Abstract
5-Formyluracil (5-foU) is a potentially mutagenic lesion of thymine produced in DNA by ionizing radiation and various chemical oxidants. The elucidation of repair mechanisms for 5-foU will yield important insights into the biological consequences of the lesion. Recently, we reported that 5-foU is recognized and removed from DNA by Escherichia coli enzymes Nth (endonuclease III), Nei (endonuclease VIII) and MutM (formamidopyrimidine DNA glycosylase). Human cells have been shown to have enzymatic activities that release 5-foU from X-ray-irradiated DNA, but the molecular identities of these activities are not yet known. In this study, we demonstrate that human hNTH1 (endonuclease III homolog) has a DNA glycosylase/AP lyase activity that recognizes 5-foU in DNA and removes it. hNTH1 cleaved 5-foU-containing duplex oligonucleotides via a β-elimination reaction. It formed Schiff base intermediates with 5-foU-containing oligonucleotides. Furthermore, hNTH1 cleaved duplex oligonucleotides containing all of the 5-foU/N pairs (N = G, A, T or C). The specific activities of hNTH1 for cleavage of oligonucleotides containing 5-foU and thymine glycol were 0.011 and 0.045 nM/min/ng protein, respectively. These results indicate that hNTH1 has DNA glycosylase activity with the potential to recognize 5-foU in DNA and remove it in human cells.
INTRODUCTION
Reactive oxygen species (ROS) are generated in living cells during normal cellular metabolism and in response to exogenous inducers such as ionizing radiation and chemical oxidants (1,2). ROS cause oxidative base damage to DNA and produce a wide variety of base modifications (3,4). Certain types of oxidized bases, such as thymine glycol (Tg) and 7,8-dihydro-8-oxoguanine, are cytotoxic and mutagenic (3). Cells have repair mechanisms that prevent the cell death and mutation caused by oxidative DNA damage (5). In most organisms, oxidative damage in DNA is primarily repaired by the base excision repair systems (3,6–8).
Exposure of DNA to ionizing radiation or chemical oxidants leads to the formation of several thymine hydroperoxides (9–11). 5-Hydroperoxymethyluracil spontaneously decomposes to form 5-formyluracil (5-foU) and 5-hydroxymethyluracil (9–11). Recent studies revealed that 5-foU is a potentially mutagenic lesion. Kasai and co-workers (12,13) found that 5-formyl-2′-deoxyuridine (5-fodUrd) is mutagenic to bacterial cells when added to the culture medium. Recently we found that the mutation frequency of plasmid pSVK3 containing site-specifically incorporated 5-foU is significantly increased compared with that of a normal T/A-containing plasmid (14). In addition, Klungland et al. (15) showed that the addition of 5-foU or 5-fodUrd to the culture medium promoted mutagenicity at the hprt locus in Chinese hamster fibroblast cells. It is inferred that 5-foU is incorporated into DNA and has base pairing properties different from thymine. These conclusions are supported by the findings that 5-fodUrd is incorporated into the DNA during cell growth and forms mispairs with G, C and T during DNA replication (16,17). In fact, it directs misincorporation of mismatched bases opposite the lesion during DNA synthesis in vitro (18,19).
It has been reported that Escherichia coil AlkA protein removes 5-foU from DNA in vitro (20). Furthermore, we recently found that E.coli enzymes Nth (endonuclease III), Nei (endonuclease VIII) and MutM form Schiff base intermediates with oligonucleotides containing 5-foU and cleave the strand at the 5-foU site (21). These proteins coordinately prevent mutations caused by 5-foU (21,22). There are some indications that mammalian cells have similar enzymatic activities that release 5-foU from oxidized DNA (23,24). However, the molecular nature of these activities has not been elucidated. The base excision repair system is highly conserved from bacteria to humans (3,6–8,25). Functional homologs of bacterial DNA glycosylases that act on oxidized bases have been identified in various kinds of eukaryotes. In humans, hOgg1 (8-oxoguanine DNA glycosylase), hMPG (N-alkylpurine glycosylase) and hNTH1 (endonuclease III homolog) show overlapping substrate specificity with E.coli MutM, AlkA and Nth, respectively (3,6–8,25–27). Recently, Hazra et al. (28) reported the presence of two human orthologs of the E.coli nei gene in the human genome database and characterized one of their products. Based on the substrate preference, they named it NEH1 (Nei homolog). The wild-type recombinant NEH1, purified to homogeneity from E.coli, excises formamidopyrimidines from damaged DNA and 8-oxoguanine and oxidized pyrimidines, such as thymine glycol, from oligonucleotides (28).
hNTH1 has a molecular mass of 36 kDa and is encoded by a gene located in chromosomal region 16p13.3 (29,30). The helix–hairpin–helix motif, [4Fe-4S] cluster and lysine residue that is necessary for the formation of the Schiff base intermediate are conserved between the bacterial and human proteins (29,30). In addition, both proteins recognize similar substrates, Tg, dihydrouracil and 5-hydroxycytosine (3,31). These facts led us to examine whether this protein has a DNA glycosylase activity that recognizes and removes 5-foU from DNA. In this report, we show that hNTH1 has 5-foU DNA glycosylase activity. Purified recombinant hNTH1 formed a Schiff base intermediate with a 5-foU-containing oligonucleotide and cleaved it by β-elimination. In addition, hNTH1 excised oligonucleotides containing all 5-foU/N pairs (N = G, A, T or C), indicating that the enzyme recognizes the lesion 5-foU. This is the first identification of a human protein that has 5-foU DNA glycosylase activity.
MATERIALS AND METHODS
Enzymes and chemicals
Phenylmethylsulfonyl fluoride, osmium tetroxide and NaBH4 were purchased from Wako Pure Chemicals (Osaka, Japan). Ampicillin was obtained from Meiji Seika (Tokyo, Japan). T4 polynucleotide kinase was obtained from New England Biolabs (Beverly, MA). Glutathione–Sepharose 4B, thrombin, Sephadex G-25 and a prepacked Mono S column for FPLC were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). [γ-32P]ATP (>148 TBq/mmol) was obtained from ICN Biomedicals (Costa Mesa, CA).
Oligonucleotide substrates
A 5-foU-containing 17mer oligonucleotide with the sequence 5′-GGTCGACTFAAGGTACC-3′ (where F represents 5-foU) was synthesized as previously described (32). In brief, an oligonucleotide containing 5-(1,2-dihydroxyethyl)uracil, the precursor of 5-foU, was synthesized by phosphoramidite chemistry using an ABI 381A DNA synthesizer (Perkin Elmer). Saturated NaIO4 was added to a solution of the oligonucleotide containing the precursor and the reaction mixture was vortex mixed for 1 min at room temperature to convert 5-(1,2-dihydroxyethyl)uracil to 5-foU (32). The presence of 5-foU in oligonucleotides was verified by HPLC and HPLC/MS analyses. A Tg-containing 17mer oligonucleotide with the sequence 5′-GGACGACAt′AAGGAACC-3′ (where t′ represents Tg) was prepared according to Dianov et al. (33). In brief, an oligonucleotide which contained a single thymine residue (5′-GGACGACATAAGGAACC-3′) was oxidized with 50 mM osmium tetroxide in the presence of 2% pyridine at room temperature. Then the oligonucleotide was purified by gel filtration on Sephadex G-25. Other oligonucleotides were purchased from Espec (Nagoya, Japan) and Takara Shuzo (Kyoto, Japan).
Expression and purification of the recombinant hNTH1 and E.coli Nth
The plasmid containing human hNTH1 cDNA was digested with BamHI and EcoRI, after which it was subcloned into BamHI + EcoRI-digested plasmid vector pGEX-4T-3. The resultant plasmid was named pGEX-4T-3-hNTH1. GST–hNTH1 fusion protein was expressed in E.coli KSR7 (nth nei mutM) (21) carrying pGEX4T-3-hNTH1 and the recombinant hNTH1 was purified according to Matsumoto et al. (34). Escherichia coli Nth was purified according to Zhang et al. (21).
DNA glycosylase/AP lyase assay for 5-foU-containing DNA
The DNA glycosylase/AP lyase assay was carried out at 37°C in a reaction mixture (10 µl) containing 20 fmol 32P-labeled oligonucleotide and purified Nth or hNTH1 in 20 mM Tris–HCl pH 8.0, 1 mM EDTA and 75 mM NaCl. The reaction was terminated by the addition of stop solution (95% formamide, 0.1% xylene cyanol and 20 mM EDTA). After heating at 95°C for 5 min, the samples were cooled on ice and then loaded onto 20% polyacrylamide gels in 90 mM Tris–borate pH 8.3, containing 7 M urea and 2 mM EDTA. After electrophoresis at 1300 V, the gels were dried and autoradiographed using Fuji RX film at –80°C. The intensity of each band was determined by image analysis using NIH Image software.
Trapping assay of protein–substrate intermediates with NaBH4
The DNA trapping assay was performed at 37°C for 5 min in a reaction mixture (10 µl) containing 50 fmol 32P-labeled oligonucleotide and purified Nth or hNTH1 in 20 mM Tris–HCl pH 8.0, 1 mM EDTA and 50 mM NaBH4. The aqueous solution of NaBH4 (0.5 M) was prepared just before use. The reaction was terminated by heating at 95°C for 5 min after the addition of an equal volume of standard loading buffer for SDS–PAGE. Trapped complexes were analyzed by 12% SDS–PAGE.
RESULTS
Cleavage of 5-foU-containing oligonucleotide by purified hNTH1
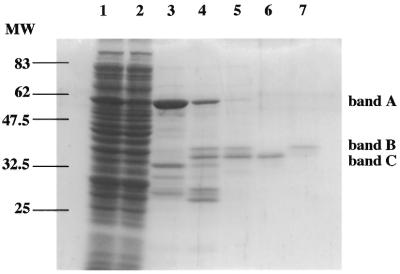
To examine whether hNTH1 has 5-foU DNA glycosylase activity, we first purified the recombinant GST–hNTH1 fusion protein from GST–hNTH1-overexpressing E.coli cells (Fig. 1). The GST–hNTH1 fusion protein (band A) was subsequently cleaved by thrombin to isolate the wild-type hNTH1. We observed two polypeptides of ∼36 and 34 kDa in addition to GST in the thrombin digest, indicating that thrombin cleaved at two sites in the fusion protein. Ikeda et al. (29) showed that the 34 kDa protein is a truncated form of hNTH1 lacking 22 residues at the N-terminus. We isolated the full-length (band B) and truncated (band C) proteins by chromatography on a Mono S column (Fig. 1). The full-length and truncated hNTH1 proteins were eluted with 390 and 470 mM NaCl, respectively. Since the truncated hNTH1 protein showed much higher 5-foU-containing oligonucleotide cleaving activity, we used the truncated form of hNTH1 for further experiments.
Figure 1.
Expression and purification of recombinant hNTH1. Proteins were assayed by 12% SDS–PAGE and stained with Coomassie blue. Lane 1, soluble fraction from E.coli induced by 0.1 mM IPTG for 4 h and disrupted by sonication; lane 2, flow-through fraction from glutathione–Sepharose 4B column; lane 3, fraction containing GST–hNTH1 fusion protein eluted with glutathione; lane 4, GST–hNTH1 fusion protein digested by thrombin (5 U/ml); lane 5, purified hNTH1 after elution from a glutathione– Sepharose 4B column. Truncated (lane 6) and full-length (lane 7) hNTH1 were separated by fractionation on a Mono S column. Bands A–C represent GST–hNTH1 fusion, full-length and truncated hNTH1, respectively.
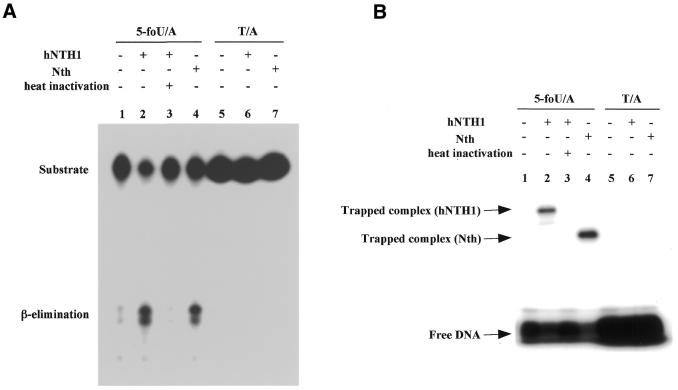
We performed the DNA glycosylase/AP lyase assay as follows. 32P-labeled oligonucleotide containing 5-foU or T was annealed to the complementary strand to construct 5-foU/A or T/A base pair-containing duplex oligonucleotides, respectively. Each duplex oligonucleotide was incubated with the recombinant hNTH1 and then the reaction mixtures were electrophoresed on denaturing polyacrylamide gels. As shown in Figure 2A, the oligonucleotide was cleaved at the site of the 5-foU by the AP lyase activity of hNTH1 and E.coli Nth. The oligonucleotide containing a T/A base pair was not cleaved by these proteins. Prolonged incubation for 1 h with 24 ng hNTH1 did not cleave the T/A-containing oligonucleotides (data not shown). Sometimes the β-elimination products generated by AP lyases, e.g. endonuclease III, give rise to several separate bands in PAGE analysis, presumably because of Tris adduct formation and/or isomerization of the 3′-hydroxypentenal terminus (21,29). These results indicate that hNTH1 has DNA glycosylase activity specific for 5-foU. The mobility of the cleavage product revealed that the enzyme cleaves the 5-foU-containing oligonucleotide by a β-elimination reaction.
Figure 2.
Cleavage activity of hNTH1 on duplex oligonucleotides with or without 5-foU. (A) Cleavage of 5-foU-containing oligonucleotides by purified hNTH1. The duplex oligonucleotide, 5-foU/A (lanes 1–4) or T/A (lanes 5–7), was incubated at 37°C for 20 min without protein (lanes 1 and 5) or with 24 ng hNTH1 (lanes 2 and 6), 25 ng E.coli Nth (lanes 4 and 7) or 24 ng heat-inactivated hNTH1 (lane 3). The inactivation of hNTH1 was performed by heating at 100°C for 10 min. After the reaction was terminated, the products were separated by denaturing 20% PAGE on gels containing 7 M urea. (B) Formation of Schiff base intermediate with 5-foU-containing oligonucleotides by hNTH1. The duplex oligonucleotide substrates were incubated with or without enzyme at 37°C for 5 min in the presence of 50 mM NaBH4. The amounts of hNTH1 and Nth in the reaction mixture were 200 and 100 ng, respectively.
It is known that hNTH1 forms a Schiff base intermediate with substrate DNA and subsequently cleaves the phosphodiester backbone of DNA via a β-elimination reaction (3,6–8). To determine whether hNTH1 forms a Schiff base intermediate with a 5-foU-containing oligonucleotide, we next performed a DNA trapping assay with NaBH4 (35–38). As shown in Figure 2B, hNTH1 was trapped by a 5-foU-containing oligonucleotide, as was E.coli Nth. These results indicate that hNTH1 recognizes and cleaves 5-foU-containing oligonucleotides as natural substrates.
Efficiency of the 5-foU DNA glycosylase/AP lyase activity of hNTH1
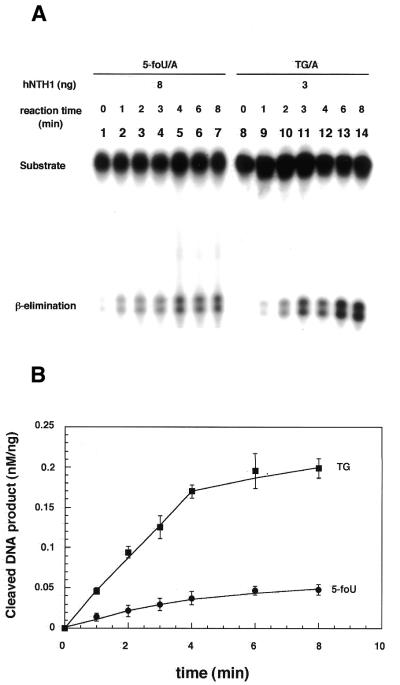
Next, we compared the activity of hNTH1 for 5-foU with that for Tg. As an indicator of the enzymatic activity, we measured the initial velocity of the formation of the cleavage product (Fig. 3). Then the intensity of the upper band was determined and the relative amount of product (nM/ng protein) is plotted in Figure 3B. Under the present conditions, hNTH1 maintained linear kinetics for ∼4 min with both substrates. The specific activities of cleavage of DNA containing 5-foU and Tg were 0.011 and 0.045 nM/min/ng protein, respectively. The DNA glycosylase activity of hNTH1 for 5-foU was about one-quarter of that for Tg. These results suggest that 5-foU is a good substrate for hNTH1 under physiological conditions.
Figure 3.
Cleavage of oligonucleotides containing 5-foU and Tg by hNTH1. (A) The duplex oligonucleotides 5-foU/A (lane 1–7) and Tg/A (lane 8–14) were incubated with hNTH1 at 37°C for 0 (lanes 1 and 8), 1 (lanes 2 and 9), 2 (lanes 3 and 10), 3 (lanes 4 and 11), 4 (lanes 5 and 12), 6 (lanes 6 and 13) or 8 (lanes 7 and 14) min. In the reaction mixture, 8 and 3 ng hNTH1 were added for 5-foU and Tg, respectively. The products were separated by denaturing 20% PAGE in gels containing 7 M urea. (B) The intensity of each band was analyzed using NIH Image software. Error bars show the standard deviations.
Effects of the base opposite 5-FoU on DNA cleavage by hNTH1
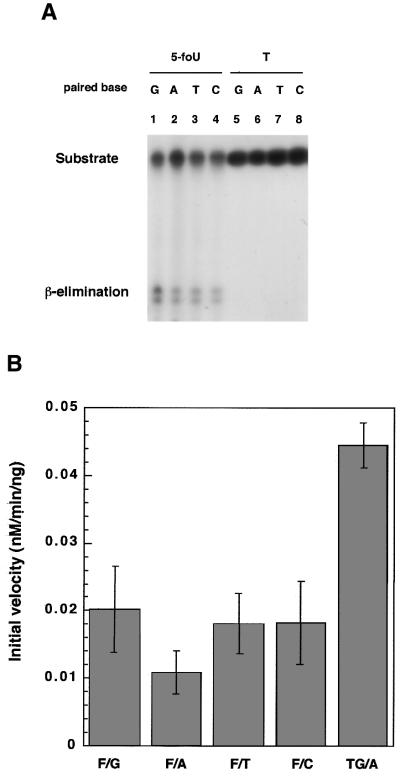
5-foU in DNA arises by the oxidation of thymine in situ. Therefore, we used a substrate duplex oligonucleotide containing 5-foU paired with A. On the other hand, 5-foU in template DNA can direct misincorporation of mismatched bases opposite the lesion during DNA synthesis in vitro (18,19) and in vivo (14–17). In addition, 5-formyl-dUTP can also be incorporated during in vitro DNA synthesis (11,12) and causes mutations in E.coli (12,13). These results suggest that 5-foU/N (N = G, A, T or C) pairs are formed during DNA replication in E.coli cells. Therefore, we examined whether hNTH1 can recognize 5-foU in all types of base pairs in the substrate DNA. As shown in Figure 4, hNTH1 cleaved oligonucleotides containing 5-foU/G, 5-foU/A, 5-foU/T and 5-foU/C, whereas none of the substrates containing T/G, A, T and C pairs were cleaved. These results indicate that hNTH1 does not recognize the 5-foU/N mismatches but the lesion itself in DNA. Next, we quantified the intensity of each band and calculated the initial velocity of the cleavage reaction (Fig. 4B). Depending on the base paired with 5-foU, the activity of hNTH1 for 5-foU varied from 0.011 to 0.02 nM/min/ng protein. The 5-FoU/A pair was recognized less efficiently than other 5-foU/N pairs.
Figure 4.
Effects of the base paired with 5-foU on the cleavage of oligonucleotides by hNTH1. (A) The duplex oligonucleotide substrate, 5-foU/N (lanes 1–4) or T/N (lanes 5–8), was incubated with hNTH1 (8 ng) at 37°C for 3 min. N represents G (lanes 1 and 5), A (lanes 2 and 6), T (lanes 3 and 7) or C (lanes 4 and 8). The products were separated by denaturing 20% PAGE in gels containing 7 M urea. (B) The intensity of the upper band was analyzed using NIH Image software. Error bars show standard deviations.
Formation of the Schiff base intermediate by hNTH1 with 5-foU-containing oligonucleotide
In Figure 2B we showed that hNTH1 forms a Schiff base intermediate with duplex oligonucleotides containing a 5-foU/A pair. Next, we examined whether hNTH1 can form Schiff base intermediates with 5-foU/N and Tg/A (Fig. 5). Bands A and C correspond to the trapped intermediates and free oligonucleotide, respectively. As shown in Figure 5, hNTH1 formed a Schiff base intermediate with all duplex oligonucleotides containing 5-foU. hNTH1 appeared to form the largest amount of the intermediate with the oligonucleotide containing Tg/A among these substrates. The amount of the intermediate formed was independent of the base paired with 5-foU.
Figure 5.
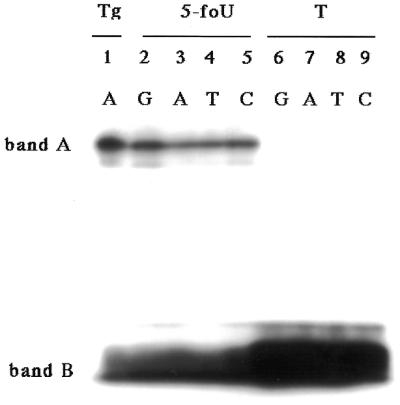
Trapping assay of purified hNTH1 with NaBH4. The duplex oligonucleotide Tg/A (lane 1), 5-foU/N (lanes 2–5) or T/N (lanes 6–9) was incubated with 100 ng hNTH1 at 37°C for 5 min in the presence of 50 mM NaBH4. N represents G (lanes 2 and 6), A (lanes 3 and 7), T (lanes 4 and 8) or C (lanes 5 and 9).
DISCUSSION
5-foU is a major oxidative damage product derived from thymine. It has been shown that 5-foU in DNA is mutagenic to E.coli (12,13,15,21). To prevent mutations induced by 5-foU in DNA, proteins AlkA, MutM, Nth and Nei recognize and remove the lesion from DNA in E.coli (21,22). Mammalian cells also possess DNA glycosylase activities that remove 5-foU from DNA (23,24). In this study, we have shown that hNTH1, a human endonuclease III homolog, recognizes and cleaves duplex oligonucleotides containing a 5-foU (Fig. 2A). The specific activities of cleavage of DNA containing 5-foU and Tg were 0.011 and 0.045 nM/min/ng protein, respectively. Thus, hNTH1 shows 4-fold higher efficiency for Tg than 5-foU in DNA (Fig. 3). Dizdaroglu et al. (39) reported that kcat values for the removal of damaged bases from oxidized DNA by hNTH1 vary by an order of magnitude. In addition, with defined oligonucleotide substrates E.coli Nth recognizes Tg with about 5-, 7-, 10- and 50-fold higher efficiency than uracil glycol, 5-hydroxycytosine, dihydrothymine and 5-hydroxyuracil, respectively (39). From these results we conclude that 5-foU is a major substrate of hNTH1.
DNA glycosylases are divided into two groups: monofunctional and bifunctional DNA glycosylases. Monofunctional DNA glycosylases have only DNA glycosylase activity, whereas bifunctional DNA glycosylases possess both DNA glycosylase and AP lyase activity, by which they cleave the phosphodiester backbone in DNA (3,5–8). In the case of bifunctional DNA glycosylases, a nucleophilic amino group derived from the protein displaces the base, forming a protonated Schiff base intermediate, and subsequently the DNA strand is cleaved as a result of β-elimination (3,6–8). Some of these enzymes catalyze a subsequent δ-elimination reaction (3,6–8). Escherichia coli Nth and human hNTH1 are bifunctional DNA glycosylases which catalyze β-elimination reactions (29,40). In this study, we have shown that hNTH1 cleaves the DNA strand via a β-elimination reaction, as does E.coli endonuclease III (Fig. 2A), and forms a Schiff base intermediate with the substrate oligonucleotides (Figs 2B and 5). These reactions were catalyzed by hNTH1 for all substrates containing 5-foU/N. Since T/N mismatched base pairs were not recognized by hNTH1, we conclude that hNTH1 does not recognize mismatched base pairs but rather the lesion 5-foU as an oxidized pyrimidine in DNA.
Both the 5-foU residue in template DNA and 5-formyl-dUTP added to the culture medium cause mutations in vivo and in vitro (11–19,21). Thus, it is likely that 5-foU in DNA is paired with bases other than A in E.coli cells. The present experimental results indicate that hNTH1 can remove 5-foU from DNA containing 5-foU/N mismatched base pairs (Fig. 4). The initial velocities (V0) for hNTH1 indicate the following preferences: Tg/A (relative V0 = 1) > 5-foU/G (0.46) > 5-foU/T (0.41) = 5-foU/C (0.41) > 5-foU/A (0.25). Therefore, we conclude that hNTH1 removes 5-foU mispaired with G, A, T and C, in that order of preference, from DNA.
Human hNTH1 is a structural and functional homolog of E.coli Nth (3,29–31). Mammalian cells have been shown to possess at least two kinds of DNA glycosylase activity that remove 5-foU from DNA (23,24). In this study, we have demonstrated that hNTH1 has 5-foU DNA glycosylase activity. Four kinds of DNA glycosylase can act on 5-foU in DNA in E.coli (20,21). Therefore, it is possible that mammalian cells also possess another 5-foU DNA glycosylase(s) in addition to hNTH1. Recently, Hazra et al. (28) reported the presence of two human orthologs of the E.coli nei gene in the human genome database and characterized one of their products. Based on the substrate preference, they named it NEH1 (Nei homolog). The wild-type recombinant NEH1, purified to homogeneity from E.coli, excises formamidopyrimidines from damaged DNA and 8-oxoguanine and oxidized pyrimidines, such as thymine glycol, from oligonucleotides (28). We also separately isolated a human homolog of E.coli Nei, named hNEI1. We found that hNEI1 recognized and efficiently cleaved oligonucleotides containing 5-foU (our unpublished results). However, further investigations will be needed to elucidate the molecular basis of the repair mechanisms for 5-foU in mammalian cells.
Acknowledgments
ACKNOWLEDGEMENTS
This study was partly supported by a Grant-in-Aid for Scientific Research on Priority Areas (C) and a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Halliwell B. and Gutteridge,J.M.C. (1990) Role of free radicals and catalytic metal ions in human disease: overview. Methods Enzymol., 186, 1–85. [DOI] [PubMed] [Google Scholar]
- 2.Ames B.N., Shigenaga,M.K. and Hagen,T.M. (1993) Oxidants, antioxidants, and the degenarative diseases of aging. Proc. Natl Acad. Sci. USA, 90, 7915–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace S.S. (1997) Oxidative damage to DNA and its repair. In Scandalios,J.G. (ed.), Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 49–81.
- 4.Cadet J., Delatour,T., Douki,T., Gasparutto,D., Pouget,J.P., Ravanat,J.L. and Sauvaigo,S. (1999) Hydroxyl radicals and DNA base damage. Mutat. Res., 424, 9–21. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg E.C., Walker,G. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- 6.David S.S. and Williams,S.D. (1998) Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev., 98, 1221–1261. [DOI] [PubMed] [Google Scholar]
- 7.McCulough A.K., Dodson,M.L. and Lloyd,R.S. (1999) Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem., 68, 255–285. [DOI] [PubMed] [Google Scholar]
- 8.Mol C.D., Parikh,S.S., Putnam,C.D., Lo,T.P. and Tainer,J.A. (1999) DNA repair mechanisms for the recognition and removal of damaged bases. Annu. Rev. Biophys. Biomol. Struct., 28, 101–128. [DOI] [PubMed] [Google Scholar]
- 9.Teebor G.W., Boorstein,R.J. and Cadet,J. (1988) The repairability of oxidative free radical mediated damage to DNA: a review. Int. J. Radiat. Biol., 54, 131–150. [DOI] [PubMed] [Google Scholar]
- 10.Tofigh S. and Frenkel,K. (1989) Effect of metals on nucleoside hydroperoxide, a product of ionizing radiation in DNA. Free Radic. Biol. Med., 7, 131–143. [DOI] [PubMed] [Google Scholar]
- 11.Bjelland S., Ånensen,H., Knævelsrud,I. and Seeberg,E. (2001) Cellular effects of 5-formyluracil in DNA. Mutat. Res., 486, 147–154. [DOI] [PubMed] [Google Scholar]
- 12.Kasai H., Iida,A., Yamaizumi,Z., Nishimura,S. and Tanooka,H. (1990) 5-Formyl-deoxyuridine: a new type of DNA damage induced by ionizing radiation and its mutagenicity to Salmonella strain TA102. Mutat. Res., 243, 249–253. [DOI] [PubMed] [Google Scholar]
- 13.Fujikawa K., Kamiya,H. and Kasai,H. (1998) The mutations induced by oxidatively damaged nucleotides, 5-formyl-dUTP and 5-hydroxyl-dCTP, in Escherichia coli. Nucleic Acids Res., 26, 4582–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyabe I., Zhang,Q.-M., Sugiyama,H., Kino,K. and Yonei,S. (2001) Mutagenic effects of 5-formyluracil on a plasmid vector during replication in Escherichia coli. Int. J. Radiat. Biol., 77, 53–58. [DOI] [PubMed] [Google Scholar]
- 15.Klungland A., Paulsen,R., Rolseth,V., Yamada,Y., Ueno,Y., Wiik,P., Matsuda,A., Seeberg,E. and Bjelland,S. (2001) 5-Formyluracil and its nucleoside derivatives confer toxicity and mutagenicity to mammalian cells by interfering with normal RNA and DNA metabolism. Toxicol. Lett., 119, 71–78. [DOI] [PubMed] [Google Scholar]
- 16.Ånensen H., Provan,F., Lian,A.T., Reinertsen,S.-H.H.S., Ueno,Y., Matsuda,A., Seeberg,E. and Bjelland,S. (2001) Mutations induced by 5-formyl-2′-deoxyuridine in Escherichia coli include base substitutions that can arise from mispairs of 5-formyluracil with guanine, cytosine and thymine. Mutat. Res., 476, 99–107. [DOI] [PubMed] [Google Scholar]
- 17.Kamiya H., Murata-Kamiya,N., Karino,N., Ueno,Y., Matsuda,A. and Kasai,H. (2002) Induction of T→G and T→A transversions by 5-formyluracil in mammalian cells. Mutat. Res., 513, 213–222. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q.-M., Sugiyama,H., Miyabe,I., Matsuda,S., Saito,I. and Yonei,S. (1997) Replication of DNA templates containing 5-formyluracil, a major oxidative lesion of thymine in DNA. Nucleic Acids Res., 25, 3969–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q.-M., Sugiyama,H., Miyabe,I., Matsuda,S., Kino,K., Saito,I. and Yonei,S. (1999) Replication in vitro and cleavage by restriction endonuclease of 5-formyluracil- and 5-hydroxymethyluracil-containing oligonucleotides. Int. J. Radiat. Biol., 75, 59–65. [DOI] [PubMed] [Google Scholar]
- 20.Bjelland S., Birkeland,N.K., Benneche,T., Volden,G. and Seeberg,E. (1994) DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J. Biol. Chem., 269, 30489–30495. [PubMed] [Google Scholar]
- 21.Zhang Q.-M., Miyabe,I., Matsumoto,Y., Kino,K., Sugiyama,H. and Yonei,S. (2000) Identification of repair enzymes for 5-formyluracil in DNA: Nth, Nei and MutM proteins of Escherichia coli. J. Biol. Chem., 275, 35471–35477. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q.-M. (2001) Role of the Escherichia coli and human DNA glycosylases that remove 5-formyluracil from DNA in the prevention of mutations. J. Radiat. Res., 42, 11–19. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q.-M., Fujimoto,J. and Yonei,S. (1995) Enzymatic release of 5-formyluracil by mammalian liver extracts from DNA irradiated with ionizing radiation. Int. J. Radiat. Biol., 68, 603–607. [DOI] [PubMed] [Google Scholar]
- 24.Bjelland S., Eide,L., Time,R.W., Store,R., Eftedal,I., Volden,G. and Seeberg,E. (1995) Oxidation of thymine to 5-formyluracil in DNA: mechanisms of formation, structural implications, and base excision by human cell free extracts. Biochemistry, 34, 14758–14764. [DOI] [PubMed] [Google Scholar]
- 25.Eisen J.A. and Hanawalt,P.C. (1999) A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res., 435, 171–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitra S. (2001) DNA glycosylases: specificity and mechanisms. Prog. Nucleic Acid Res. Mol. Biol., 68, 189–205. [DOI] [PubMed] [Google Scholar]
- 27.Jiang D., Hatahet,Z., Melamede,R.J., Kow,Y.W. and Wallace,S.S. (1997) Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem., 272, 32230–32239. [DOI] [PubMed] [Google Scholar]
- 28.Hazra T.K., Izumi,T., Boldogh,I., Imhoff,B., Kow,Y.W., Jaruga,P., Dizdaroglu,M. and Mitra,S. (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA, 99, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeda S., Biswas,T., Roy,R., Izumi,T., Boldogh,I., Kurosky,A., Sarker,A.H., Seki,S. and Mitra,S. (1998) Purification and characterization of a human NTH1, a homolog of Escherichia coli endonuclease III. J. Biol. Chem., 273, 21585–21593. [DOI] [PubMed] [Google Scholar]
- 30.Hilbert T.P., Chaung,W., Boorstein,R.J., Cunnigham,R.P. and Teebor,G.W. (1997) Cloning and expression of the cDNA encoding the human homologue of the DNA repair enzyme, Escherichia coli endonulease III. J. Biol. Chem., 272, 6733–6740. [DOI] [PubMed] [Google Scholar]
- 31.Eide L., Luna,L., Gustad,E.C., Henderson,P.T., Essigmann,J.M., Demple,B. and Seeberg,E. (2001) Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry, 40, 6653–6659. [DOI] [PubMed] [Google Scholar]
- 32.Sugiyama H., Matsuda,S., Zhang,Q.-M., Yonei,S. and Saito,I. (1996) New synthetic method of 5-formyluracil-containing oligonucleotides and their melting behavior. Tetrahedron Lett., 37, 9067–9070. [Google Scholar]
- 33.Dianov G.L., Thybo,T., Dianova,I.I., Lipinski,L.J. and Bohr,V.A. (2000) Single nucleotide patch base excision repair is the major pathway for removal of thymine glycol from DNA in human cell extracts. J. Biol. Chem., 275, 11809–11813. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto Y., Zhang,Q.-M., Takao,M., Yasui,A. and Yonei,S. (2001) Escherichia coli Nth and human hNTH1 DNA glycosylases are involved in removal of 8-oxoguanine from 8-oxoguanune/guanine mispairs in DNA. Nucleic Acids Res., 29, 1975–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailly V. and Verly,W.G. (1987) Escherichia coli endonuclease III is not an endonuclease but a β-elimination catalyst. Biochem. J., 242, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tchou J. and Grollman,A.P. (1995) The catalytic mechanism of Fpg protein. Evidence for a Schiff base intermediate and amino terminus localization of the catalytic site. J. Biol. Chem., 270, 11671–11677. [DOI] [PubMed] [Google Scholar]
- 37.Rabow L.E. and Kow,Y.W. (1997) Mechanism of action of base release by Escherichia coli Fpg protein: role of lysine 155 in catalysis. Biochemistry, 36, 5084–5096. [DOI] [PubMed] [Google Scholar]
- 38.Nash H.M., Bruner,S.D., Schärer,O.D., Kawate,T., Addona,T.A., Spooner,E., Lane,W.S. and Verdine,G.L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol., 6, 968–980. [DOI] [PubMed] [Google Scholar]
- 39.Dizdaroglu M., Karahalil,B., Sentürker,S., Buckley,T.J. and Roldán-Arjona,T. (1999) Excision of products of oxidative DNA base damage by human NTH1 protein. Biochemistry, 38, 243–246. [DOI] [PubMed] [Google Scholar]
- 40.Aspinwall R., Rothwell,D.G., Roldan-Arjona,T., Anselmino,C., Ward,C.J., Cheadle,J.P., Sampson,J.R., Lindahl,T., Harris,P.C. and Hickson,I.D. (1997) Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl Acad. Sci. USA, 94, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]