Abstract
Accumulating evidence suggests that altered RNA editing of the serotonin 2C receptor (HTR2C) is involved in the pathophysiology of mental disorders and the action of antidepressants. Estimating RNA editing of HTR2C in various samples is a first step to understanding its pathophysiological roles. Here, we developed a high-throughput quantification method of RNA editing efficiency by pyrosequencing. By optimizing the dispensation order, the RNA editing efficiency of all five RNA editing sites including consecutively ordered sites in HTR2C was obtained. More importantly, our method made it possible to determine the content of partial HTR2C isoforms, which enabled us to monitor possible functional changes of HTR2C. This method was validated in both oligonucleotide and RT-PCR product templates, and showed good correlation with conventional cloning-sequencing analysis. Our method could be a valuable tool in the rapid assessment of RNA editing status, including assessment of natural variations, alterations in disease tissues, and responses to drugs.
Keywords: HTR2C, mental disorder, antidepressant
INTRODUCTION
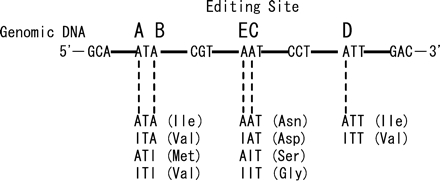
Pre-mRNA of serotonin 2C receptor (HTR2C) undergoes adenosine-to-inosine (A-to-I) RNA editing, by which specific adenosines are converted into inosines by adenosine deaminases that act on RNA (ADARs) (Reenan 2001; Bass 2002; Maas et al. 2003). Since inosine is read as guanosine by translation machinery, A-to-I RNA editing leads to amino acid substitutions. To date, five adenosine residues (termed sites A to E) in the second intracellular loop of HTR2C have been found to be edited (Burns et al. 1997), and amino acid substitution occurs at three sites (I157 to V or M; N159 to S, D, or G; and I161 to V). This results in the generation of up to 24 different HTR2C isoforms. Importantly, different HTR2C isoforms exhibit considerably different G-protein coupling efficiencies, and the combination of isoforms is regulated in a brain region–specific manner (Burns et al. 1997; Fitzgerald et al. 1999; Herrick-Davis et al. 1999; Niswender et al. 1999; Wang et al. 2000).
Genetic, pharmacological, animal model, and postmortem studies suggest the involvement of HTR2C in neuropsychiatric disorders. Functional polymorphism of Cys23Ser polymorphism of the serotonin 2C receptor (HTR2C) (Okada et al. 2004) is associated with depression and bipolar disorder (Oruc et al. 1997; Gutierrez et al. 2001; Lerer et al. 2001), tardive dyskinesia in schizophrenia (Segman et al. 2000), psychotic symptoms in late-onset Alzheimer’s disease (Holmes et al. 1998), and migraine with aura (Kusumi et al. 2004). HTR2C-deficient mice exhibited abnormal control of feeding behavior and enhanced seizure susceptibility (Tecott et al. 1995). In addition, down-regulation of HTR2C was found in postmortem brains of patients with bipolar disorder and schizophrenia (Castensson et al. 2003; Iwamoto et al. 2004).
RNA editing of HTR2C is also thought to be involved in the pathophysiology of mental disorders and the action of antidepressants (Seeburg 2002). In animal models, it has been shown that differential HTR2C RNA editing among inbred strains of mice may underlie differences in stress reactivity (Englander et al. 2005). Fluoxetine, a selective serotonin reuptake inhibitor (SSRI), alters RNA editing of HTR2C (Gurevich et al. 2002; Englander et al. 2005), whereas some drugs such as cocaine or reserpine do not appear to affect RNA editing (Iwamoto and Kato 2002). In cellular models, increased RNA editing of HTR2C has been reported in human glioblastoma cells treated with interferon-α (IFN-α) (Yang et al. 2004). In postmortem brains, altered RNA editing of HTR2C has been reported in patients with schizophrenia, depression, and those who committed suicide (Niswender et al. 2001; Sodhi et al. 2001; Gurevich et al. 2002; Iwamoto and Kato 2003), although the data remain inconsistent (Niswender et al. 2001; Sodhi et al. 2001; Gurevich et al. 2002; Dracheva et al. 2003; Iwamoto and Kato 2003).
Estimating the RNA editing status of HTR2C in various samples is a first step to understanding its role in complex neuropsychiatric disorders and the mechanism of action of psychiatric drugs. However, determining RNA editing efficiency of HTR2C is laborious work. Typically, after the RT-PCR amplification of the edited region, products are subjected to cloning and sequence analysis, or radioisotope (RI)-based primer extension analysis. To reduce the time and effort involved, we previously devised a non-RI method for estimating RNA editing efficiency by primer extension combined with denaturing high-performance liquid chromatography using the RT-PCR product as template (Iwamoto and Kato 2002). Although this method is rapid and accurate, a disadvantage is that it is limited to estimating the RNA editing efficiency of sites A and D, which are located at either end of the RNA editing region of HTR2C. Information for the other three sites, which are located downstream of site A and upstream of site D, is unavailable by this method.
Pyrosequencing is a well-established genetic analysis method based on the principle of sequencing by synthesis using four enzymes (Ronaghi et al. 1998; Agah et al. 2004). In this technique, after each dNTP is dispensed, DNA polymerase incorporates it into the sequencing primer that was pre-annealed to the single-stranded template DNA, and pyrophosphate (PPi) is released. ATP sulfurylase converts adenosine-5′-phosphosulfate (APS) to ATP with PPi. Luciferase then produces light by using ATP. The light is detected by a charge coupled device camera, and presented as a peak in a pyrogram. Each peak height is proportional to the number of dNTPs incorporated, allowing quantitative measurements. After the apyrase degrades unincorporated dNTPs and excess ATP, the next dNTP is added. Here, we developed a method to estimate the efficiencies of all five RNA editing sites and partial isoform contents of HTR2C by using pyrosequencing technology.
RESULTS
Quantification of sites A, B, and D of HTR2C
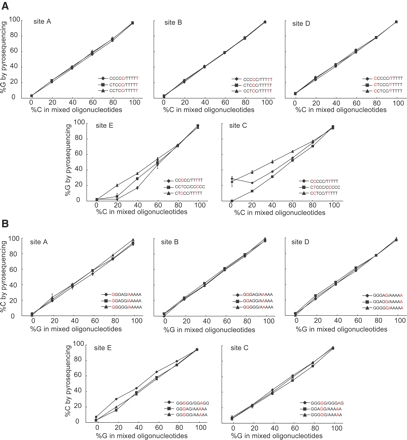
Among the five edited sites, sites A, B, and D were located either at both ends (sites A and D) or in a nonconsecutive order (site B) (Fig. 1). Therefore, estimation of RNA editing efficiencies of these sites is expected to be done by standard allele quantification protocols provided by manufacturers. Indeed, the ratios of adenine to guanine (forward orientation, Fig. 2A) and thymine to cytosine (reverse orientation, Fig. 2B) in these three sites were accurately estimated regardless of the combination of oligonucleotide templates.
FIGURE 1.
Amino acid changes associated with RNA editing of the HTR2C.
FIGURE 2.
Estimation of RNA editing efficiencies using oligonucleotide templates. The results of pyrosequencing in the forward orientation (A) and the reverse orientation (B). In each experiment, two oligonucleotides were mixed and used as templates. Combinations of two oligonucleotides are shown for each site (see graphs for symbol keys). The percentage of C or G in the mixed templates is plotted on the X-axis. The observed values by pyrosequencing analysis are plotted on the Y-axis. Experiments were done in triplicate, and data are given as the mean ± SD. The observed values in sites A, B, and D were derived from the AQ mode in the software. Those in sites E and C were determined from the sum of partial isoform contents (see Fig. 4).
Quantification of sites E and C of HTR2C
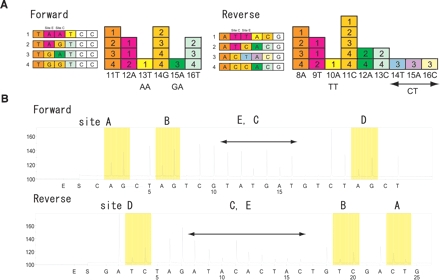
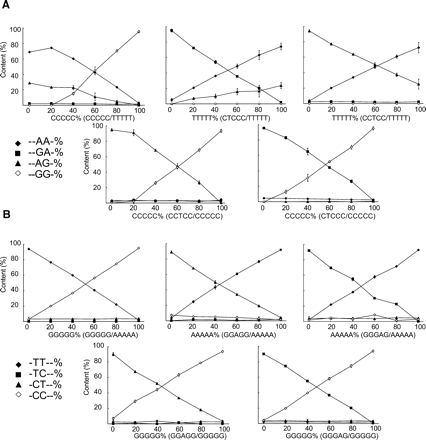
Because of the nature of the primer extension reaction, the dispensation order (the order in which they should be dispensed in a pyrosequencing run) is important for determining the ratios of consecutive polymorphic bases. In HTR2C, sites E and C are consecutively ordered (Fig. 1). For example, since dATPαS would be incorporated into two consecutive adenines of “- -AA-” and the first adenine of “- -AG-” isoforms, the resultant intensity of emitted light (given in a peak height) by injection of dATPαS is a sum of the two isoforms. Therefore, we cannot estimate the RNA editing efficiencies by the typical dispensation order determined by software. To overcome this, we manually determined the dispensation order so as to determine the content of one partial isoform at first (Fig. 3A). After that, the contents of the other three partial isoforms were sequentially determined. Finally, RNA editing efficiency of sites E and C was assessed by the sum of the relative content of isoforms. To evaluate this method, we calculated the content of each isoform in both orientations with oligonucleotide templates. When equal amounts of four oligonucleotides, each of which corresponds to one partial isoform, were mixed and used as templates, almost identical pyrograms were obtained, as predicted (Fig. 3B). In addition, we efficiently estimated the isoform contents and editing efficiencies by our methods (data not shown).
FIGURE 3.
Strategy for the estimation of RNA editing efficiencies of sites E and C. (A) Four possible isoforms regarding sites E and C (partial isoforms) are shown at the left. The dispensation order and theoretical peak height in each dispensation are shown at the right. Theoretical peak height was determined by assuming that equal amounts of four partial isoforms were present in templates. In the forward strand, the 11th dNTP dispensed is dTTP (11T). The peak height of 11T consists of content from all four partial isoforms. On the other hand, the peak heights of 13T and 15A exclusively correspond to the content of a single isoform of “- -AA-” and “- -GA-,” respectively. The content of an “- -AG-” isoform was determined by the equation as follows: [- -AG-] = 12A − 2 * 13T. The content of “- -GG-” was determined by the following equation: [- -GG-] = 16T − 15A − [- -AG-]. In the reverse strand, the peak heights of 10A and each of 14T, 15A, and 16C exclusively correspond to the content of single isoform of “-TT- -” and “-CT- -,” respectively. To ensure robustness, the content of “-CT- -” was determined by averaging peak heights of 14T, 15A, and 16C. The content of “-TC- -” and “-CC- -” was determined using the following equations: [-TC- -] = 9T − 2 − 10A, and [-CC- -] = {(12A + 13C) − 2 * [-TC- -]}/2, respectively. In both strands, negative values are set as 0. Finally, each isoform content was normalized so that the total content of the four isoforms was equal to 1. (B) Representative pyrograms. Four oligonucleotides (forward: CCCCC, CCTCC, CTCCC, and TTTTT; reverse: GGGGG, GGAGG, GGGAG, and AAAAA) (see Table 1) were mixed in equal proportions and used as templates. Arrows indicate the peaks corresponding to the dispensation of dNTP shown in Figure 1A (from 11T to 16T in forward, and from 8A to 16C in reverse). Note that the peak height of A yielded higher (110%) emission light than did natural dNTPs because of the use of dATPαS. We divided the value of the peak height of A by 1.1 before manual calculation.
We found that accuracy of this method is highly dependent on the composition of oligonucleotides in the forward orientation (Fig. 4A). In particular, when a TTTTT oligo-nucleotide was used for a mixture of oligonucleotide templates, considerable deviations from theoretical values were observed. Failure of estimation of isoform content resulted in the underestimation of total %G in site E and the overestimation of total %G in site C (Fig. 2A). Although some degree of deviation was also observed in the reverse orientation, the estimation of isoform contents (Fig. 4B) and %C in sites E and C (Fig. 2B) showed good accordance with the theoretical values. Therefore, we applied our methods in the reverse orientation to estimate RNA editing frequencies.
FIGURE 4.
Estimation of partial isoform contents using oligonucleotide templates. The results of pyrosequencing in the forward orientation (A) and the reverse orientation (B). In each experiment, two oligonucleotides were mixed and used as templates. Combinations of two oligonucleotides are shown, and the proportion of one oligonucleotide in the mixed templates is plotted on the X-axis. The proportion of each isoform determined by pyrosequencing analysis is plotted on the Y-axis. Experiments were done in triplicate, and data are given as the mean ± SD.
Quantification of RNA editing efficiencies in rat samples
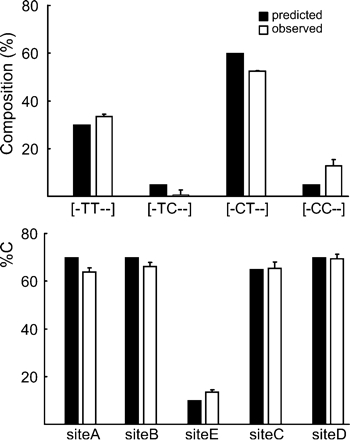
Before starting quantification of rat HTR2C RNA editing efficiencies, we re-estimated partial isoform contents and %C with oligonucleotide templates (Fig. 5). In this case, four oligonucleotides were mixed so as to resemble the natural RNA editing pattern, which included RNA editing efficiency of five sites and composition of partial isoforms. We found that our estimation method yielded consistent results with theoretical values (Fig. 5).
FIGURE 5.
Estimation of partial isoform contents and %C using the oligonucleotide template that resembles the natural RNA editing pattern. Oligonucleotides were mixed as follows: GGAGG (60%), AAAAA (30%), GGGGG (5%), and GGGAG (5%). Predicted and observed values are given. Experiments were done in triplicate, and data are given as the mean ± SD.
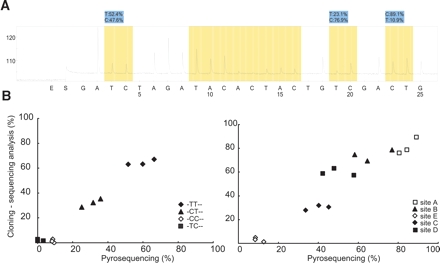
When we used RT-PCR products as templates, pyrograms generally showed unstable base lines and lower peak heights compared to those using oligonucleotide templates (Fig. 6A). However, analyses were normally flagged as “successful” by the software in sites A, B, and D (Fig. 6A). We also performed manual calculation at these three sites, which yielded almost identical estimation values as those that were automatically determined by the software (data not shown). These suggest that the unstable base lines and lower peak heights found in RT-PCR product templates were likely to be at negligible levels and they did not considerably affect our calculation method. The results derived from the pyrosequencing method were in good accordance with those derived from cloning-sequencing methods in both partial isoform contents (R = 0.984) and RNA editing efficiencies of the five sites (R = 0.952) (Fig. 6B).
FIGURE 6.
Estimation of RNA editing efficiencies of HTR2C in rat prefrontal cortices. (A) Representative pyrogram using rat RT-PCR product as a template. (B) Comparison of cloning-sequencing analysis and pyrosequencing. Partial isoform content (left) and RNA editing efficiencies of each site (right) are given. Cloning-sequencing analysis was performed by sequencing ~50 colonies per rat sample.
DISCUSSION
Compared with conventional methods, pyrosequencing provides a rapid and accurate non-RI method for quantification of polymorphic sites (Fakhrai-Rad et al. 2002; Ronaghi 2003; Elahi and Ronaghi 2004; Shendure et al. 2004). To date, this technology has been used for the purpose of quantifying entities such as allele frequency in pooled DNA samples, and DNA methylation of CpG sites in bisulfite-modified DNA. Here we provide a method for quantifying the RNA editing efficiencies of five editing sites in HTR2C in one reaction by pyrosequencing. Contrary to the above applications, this involves the quantification of two consecutive polymorphic sites, and therefore a different strategy for quantification needed to be developed. We were able to estimate RNA editing efficiencies of these consecutive sites via sequential determination of the partial isoform contents.
However, we could not optimize estimation of sites E and C in the forward orientation. Considering the relatively consistent results between observed and theoretical values in the forward orientation using the CCCCC type of oligonucleotide, and the consistent results in the reverse orientation, we assumed that the peak heights of consecutive A sites were not correctly estimated by our method. The reason for this is not clear. Our attempts at determining the correction coefficient failed to ensure the robustness in estimation using oligonucleotide templates (data not shown). Therefore, in the present study, we applied our method only in the reverse orientation and validated our method in both oligonucleotide and RT-PCR product templates. Other strategies, such as multiplex pyrosequencing approach, might improve the accuracy of the estimation since adjacent peaks would be used to confirm the quantity of the peak to be analyzed (Langaee and Ronaghi 2005).
It should also be noted that since pyrosequencing is based on primer extension, our method has some of the same technical limitations as other primer extension-based methods. First, it is difficult to estimate the instances when the polymorphic sites are completely or nearly completely composed of one base. In such cases, pyrosequencing analysis often yields the deviated results. This is probably due to the nonspecific incorporation of dNTP, and sensitivity to background noise variations. Second, although our method estimates partial isoform content, the editing pattern of five sites was not obtained, impeding the assessment of overall functional changes of HTR2C. Therefore, several conventional approaches such as cloning-sequencing analysis and digestion by restriction enzymes of RT-PCR product would still help to better understand the RNA editing.
Despite the above limitations, pyrosequencing is a superior alternative method to other primer extension-based methods, and our method provides additional information on partial isoform content. Two consecutive sites in HTR2C, E and C, correspond to the first and second codons of AAC, respectively, which encodes the 159th asparagine residue (N159). RNA editing of these sites always resulted in the amino acid change of N159 to S (AGC), D (GAC), or G (GGC). Therefore, information about these partial isoform contents would be useful for monitoring the possible functional changes of HTR2C at the amino acid level. In addition, using a strategy similar to that used in this study, partial isoforms involving sites A and B can also be examined, which reveals RNA editing status at the amino acid level. Given the high sequence identity around the edited region of mammalian HTR2C, our method can be applied to other mammalian species such as human and mouse without changing the reading sequence and dispensation order (but a minor modification of primer pairs would be needed for RT-PCR) (Iwamoto and Kato 2003). Therefore, our method described here would help the rapid assessment of RNA editing efficiencies of HTR2C in various aspects, such as assessment of natural variations, alterations in disease tissues, and responses to drugs.
MATERIALS AND METHODS
RT-PCR
For the purpose of validating the pyrosequencing analysis, total RNA extracted from the prefrontal cortices of rats (male Sprague-Dawley, 5–6 wk old, N = 3) was used. They were prepared in our previous study and already determined RNA editing status by cloning and sequencing analysis (Iwamoto et al. 2005). Briefly, a portion of RT-PCR product was TA-cloned into the pCR2.1 vector (Invitrogen), and the reaction mixture was used for transformation of Escherichia coli. The cDNA clones derived from single bacterial colonies were sequenced with an ABI3700 DNA sequencer (Applied Biosystems). cDNA synthesis and RT-PCR were performed in each animal sample, and at least 50 colonies were sequenced per sample.
For pyrosequencing analysis in the present study, RT-PCR amplification of the edited region of HTR2C was independently performed as described previously (Iwamoto and Kato 2003), with the modification of using a biotinylated primer (Table 1) in the second PCR step. RT-PCR products were purified with a PCR96 Cleanup Plate according to the manufacturer’s protocol (Millipore).
TABLE 1.
Oligonucleotides, reading sequences, and dispensation orders used in this study
| Sequences | |
| 1st RT-PCR primers | 5′-TGGATTTCACTAGATGTGCT-3′ |
| 5′-GTCCCTCAGTCCAATCACAG-3′ | |
| 2nd RT-PCR primers (reverse orientation) | Bio-5′-TGGATTTCACTAGATGTGCT-3′ |
| 5′-TTGATATTGCCCAAACGATG-3′ | |
| Pyrosequencing in forward orientationa | |
| CCCCC | 5′-ACAGGACCACGCACTGCTACATACCGGTCCAGCG-3′ |
| CCTCC | 5′-ACAGGACTACGCACTGCTACATACCGGTCCAGCG-3′ |
| CTCCC | 5′-ACAGGATCACGCACTGCTACATACCGGTCCAGCG-3′ |
| CTTCC | 5′-ACAGGATTACGCACTGCTACATACCGGTCCAGCG-3′ |
| TTTTT | 5′-ATAGGATTACGTATTGCTACATACCGGTCCAGCG-3′ |
| Sequencing primer | 5′-CGCTGGACCGGTATGTAGCA-3′ |
| Reading sequence | 5′-A/GTA/GCGTA/GA/GTCCTA/GTTG-3′ |
| Dispensation order | CAGCTAGTCGTATGATGTCTAGCT |
| Pyrosequencing in reverse orientationa | |
| GGGGG | 5′-GTGCGTGGTCCTGTTGAGCATAGCCGGTTCAATTC-3′ |
| GGAGG | 5′-GTGCGTAGTCCTGTTGAGCATAGCCGGTTCAATTC-3′ |
| GGGAG | 5′-GTGCGTGATCCTGTTGAGCATAGCCGGTTCAATTC-3′ |
| GGAAG | 5′-GTGCGTAATCCTGTTGAGCATAGCCGGTTCAATTC-3′ |
| AAAAA | 5′-ATACGTAATCCTATTGAGCATAGCCGGTTCAATTC-3′ |
| Sequencing primer | 5′-GAATTGAACCGGCTATGCTC-3′ |
| Reading sequence | 5′-AAT/CAGGAT/CT/CACGT/CAT/CTG-3′ |
| Dispensation order | GATCTAGATACACTACTGTCGACTG |
aBases that correspond to RNA editing sites are underlined.
Pyrosequencing
The RT-PCR product was processed for pyrosequencing analysis according to the manufacturer’s standard protocol (Biotage). Briefly, 4 μL of streptavidin-Sepharose beads (Amersham Biosciences) and 29 μL of binding buffer (10 mM Tris-HCl, 1 mM EDTA, 2 M NaCl, 0.1% Tween 20 at pH 7.6) were mixed with 25 μL of RT-PCR product for 10 min at room temperature. The reaction mixture was placed onto a MultiScreen-HV, Clear Plate (Millipore). After applying the vacuum, the beads were treated with a denaturation solution (0.2 N NaOH) for 1 min, and washed twice with washing buffer (10 mM Tris-Acetate at pH 7.6). The beads were then suspended with 50 μL of annealing buffer (20 mM Tris-Acetate, 2 mM Mg-Acetate at pH 7.6) containing 10 pmol of sequencing primer. When oligonucleotides were used as templates, a total of 2 pmol of oligonucleotides was mixed in 50 μL of annealing buffer containing 10 pmol of sequencing primer. The template-sequencing primer mixture was transferred onto a PSQ 96 Plate (Biotage), heated to 90°C for 2 min, and cooled to room temperature. Sequencing reactions were performed with a PSQ 96 SNP Reagent Kit (Biotage) using a PSQ96MA (Biotage) according to the manufacturer’s instructions. Experiments were done in triplicate. The primer pairs, the order of nucleotide dispensation, and the analyzed sequences are shown in Table 1.
Data analysis
RNA editing efficiencies of sites A, B, and D were estimated using the Allele Frequency Quantification (AQ) mode in the SNP software (Biotage). RNA editing efficiencies of sites E and C were manually calculated using the peak height information by the method described in Figure 3. In the pyrosequencing reaction, deoxyadenosine alfa-thio triphosphate (dATPαS), which is efficiently used by DNA polymerase, was used. To compensate for the higher emission of dATPαS, we divided the value of the peak height of A by 1.1 before manual calculation.
Acknowledgments
We thank Yoko Aida and Shinichiro Takeshima at Retrovirus Research Unit, RIKEN, for their suggestions in experiments. We thank Asaho Adachi and Ryoko Matsuda at SC BioSciences Corporation for their help with data analysis and critical comments on the manuscript. This work was supported by a Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2114505.
REFERENCES
- Agah, A., Aghajan, M., Mashayekhi, F., Amini, S., Davis, R.W., Plummer, J.D., Ronaghi, M., and Griffin, P.B. 2004. A multi-enzyme model for Pyrosequencing. Nucleic Acids Res. 32: http://nar.oxfordjournals.org/cgi/content/full/32/21/e166. [DOI] [PMC free article] [PubMed]
- Bass, B.L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71: 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., and Emeson, R.B. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308. [DOI] [PubMed] [Google Scholar]
- Castensson, A., Emilsson, L., Sundberg, R., and Jazin, E. 2003. Decrease of serotonin receptor 2C in schizophrenia brains identified by high-resolution mRNA expression analysis. Biol. Psychiatry 54: 1212–1221. [DOI] [PubMed] [Google Scholar]
- Dracheva, S., Elhakem, S.L., Marcus, S.M., Siever, L.J., McGurk, S.R., and Haroutunian, V. 2003. RNA editing and alternative splicing of human serotonin 2C receptor in schizophrenia. J. Neurochem. 87: 1402–1412. [DOI] [PubMed] [Google Scholar]
- Elahi, E. and Ronaghi, M. 2004. Pyrosequencing: A tool for DNA sequencing analysis. Methods Mol. Biol. 255: 211–219. [DOI] [PubMed] [Google Scholar]
- Englander, M.T., Dulawa, S.C., Bhansali, P., and Schmauss, C. 2005. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J. Neurosci. 25: 648–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhrai-Rad, H., Pourmand, N., and Ronaghi, M. 2002. Pyrosequencing: An accurate detection platform for single nucleotide polymorphisms. Hum. Mutat. 19: 479–485. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, L.W., Iyer, G., Conklin, D.S., Krause, C.M., Marshall, A., Patterson, J.P., Tran, D.P., Jonak, G.J., and Hartig, P.R. 1999. Messenger RNA editing of the human serotonin 5-HT2C receptor. Neuropsychopharmacology 21: 82S–90S. [DOI] [PubMed] [Google Scholar]
- Gurevich, I., Tamir, H., Arango, V., Dwork, A.J., Mann, J.J., and Schmauss, C. 2002. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34: 349–356. [DOI] [PubMed] [Google Scholar]
- Gutierrez, B., Arias, B., Papiol, S., Rosa, A., and Fananas, L. 2001. Association study between novel promoter variants at the 5-HT2C receptor gene and human patients with bipolar affective disorder. Neurosci. Lett. 309: 135–137. [DOI] [PubMed] [Google Scholar]
- Herrick-Davis, K., Grinde, E., and Niswender, C.M. 1999. Serotonin 5-HT2C receptor RNA editing alters receptor basal activity: Implications for serotonergic signal transduction. J. Neurochem. 73: 1711–1717. [DOI] [PubMed] [Google Scholar]
- Holmes, C., Arranz, M.J., Powell, J.F., Collier, D.A., and Lovestone, S. 1998. 5-HT2A and 5-HT2C receptor polymorphisms and psychopathology in late onset Alzheimer”s disease. Hum. Mol. Genet. 7: 1507–1509. [DOI] [PubMed] [Google Scholar]
- Iwamoto, K. and Kato, T. 2002. Effects of cocaine and reserpine administration on RNA editing of rat 5-HT(2C) receptor estimated by primer extension combined with denaturing high-performance liquid chromatography. Pharmacogenomics J. 2: 335–340. [DOI] [PubMed] [Google Scholar]
- ———. 2003. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci. Lett. 346: 169–172. [DOI] [PubMed] [Google Scholar]
- Iwamoto, K., Kakiuchi, C., Bundo, M., Ikeda, K., and Kato, T. 2004. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol. Psychiatry 9: 406–416. [DOI] [PubMed] [Google Scholar]
- Iwamoto, K., Nakatani, N., Bundo, M., Yoshikawa, T., and Kato, T. 2005. Altered RNA editing of serotonin 2C receptor in a rat model of depression. Neurosci. Res. 53: 69–76. [DOI] [PubMed] [Google Scholar]
- Kusumi, M., Araki, H., Ijiri, T., Kowa, H., Adachi, Y., Takeshima, T., Sakai, F., and Nakashima, K. 2004. Serotonin 2C receptor gene Cys23Ser polymorphism: A candidate genetic risk factor of migraine with aura in Japanese population. Acta Neurol. Scand. 109: 407–409. [DOI] [PubMed] [Google Scholar]
- Langaee, T. and Ronaghi, M. 2005. Genetic variation analyses by Pyrosequencing. Mutat. Res. 573: 96–102. [DOI] [PubMed] [Google Scholar]
- Lerer, B., Macciardi, F., Segman, R.H., Adolfsson, R., Blackwood, D., Blairy, S., Del Favero, J., Dikeos, D.G., Kaneva, R., Lilli, R., et al. 2001. Variability of 5-HT2C receptor Cys23Ser polymorphism among European populations and vulnerability to affective disorder. Mol. Psychiatry 6: 579–585. [DOI] [PubMed] [Google Scholar]
- Maas, S., Rich, A., and Nishikura, K. 2003. A-to-I RNA editing: Recent news and residual mysteries. J. Biol. Chem. 278: 1391–1394. [DOI] [PubMed] [Google Scholar]
- Niswender, C.M., Copeland, S.C., Herrick-Davis, K., Emeson, R.B., and Sanders-Bush, E. 1999. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem. 274: 9472–9478. [DOI] [PubMed] [Google Scholar]
- Niswender, C.M., Herrick-Davis, K., Dilley, G.E., Meltzer, H.Y., Overholser, J.C., Stockmeier, C.A., Emeson, R.B., and Sanders-Bush, E. 2001. RNA editing of the human serotonin 5-HT2C receptor. Alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology 24: 478–491. [DOI] [PubMed] [Google Scholar]
- Okada, M., Northup, J.K., Ozaki, N., Russell, J.T., Linnoila, M., and Goldman, D. 2004. Modification of human 5-HT(2C) receptor function by Cys23Ser, an abundant, naturally occurring aminoacid substitution. Mol. Psychiatry 9: 55–64. [DOI] [PubMed] [Google Scholar]
- Oruc, L., Verheyen, G.R., Furac, I., Jakovljevic, M., Ivezic, S., Raeymaekers, P., and Van Broeckhoven, C. 1997. Association analysis of the 5-HT2C receptor and 5-HT transporter genes in bipolar disorder. Am. J. Med. Genet. 74: 504–506. [PubMed] [Google Scholar]
- Reenan, R.A. 2001. The RNA world meets behavior: A → I pre-mRNA editing in animals. Trends Genet. 17: 53–56. [DOI] [PubMed] [Google Scholar]
- Ronaghi, M. 2003. Pyrosequencing for SNP genotyping. Methods Mol. Biol. 212: 189–195. [DOI] [PubMed] [Google Scholar]
- Ronaghi, M., Uhlen, M., and Nyren, P. 1998. A sequencing method based on real-time pyrophosphate. Science 281: 363–365. [DOI] [PubMed] [Google Scholar]
- Seeburg, P.H. 2002. A-to-I editing: New and old sites, functions and speculations. Neuron 35: 17–20. [DOI] [PubMed] [Google Scholar]
- Segman, R.H., Heresco-Levy, U., Finkel, B., Inbar, R., Neeman, T., Schlafman, M., Dorevitch, A., Yakir, A., Lerner, A., Goltser, T., et al. 2000. Association between the serotonin 2C receptor gene and tardive dyskinesia in chronic schizophrenia: Additive contribution of 5-HT2Cser and DRD3gly alleles to susceptibility. Psychopharmacology (Berl) 152: 408–413. [DOI] [PubMed] [Google Scholar]
- Shendure, J., Mitra, R.D., Varma, C., and Church, G.M. 2004. Advanced sequencing technologies: Methods and goals. Nat. Rev. Genet. 5: 335–344. [DOI] [PubMed] [Google Scholar]
- Sodhi, M.S., Burnet, P.W., Makoff, A.J., Kerwin, R.W., and Harrison, P.J. 2001. RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Mol. Psychiatry 6: 373–379. [DOI] [PubMed] [Google Scholar]
- Tecott, L.H., Sun, L.M., Akana, S.F., Strack, A.M., Lowenstein, D.H., Dallman, M.F., and Julius, D. 1995. Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature 374: 542–546. [DOI] [PubMed] [Google Scholar]
- Wang, Q., O’Brien, P.J., Chen, C.X., Cho, D.S., Murray, J.M., and Nishikura, K. 2000. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J. Neurochem. 74: 1290–1300. [DOI] [PubMed] [Google Scholar]
- Yang, W., Wang, Q., Kanes, S.J., Murray, J.M., and Nishikura, K. 2004. Altered RNA editing of serotonin 5-HT2C receptor induced by interferon: implications for depression associated with cytokine therapy. Brain Res. Mol. Brain Res. 124: 70–78. [DOI] [PubMed] [Google Scholar]