled. Considering that each bead contains on average a single gene and that in vitro transcription reactions will likely not yield more than ~1000 copies of RNA per dsDNA template (Milligan et al. 1987; Sampson and Uhlenbeck 1988), it seems likely that the encapsulated ligase ribozymes are in fact turning over their substrates. In fact, when we measured the amount of parental ribozyme b1-207t RNA that was produced in a solution-based assay, we found that only ~150 copies of RNA were generated per dsDNA template. When compared to the amount of ligated product produced in these same reactions, each ribozyme was calculated to be turning over 10–40 times (data not shown).
The comparison between compartmentalized and solution phase reactions (Fig. 2A,B) suggests that efficient transcription may be occurring in only a portion of the population. Consistent with these results, when lower gene-to-bead ratios (1:10 and 1:50) are used, the size of the “reacted” population is proportionally reduced (Fig. 2C,D). It should also be noted that the “reacted,” compartmentalized beads are much more fluorescent than the “reacted” beads in solution. These results are consistent with previous experiments carried out with compartmentalized protein enzymes, and most likely reflects the increase in the local concentration of active catalyst in single compartments as compared to bulk phase solution (Griffiths and Tawfik 2003). The fluorescence of the “reacted” population is consistent with >10,000 immobilized fluorescent oligonucleotides.
The slight increase (approximately fivefold) in the fluorescence intensity of the “unreacted” population observed at higher gene-to-bead ratios appears to be the result of partial equilibration of the biotinylated ligation product between “reacted” and “unreacted” beads during antibody labeling. In the case of reactions conducted at a 1:1 ratio of gene-to-bead, the loss of only 10% of the “reacted” signal is sufficient to cause the observed increase in background (Fig. 2, cf. “unreacted” populations in B and D). In order to test the equilibration hypothesis, beads were heated (5 min at 70°C) prior to antibody labeling. This resulted in an almost complete equilibration of label and the production of a single, uniformly labeled population (data not shown). In order to eliminate equilibration during selection experiments, an excess of free biocytin was included in all of the wash steps subsequent to breaking the emulsion.
Additionally, we conducted control experiments in which the microbeads were loaded such that there was one functional ribozyme and 100 nonfunctional ribozymes per bead. Overloading the beads at this level had no effect on the size of the “reacted” population (data not shown). Thus, it should be possible to identify functional ribozymes even if they are diluted on beads in the first or subsequent rounds of selection.
Selection for ligase ribozymes
A ribozyme library was generated based on the sequence of the optimized trans-acting Bartel Class I ligase b1-207t (Bergman et al. 2000). A single-stranded DNA pool was chemically synthesized such that 90 positions in the ribozyme contained 70% wild-type and 30% non-wild-type residues (see below, Fig. 4, uppercase). In order to more completely randomize the ribozyme sequences that were originally selected, we added a 3′ constant region at the end of the pool that could serve as a primer site for PCR. The pool was subsequently PCR-amplified with a 5′ biotinylated primer so that ribozyme templates could be immobilized on the microbeads.
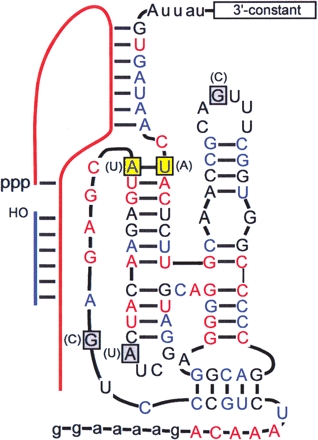
FIGURE 4.
Active variant (r4.dom) isolated from the selection. The predicted secondary structure is based on that of the wild-type b1-207t ribozyme. All residues shown in uppercase were doped at a level of 70% during the selection, while those residues shown in lowercase were held constant. Invariant (red) and semi-conserved (blue) residues identified by Ekland and Bartel (1995) are indicated. Residues that differed from those originally found in the parental ribozyme are boxed in grey, with the inverted base pair boxed in yellow. The original sequence identity of each position is indicated in parentheses. The sequences of the 3′ constant region and the substrates PPPS (red line) and SOH (blue line) are given in Materials and Methods.
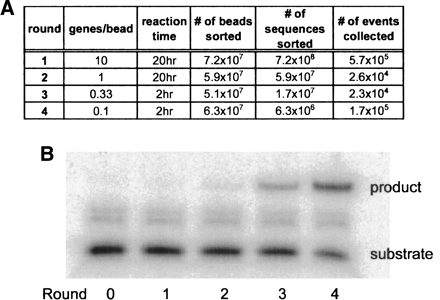
The selection for trans-acting ribozyme ligases was carried out in a manner similar to the trial assays described above. In order to sample a larger population during the first round, the beads were overloaded with 10 genes per bead. The initial library size was ~2 × 109 variants. As with the control reactions, each bead was also loaded with ~75,000 substrate (SOH) molecules. Each round of selection was carried out in parallel with an emulsified positive control reaction that contained a non-amplifiable, wild-type b1-207t ribozyme at a 1:2 gene-to-bead ratio. This control ensured proper reaction assembly and was used to adjust the sorting gate on the flow cytometer. For all rounds, the top ~0.1% fluorescent beads was collected. This level of fluorescence corresponds to the signal observed for beads labeled with >1000 fluorescent labels, and therefore the IVC protocol should promote the selection of ribozymes that are capable of multiple turnover. The ribozyme templates on the collected microbeads were subsequently amplified by PCR and the process was repeated. The stringency of the selection was increased by decreasing the reaction time, and the selection of false positives was reduced by decreasing the gene-to-bead ratio (Fig. 3A).
FIGURE 3.
Progress of the compartmentalized in vitro selection. (A) Description of selection stringency and progress, by round. (B) Ligation activity of the pool. Activity was determined using a coupled transcription–ligation assay. A 5′ P32-labeled RNA substrate (SOH) was used in solution, without emulsification. Ligation products and substrates were at the correct molecular weight, as determined by their mobility relative to standards. The bands located between the “product” and the “substrate” were artifacts of T7 RNA polymerase acting on RNA substrates; these bands were also observed in the absence of a dsDNA template.
The progress of the selection was monitored by performing coupled transcription and ligation reactions following each round of selection. Assays were conducted under selection-like conditions but without emulsification using a 5′ radiolabeled SOH, a dsDNA template amplified from each round of the selection, PPPS, and the components an in vitro transcription reaction (buffer, NTPs, and T7 RNAP). Enhanced ligation activity in the pool was apparent by Round 2 and improved ~90-fold by Round 4 (Fig. 3B). Similar results were observed when the assays were carried out with purified RNA rather than dsDNA templates. Importantly, when these reactions were conducted in the presence of a 10-fold excess of both substrates, the reactions went to near completion, demonstrating that substrate turnover was occurring (data not shown).
Analysis of selected ligase clones
The selected populations from Rounds 3 and 4 were cloned and sequenced. Of the 34 clones isolated from Round 3, three sequences occurred three times and three occurred twice. Surprisingly, when these clones were assayed in a coupled transcription–ligation reaction, no ligation activity was observed. Similarly, when assays were conducted using purified RNA as the input in the reaction, no activity was observed. Analysis of the remaining 19 clones from Round 3 yielded a single active clone which was only five mutations away from the sequence of the parental ribozyme b1-207t (Fig. 4, r4.dom). Following Round 4, this clone was found 14 times in the sequenced population, along with 11 additional clones. Analysis of the remaining clones revealed no active species. The winnowing of a functional population is generally incomplete in the early rounds of a selection, and the isolation of inactive species, whether aptamers or ribozymes, in parallel with active ones is frequently observed (Robertson et al. 2004). In the case of the current selection, the co-isolation of inactive variants may have been facilitated by the equilibration or release of dsDNA templates during the labeling procedure prior to sorting.
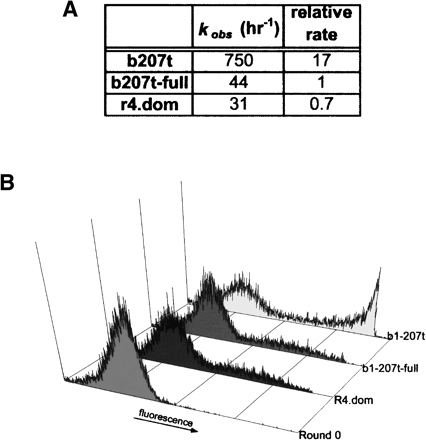
Rates of ligation were measured in solution for the active clone using purified RNA. Rate measurements were conducted under multiple turnover conditions (240 nM SOH, 2 μM PPPS, and either 2.4 or 24 nM ribozyme). For comparison, rates of ligation were also measured for the wild-type ribozyme b1-207t and for a modified version on this ribozyme denoted b1-207t-full that contained the same 3′ constant region used to amplify the pool. The addition of the constant region to the 3′ end of the parental ribozyme resulted in a 17-fold reduction in the rate of ligation. As shown in Figure 5A, the dominant clone r4.dom is almost as active as the comparable wild-type ribozyme b1-207t-full.
FIGURE 5.
Ligation kinetics for the selected clone r4.dom. (A) Rates of ligation. The observed rate of ligation (kobs) was determined from purified RNA under selection-like conditions, without emulsification. For comparison, the rate of ligation of the parental ribozyme b1-207t and the parental ribozyme bearing the 3′ constant extension (b1-207t-full) are also shown. (B) Emulsified ligation reactions. Assays were conducted under the conditions employed during the selection and analyzed by FACS. Histograms have been normalized to the number of single bead events collected.
To compare the rates of ligation under actual selection conditions with the rates of ligation in solution, we performed emulsion-based assays with the selected clone and the control ribozymes (b1-207t and b1-207t-full). The dsDNA genes for each clone were loaded onto microbeads at a 1:1 gene-to-bead ratio. After emulsification, the beads were incubated for 20 h at room temperature. The emulsion was then broken, and the beads were labeled with antibodies and scanned. There is good qualitative agreement between the rate of ligation observed in solution-based assays (Fig. 5A) and those observed in emulsions (Fig. 5B), with b1-207t showing greater activity than b1-207t-full and r4.dom. Again, the maximal level of fluorescence observed with the active clone and the control ribozyme should only have been possible if ribozymes within the emulsion were turning over their oligonucleotide substrates.
DISCUSSION
We have shown that it is possible to develop methods for trans as opposed to cis selection. Such methods should prove useful in a variety of ribozyme selections and should lead to the identification and improvement of multiple turnover ribozymes.
The IVC selection proved to be extremely efficient at finding functional catalysts. Our starting pool averaged 27 mutations per ribozyme, a relatively high mutational load, especially for a ribozyme that contains 28 invariant positions and 29 semi-conserved positions within the 90-nucleotide randomized region. This fact coupled with the relatively small starting pool, ~2.4 × 108 variants (7.2 × 108 × 33% emulsion efficiency), made it difficult to isolate ribozymes that were similar to the parental ribozyme sequence. Nonetheless, we identified a single variant that contained only five noncritical mutations (Fig. 4). Using the binomial distribution, we calculate that the probability of finding a variant that contains any five mutations is one in ~1.4 × 108. Thus, this may have been only one wild-type-like ribozyme in the starting population. However, a more pertinent question may be: How frequently should an active ribozyme have occurred in our population? An upper estimate for this value can be calculated by assuming that the 22 invariant and semi-conserved residues in single-stranded regions of the ribozyme (including seven positions that pair with substratePPPS) must be conserved, as must all 26 base pairs (but not the identity of the bases). Allowing for the formation of G:U wobble pairings, we calculate the probability of finding a sequence that conforms to these specifications as 0.722 × 0.626 = 6.7 × 10−10. This corresponds to one wild-type-like variant in ~1.5 × 109 sequences, or about one in every six selection experiments.
The rarity of the isolated ribozyme and the stringency of selection are further confirmed by the fact that two of the five mutations invert an otherwise conserved base-pairing. Thus, these results confirm that the IVC selection method is as robust for ribozymes as it has previously proven to be for protein enzymes.
However, in the current instance, although multiple turnover did occur in the emulsions, there was not a selection for improved turnover relative to the wild-type enzyme. This result likely reflects the fact that the Bartel ligase was extremely well optimized to begin with. The ribozyme ligase b1-207 was generated by a combination of in vitro selection and engineering. A doped sequence selection with the parental ligase was carried out at a 20% mutation rate and identified 33 invariant positions, 29 semi-conserved residues, and 36 positions whose identity always co-varied (Ekland and Bartel 1995). The knowledge obtained from this selection was then used to engineer ribozymes with significantly higher ligation rates than the parental ligase. The best construct (b1-207) contained a number of deletions in noncritical structural regions, had 10 mutations relative to the parental ligase, and displayed a self-ligation rate that was 50 times higher.
These and other experiments have revealed that the Bartel ligase is relatively isolated in sequence space. Selections in which the ribozyme was converted from a ligase to a polymerase left the core of the ribozyme untouched (Johnston et al. 2001). The ligase was also adapted to a continuous selection scheme and evolved over the equivalent of 300 rounds of selections. As many as 29 neutral mutations accrued, but the core residues remained unchanged (Wright and Joyce 1997). Similarly, when the continuous selection scheme was employed to isolate variants that functioned under extreme pH conditions, the ribozyme core again remained unchanged (Kuhne and Joyce 2003). Finally, Vaish et al. (2003) removed almost half of the ribozyme and mutagenized the remaining 62 nucleotides at a rate of 30%, then selected a “half-ribozyme” whose activity was dependent on a target RNA. Yet even in this instance, only three mutations in invariant residues were observed (Vaish et al. 2003).
While we randomized the previously optimized b1-207 variant at a rate of 30% (rather than 20%), we also examined a much smaller initial population (108 vs. 1014). Given that we recovered only a single variant whose activity was similar to that of the parental ligase, it seems likely that we did not explore either a sufficiently diverse (Hamming distance) or sufficiently large (pool size) sequence space to identify a ribozyme ligase that was more efficient than b1-207 had already become.
To the extent that IVC is inherently limited in the numbers of sequences that can be assessed relative to conventional in vitro selection experiments, it remains problematic whether it is better to sieve a larger pool and then adapt ribozymes to function in trans or to accept a smaller pool size and directly select for function in trans. To further improve the IVC method, it should be possible to increase the gene-to-bead ratio at the outset of the selection. We have observed that ratios as high as 100:1 can be achieved with little or no effect on signal detection. Thus, library size can readily be increased to 109, and higher loading levels may be possible with further optimizations. Moreover, it may be possible to eliminate the use of FACS entirely (or at least during the early rounds of selection) by employing a bead capture method. The IVC selection method would then become more analogous to more traditional methods of in vitro selection in which cis-acting ligases are captured via biotin-tagged substrates. However, the ability to select for ribozymes that functioned in trans would remain, and following capture, trans-acting ligases, capable of multiple turnover could be again be identified by FACS.
MATERIALS AND METHODS
Oligonucleotide sequences, pools, and primers
All oligonucleotides were synthesized in our laboratory using standard phosphoramidite chemistry on an Expedite 8909 DNA synthesizer (PE Biosystems) or purchased from IDT. Synthesis reagents were purchased from Glen Research. All oligonucleotides were gel-purified prior to use.
The sequence of RNA substrate SOH was 5′-aaaaaaaCCAGUC, where the lowercase letters indicate DNA. A 5′ biotinylated version of the substrate was employed for FACS and in vitro selection experiments. The RNA substrate PPPS was generated by in vitro transcription (Epicentre) from a dsDNA template. The sequence of PPPS was 5′-GGAACACUAUACGACUGGCACCA.
The sequence of the template for the Bartel Class I ligase b1-207t was 5′-TTCTAATACGACTCACTATAGGAAAAGAC AATCTGCCCTCAGAGCTTGAGAACATCTTCGGATGCAGGG GAGGCAGCCCCCGGTGGCTTTAACGCCAACGTTCTCAACA ATAGTGA (T7 promoter region underlined). Biotinylated dsDNA was amplified by PCR using a biotinylated 5′ primer 5B.9n20.t7 (5′-B-TTTACGTACTTCTAATACGACTCACTATA) and a 3′ primer 3.b1.207t (5′-TCACTATTGTTGAGAACGTT).
The sequence of the template for the degenerate pool based on b1-207t was 5′ TTCTAATACGACTCACTATAGGAAAAGa caaatctgccctcagagcttgagaacatcttcggatgcaggggaggcagcccccggtggctt taacgccaacgttctcaacaatagtgaTATTCAGAGATGGTAGAGAACACA. The sequence is identical to b1-207t except for a 3′ constant extension of 24 residues. The doped positions (lowercase) were mutagenized at a rate of 30% and were generated by mixing phosphoramidites as previously described (Robertson and Ellington 1999). The double-stranded pool was amplified by PCR using the biotinylated 5′ primer 5B.9n20.t7 and the 3′ primer 3.bl.dope (5′-GTGTTCTCTACCATCTCTG). The degree of degeneracy of the population was confirmed by sequencing 20 clones following template amplification.
The template for the control ribozyme b1-207t-full was generated by PCR amplification of the b1-207t template with the biotinylated 5′ primer 5B.9n20.t7 and the 3′ primer 3.b207-full (5′-TGTGTTCTCTACCATCTCTGAATATCACTATTGTTGAGAAC GTT).
Compartmentalized reactions
The compartmentalization selection strategy was adapted from Griffiths and Tawfik (2003) and Ghadessy et al. (2001). Gene immobilization and compartmentalization were carried out as follows. Twenty microliters of 0.98-micron streptavidin-coated beads (1.9 × 1010/mL, lot number 6382, Bangs Laboratories, Inc.) were washed with 180 μL PBSTE (10 mM sodium phosphate, pH 7.4, 150 mM NaCl, 0.1% Tween-20, 10 mM EDTA). The beads were pelleted by centrifugation at 6000g for 5 min and then resuspended in 50 μL of PBSTE along with 52 pg of dsDNA (1:1 gene-to-bead ratio), 5.2 pg (1:10), or 1 pg (1:50). After incubating for 15 min, 48 pmol (~75,000/bead) of the RNA substrate 5B.SOH were added and the beads were incubated for an additional 45 min. Following incubation the beads were pelleted, washed once with 100 μL of PBSTE and twice with 100 μL of transcription wash buffer (transcription buffer with 0.1% BSA). Pelleted beads were resuspended in a 200 μL in vitro transcription reaction containing transcription buffer (50 mM EPPS, pH 8.0, 2 mM spermidine, 10 mM DTT, 50 mM KCl, 30 mM MgCl2), 4 U yeast pyrophosphatase (Sigma), 160 U RNAsin Plus (Promega), 2 mM NTPs (Epicentre), and 2 μM PPPS. The transcription reactions were assembled on ice and the enzyme T7 RNA polymerase (20 μL at 50 U/μL; Strategene) was added just prior to emulsification.
Emulsification was achieved by adding the 200 μL transcription reaction containing the microbeads to 500 μL of an oil mixture containing mineral oil (Sigma), 5.5% Span-80 (Sigma), 0.5% Tween-80 (Sigma), and 0.1% Triton X-100 (Sigma) (Ghadessy et al. 2001). The mixture was stirred for 5 min on ice, the emulsion was transferred to a 1.5-mL Eppendorf tube, and reactions were allowed to proceed for 20 h at ~23°C. The reaction was terminated by the addition of 40 μL of 500 mM EDTA and 160 μL PBSTEBB (PBSTE with 0.1% BSA and 100 μM biocytin) and then extracted with mineral oil (3 × 1 mL) and hexane (3 × 1 mL). During the extractions, all centrifugation steps were conducted at 13,000g and 4°C. After the removal of residual hexane by vacuum centrifugation, the beads were again pelleted and subsequently washed with PBSTEBB (3 × 100 μL) to remove any unligated substrates, ribozymes, and components of the transcription reaction.
Non-emulsified control reactions or assays were conducted as described above with the exception that after immobilization of the gene and substrate, the beads were resuspended in a 20 μL transcription reaction. Reactions were allowed to proceed for 20 h and stopped by the addition of 4 μL of 500 mM EDTA. After washing, the beads were resuspended in 200 μL PBSTE. Labeling of the beads is described below.
Bead-labeling and FACS analysis
Initial control reactions were performed using the biotinylated dsDNA encoding the b1-207t ribozyme. Following emulsification and reaction, the microbeads were labeled using a 5′ fluorescently labeled oligonucleotide complementary to the 3′ end of the RNA substrate PPPS (5F.RC.PPPS; 5′-6-FAM-TTTGGTGCCAGTCGTATAGTGT TC). Fluorescent oligonucleotide hybridization was achieved by incubating the beads in PBSTE containing 2.5 μM 5F.RC.PPPS for 30 min at room temperature. Following hybridization, the beads were washed twice with 100 μL PBSTEBB. The fluorescent signal resulting from oligonucleotide hybridization proved too weak to effectively detect and sort the microbeads, so the labeling reaction was subsequently amplified using fluorescently labeled antibodies (AlexFlour488 Signal Amplification Kit; Molecular Probes). Antibody labeling was conducted in 200 μL of PBSTE containing 100 μM biocytin, 1% BSA, 100 μg tRNA, 80 U RNAsin, and 10 μg antibody.
Following labeling, the beads were washed twice with 100 μL of PBSTEBB and resuspended in 100 μL PBS on ice and sonicated for 1 min in a Branson200 ultrasonic cleaner (Branson). Approximately 5 μL of the sonicated, resuspended beads were diluted in 500 mL PBSTE and scanned on a FACScalibur flow cytometer (BD). Each scan consisted of 10,000 events. It should be noted that the actual FACS-based selections (below) involved many more events.
Compartmentalized selection
Compartmentalized selections were carried out as described above using the doped pool based on the b1-207t sequence. During the first round of selection, 2 × 108 microbeads were loaded with DNA templates at a ratio of 10:1 genes-to-beads. After labeling with antibodies, the beads were diluted in 1.5 mL of PBSTE and sorted on a MoFlo flow cytometer at ~30,000/sec (the MoFlo can be used to sort many more events than the FACScalibur). The top ~0.1% of the population was collected after each sort. The sorting gate was initially set based on the fluorescent signal observed with a positive control reaction involving the parental ribozyme b1-207t. Only single bead signals were isolated (some signals were clearly due to aggregated beads, and were discarded). The genes on the collected beads were subsequently amplified by PCR using the pool-specific primers 5B.9n20.t7 and 3.bl.dope. The dsDNA was gel-purified on a 3% agarose gel. Gel purification was facilitated by using a Qiaquick gel purification kit (Qiagen), and the purified dsDNA was quantitated using a NanoDrop ND-1000 spectrophotometer (NanoDrop). DNA quantitation was confirmed by comparison with a known standard following electrophoresis on a 4% agarose gel. The stringency of the selection was progressively increased as described in the text and as shown in Figure 4.
Cloning and sequencing
dsDNA from Rounds 3 and 4 of the selection was re-amplified using a non-biotinylated 5′ primer. The PCR products were cloned using the Topo TA cloning kit (PCR4.1; Invitrogen). Isolated clones were sequenced using the BigDye terminator mix and a AB3730 DNA Analyzer (Applied Biosystems).
Kinetic analysis
Coupled transcription–ligation reactions (10 μL) were assembled in a manner similar to the selection, but without immobilization of the gene and SOH. Reactions contained transcription buffer, 0.2 U yeast pyrophosphatase, 0.8 U RNAsin Plus, 2 mM NTPs, 500 nM 5′-radiolabeled SOH, 2 μM PPPS, and 10 ng of a given dsDNA template, and were initiated by the addition of 2 μL T7 RNAP. Reactions were allowed to proceed for 1 h at room temperature and were stopped by the addition of 15 μL stop dye (95% form-amide saturated with urea, 20 mM EDTA, 0.05% bromophenol blue). Samples were heat denatured for 3 min at 70°C and analyzed on a denaturing (7 M urea) 8% polyacrylamide gel. After drying the gel, the amounts of radiolabel in individual bands were quantitated using a Phosphorimager (Molecular Dynamics).
Ligation rates for individual clones were determined using gel-purified RNA. Mock transcription reactions (10 μL) were assembled as described above but did not include T7 RNAP. The reactions contained either 2.4 nM or 24 nM ribozyme in order to identify conditions in which initial velocities were linear. The value of kobs was determined by fitting three time points that occurred within the first ~5% of the reaction.
Acknowledgments
We thank the labs of George Georgiou and Brent L. Iverson for the use of their FACScalibur and MoFlo cytomometers. In addition, M.L. would like to thank Barret R. Harvey for his assistance with this project. This project was supported by a grant from the National Institutes of Health (8R01 EB002043) and by the DoD.
Published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2121705.
REFERENCES
- Bartel, D.P. and Szostak, J.W. 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418. [DOI] [PubMed] [Google Scholar]
- Bergman, N.H., Johnston, W.K., and Bartel, D.P. 2000. Kinetic framework for ligation by an efficient RNA ligase ribozyme. Biochemistry 39: 3115–3123. [DOI] [PubMed] [Google Scholar]
- Buskirk, A.R., Kehayova, P.D., Landrigan, A., and Liu, D.R. 2003. In vivo evolution of an RNA-based transcriptional activator. Chem. Biol. 10: 533–540. [DOI] [PubMed] [Google Scholar]
- Ekland, E.H. and Bartel, D.P. 1995. The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucleic Acids Res. 23: 3231–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland, E.H., Szostak, J.W., and Bartel, D.P. 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269: 364–370. [DOI] [PubMed] [Google Scholar]
- Ferber, M.J. and Maher 3rd, L.J. 1998. Combinatorial selection of a small RNA that induces amplification of IncFII plasmids in Escherichia coli. J. Mol. Biol. 279: 565–576. [DOI] [PubMed] [Google Scholar]
- Ghadessy, F.J., Ong, J.L., and Holliger, P. 2001. Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl. Acad. Sci. 98: 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadessy, F.J., Ramsay, N., Boudsocq, F., Loakes, D., Brown, A., Iwai, S., Vaisman, A., Woodgate, R., and Holliger, P. 2004. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat. Biotechnol. 22: 755–759. [DOI] [PubMed] [Google Scholar]
- Griffiths, A.D. and Tawfik, D.S. 2003. Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J. 22: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger, L. 1997. The new world of ribozymes. Curr. Opin. Struct. Biol. 7: 324–335. [DOI] [PubMed] [Google Scholar]
- Jaeger, L., Wright, M.C., and Joyce, G.F. 1999. A complex ligase ribozyme evolved in vitro from a group I ribozyme domain. Proc. Natl. Acad. Sci. 96: 14712–14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena, V.K. and Gold, L. 1997. In vitro selection of self-cleaving RNAs with a low pH optimum. Proc. Natl. Acad. Sci. 94: 10612–10617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, W.K., Unrau, P.J., Lawrence, M.S., Glasner, M.E., and Bartel, D.P. 2001. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292: 1319–1325. [DOI] [PubMed] [Google Scholar]
- Kuhne, H. and Joyce, G.F. 2003. Continuous in vitro evolution of ribozymes that operate under conditions of extreme pH. J. Mol. Evol. 57: 292–298. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, T. 1997. Novel and variant ribozymes obtained through in vitro selection. Curr. Opin. Chem. Biol. 1: 17–25. [DOI] [PubMed] [Google Scholar]
- Pan, T. and Uhlenbeck, O.C. 1992. In vitro selection of RNAs that undergo autolytic cleavage with lead(2+). Biochemistry 31: 3887–3895. [DOI] [PubMed] [Google Scholar]
- Robertson, M.P. and Ellington, A.D. 1999. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 17: 62–66. [DOI] [PubMed] [Google Scholar]
- Robertson, M.P., Knudsen, S.M., and Ellington, A.D. 2004. In vitro selection of ribozymes dependent on peptides for activity. RNA 10: 114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, J. and Joyce, G.F. 2001. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA 7: 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, J.R. and Uhlenbeck, O.C. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 85: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup, G.A. and Maher 3rd, J.J. 1998. Selection and characterization of RNAs that relieve transcriptional interference in Escherichia coli. Nucleic Acids Res. 26: 2715–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, J. and Breaker, R.R. 2000. Structural diversity of self-cleaving ribozymes. Proc. Natl. Acad. Sci. 97: 5784–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaish, N.K., Jadhav, V.R., Kossen, K., Pasko, C., Andrews, L.E., McSwiggen, J.A., Polisky, B., and Seiwert, S.D. 2003. Zeptomole detection of a viral nucleic acid using a target-activated ribozyme. RNA 9: 1058–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kiedrowski, G. 1986. A self replicating hexadeoxynucleotide. Angew Chem. Int. Ed. Engl. 25: 932–935. [Google Scholar]
- ———. 1993. Minimal replicator theory. I. Parabolic versus exponential growth. Bioorg. Chem. Frontiers 3: 113–146. [Google Scholar]
- Wadhwa, R., Yaguchi, T., Kaur, K., Suyama, E., Kawasaki, H., Taira, K., and Kaul, S.C. 2004. Use of a randomized hybrid ribozyme library for identification of genes involved in muscle differentiation. J. Biol. Chem. 279: 51622–51629. [DOI] [PubMed] [Google Scholar]
- Wright, M.C. and Joyce, G.F. 1997. Continuous in vitro evolution of catalytic function. Science 276: 614–617. [DOI] [PubMed] [Google Scholar]
- Zimmerman, J.M. and Maher 3rd, L.J. 2002. In vivo selection of spectinomycin-binding RNAs. Nucleic Acids Res. 30: 5425–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]