Abstract
To test whether the ribosome displays specificity for the esterified amino acid and the tRNA body of an aminoacyl-tRNA (aa-tRNA), the stabilities of 4 correctly acylated and 12 misacylated tRNAs in the ribosomal A site were determined. By introducing the GAC (valine) anticodon into each tRNA, a constant anticodon·codon interaction was maintained, thus removing concern that different anticodon·codon strengths might affect the binding of the different aa-tRNAs to the A site. Surprisingly, all 16 aa-tRNAs displayed similar dissociation rate constants from the A site. These results suggest that either the ribosome is not specific for different amino acids and tRNA bodies when intact aa-tRNAs are used or the specificity for the amino acid side chain and tRNA body is masked by a conformational change upon aa-tRNA release.
Keywords: tRNA, ribosome, uniformity, misacylated tRNA, A site, ribosome binding
Several studies indicate that the rate of incorporation of an amino acid into a growing polypeptide chain in Escherichia coli is roughly uniform when the substrate, aa-tRNA·EF-Tu·GTP ternary complex, is saturating. For example, Curran and Yarus (1989) evaluated 18 elongator aminoacyl-tRNAs (aa-tRNAs) in their ability to compete with a frameshift and concluded that the rates of incorporation of aa-tRNAs belonging to codons used frequently in highly expressed genes were all near the average aa-tRNA selection rate, while aa-tRNAs that were not preferred in highly expressed genes were incorporated faster or slower than the mean. This observation supports the proposal made by Grosjean and Fiers (1982), who suggested that codons used in highly expressed genes are selected for maintenance of a uniform anticodon·codon interaction strength. Furthermore, Pedersen (1984) found that, in three different growth media, the translation rates of several different mRNAs varied by less than twofold, and that the small differences observed were due to the low concentration of aa-tRNAs belonging to rare codons. Thus, while the lower concentration of rare aa-tRNAs (and thus ternary complexes) can lead to slower translation rates, it appears that all aa-tRNAs are used in an equivalent manner during the translation of highly expressed genes.
Not only does the overall rate of translation appear to be uniform, but there is accumulating biochemical evidence that several of the individual steps of translation elongation also display uniformity. It has been shown that the GTP-bound form of elongation factor Tu (EF-Tu·GTP) binds all elongator aa-tRNAs with similar affinities (Louie et al. 1984; Ott et al. 1990). In addition, recent studies by Fahlman et al. (2004) have demonstrated that eight different purified aa-tRNAs bind the ribosomal A and P sites with a very small range of KD values. Finally, kinetic studies have shown that Phe-tRNAPhe and Trp-tRNATrp have similar rates of GTP hydrolysis and peptide bond formation (Pape et al. 1998; Cochella and Green 2005).
The manner in which uniformity is achieved is difficult to understand given the substantial diversity among aa-tRNAs. Each tRNA has a unique anticodon, and the strengths of different anticodon·codon interactions can vary by at least 30-fold (Grosjean et al. 1978). In addition, the anticodon·codon interaction is buttressed by A minor interactions formed with 30S ribosomal bases A1492 and A1493, and this type of motif has been shown to display some specificity for different receptor base pairs (Battle and Doudna 2002). Also, each tRNA is esterified with a cognate amino acid, and the chemical and physical properties of the side chains can be quite different. Furthermore, while the different tRNA bodies have a similar overall shape, their structures vary significantly at the atomic level and thus have the potential to interact differently with the translational machinery. Finally, each tRNA has a unique cluster of post-transcriptional modifications. This raises the question: How can such a diverse set of molecules behave as equivalent substrates of the highly conserved translational machinery?
An understanding of how aa-tRNAs bind uniformly to EF-Tu resulted from analysis of the binding properties of misacylated tRNAs (LaRiviere et al. 2001). In this study four tRNA bodies were each acylated with their cognate amino acid and three noncognate amino acids. When the resulting 16 aa-tRNAs were tested in their ability to bind EF-Tu, it was found that, while all of the cognate aa-tRNAs bound with similar affinities, the misacylated tRNAs bound either tighter or weaker. Analysis of the data revealed that the amino acid side chain and tRNA body each make independent, highly variable thermodynamic contributions to EF-Tu binding, but that these contributions compensate for one another such that the overall binding of cognate aa-tRNAs is uniform. In other words, EF-Tu binds differentially to each amino acid side chain, but the sequence of each tRNA body has evolved to modify its thermodynamic contribution in a way that results in overall uniform binding for cognate aa-tRNAs. This binding phenomenon was termed thermodynamic compensation and has subsequently been substantiated using many other misacylated tRNAs (Asahara and Uhlenbeck 2002, 2005; Dale et al. 2004).
Several lines of evidence suggest that a similar compensatory mechanism may explain the observed uniform binding of aa-tRNAs to the ribosomal A site. A comparison of X-ray crystal structures of the 50S subunit of Haloarcula marismortui ribosomes complexed with either a minihelix conjugated with a puromycin at its 3′ end (Nissen et al. 2000), the Yarus inhibitor CCdA-phosphate-puromycin (Nissen et al. 2000), or CC-puromycin-phenylalanine-caproic acid-biotin (Schmeing et al. 2002) in the A site shows that all three molecules interact nearly identically with the peptidyl transferase center (Hansen et al. 2002). In each case, the moiety corresponding to the amino acid side chain is located in a hydrophobic cleft formed by the stacking of ribosomal RNA nucleotides, including the highly conserved A2486 and C2487 (A2451 and C2452, E. coli numbering). Because several antibiotics analogous to aminoacylated nucleosides (e.g., anisomysin, sparsomycin, and chloramphenicol) have also been shown to interact with this cleft, Hansen et al. (2003) labeled it the A site crevice and noted that it was large enough to accommodate the esterified amino acids of all aa-tRNAs. Several biochemical experiments suggest that this amino acid binding pocket shows specificity for the identity of the amino acid. Derivatives of the dinucleotide CpA esterified with different amino acids display a 90-fold range of apparent Kms in a peptidyltransferase assay (Bhuta et al. 1981), and derivatives of puromycin with altered amino acid side chains inhibit in vitro translation to varying extents (Starck et al. 2003). In addition, Fahlman et al. (2004) showed that the removal of the esterified amino acid from each of eight different purified aa-tRNAs reduces the binding affinity for the A site by 1.2- to 17-fold, depending on the tRNA. While these amounts are less than the nine- to 2250-fold loss in affinity of different tRNAs to EF-Tu when the esterified amino acid is removed (Janiak et al. 1990; Asahara and Uhlenbeck 2005), the contacts between tRNA and the ribosome are more extensive than with EF-Tu, resulting in a more stable complex and a smaller relative contribution of the esterified amino acid. Nevertheless, the presence of the esterified amino acid in the A site significantly affects the koff value in five of the eight aa-tRNAs previously tested (Fahlman et al. 2004). Thus, as in the case with EF-Tu, the sequences of the tRNA bodies may compensate for the thermodynamic contribution of the amino acid in a way that results in overall uniform binding to the ribosomal A site.
The possibility that the ribosome shows specificity for different amino acid side chains and tRNA bodies prompted us to evaluate the stability of four cognate and 12 misacylated aa-tRNAs in the ribosomal A site. This experiment was needed to test whether, in the context of an intact aa-tRNA, the amino acid specificity is really due to the amino acid side chain and not to some other feature of the amino acid, such as the α-amino group. Furthermore, if steric clash occurs between certain amino acid side chains and the ribosome, the effect of changing an amino acid may be much larger than simply removing it. For example, in the case of EF-Tu, the largest observed effect of amino acid removal from a tRNA is 2250-fold, but misacylated tRNAs have experimentally shown a 5200-fold range in binding (LaRiviere et al. 2001), and a 1.4 × 106-fold range in KD has recently been predicted (Asahara and Uhlenbeck 2005). Thus, the effect of misacylation on binding could be substantially larger than effects of simply removing the amino acid.
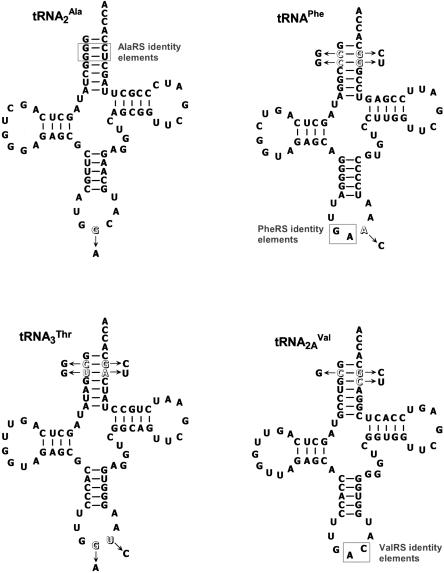
Following the experimental design in LaRiviere et al. (2001), four amino acids varying in size and hydrophobicity (alanine, phenylalanine, valine, and threonine) were each esterified to E. coli tRNA2Ala, tRNAPhe, tRNA2AVal, and tRNA3Thr. To facilitate misacylation, the G2-C71 and G3-U70 base pairs and the GAC (valine) anticodon were introduced into each tRNA (Fig. 1). The base pair swaps in the acceptor stem permitted the recognition of each tRNA by alanyl-tRNA synthetase (AlaRS), thus allowing all four tRNAs to be esterified with alanine (Ala). Aminoacylation with phenylalanine (Phe) and valine (Val) was possible because both E. coli ValRS and yeast PheRS recognize tRNAs containing the GAC anticodon (Asahara and Uhlenbeck 2002), and tRNAs could be esterified with threonine (Thr) using a mutant E. coli ValRS containing an amino acid substitution in its editing site (Döring et al. 2001). An additional advantage of introducing the GAC anti-codon into all four tRNAs was that it allowed a constant anticodon·codon interaction to be maintained for all 16 aa-tRNAs. This design eliminated the concern that different anticodon·codon pairs are expected to contribute differently to A site stability (Grosjean and Fiers 1982) and permitted the specific assessment of the relative contributions of the tRNA body and amino acid side chain to aa-tRNA stability in the A site. Note that since these hybrid tRNAs were made by in vitro transcription, they lacked post-transcriptional modifications; however, the four tRNA bodies that were chosen were ones where the effect of these modifications was relatively small (Fahlman et al. 2004; this paper, Fig. 2).
FIGURE 1.
Cloverleafs of E. coli tRNAs used in the study. Isoacceptor numbers are as given in Komine et al. (1990). Identity elements for AlaRS, PheRS, and ValRS (Giegé et al. 1998 and references therein) utilized in this study are boxed in each appropriate tRNA. Arrows indicate the nucleotides that were substituted for the parent nucleotides (outlined) in order to facilitate misacylation. tRNAs were prepared by in vitro transcription (Sampson and Uhlenbeck 1988), as described by Dale et al. (2004) with the following exceptions: Templates were made by primer extension of two overlapping DNA oligomers, and 16 mM GMP was used rather than 20 mM GMP. After purification on 10% polyacrylamide gels, tRNAs were 3′-[32P]-labeled using [α-32P]-ATP (3000 Ci/mmol; Amersham) and tRNA nucleotidyl transferase, according to Wolfson and Uhlenbeck (2002), except 2 μM CTP was also added to the reaction mix and reactions were incubated 2 min after addition of pyrophosphatase. Hexahistidine-tagged AlaRS was purified as described (Wolfson et al. 1998; Wolfson and Uhlenbeck 2002), purified ValRS was a gift from J. Perona (University of California, Santa Barbara), T222P mutant ValRS was a gift from P. Schimmel (Scripps Institute), and hexahistidine-tagged yeast PheRS was purified from a plasmid kindly provided by D. Tirrell (California Institute of Technology). tRNAs were aminoacylated, purified, and stored as described by Dale et al. (2004), except 300 μM unlabeled amino acid was used instead of [3H]amino acid.
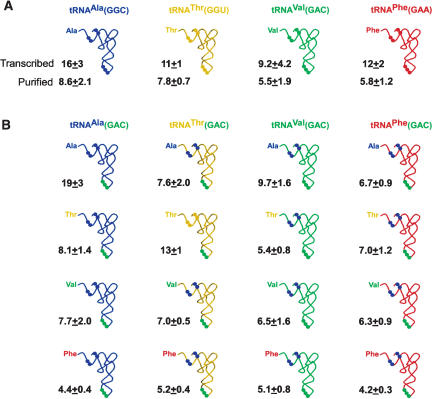
FIGURE 2.
Dissociation rate constants (koff s) of cognate and misacylated tRNAs from the ribosomal A site. Nucleotides important for misacylation are highlighted as dots. Values given are koffs (min−1 × 103) and represent the mean of at least three independent experiments. Errors given are the standard deviation of the mean. (A) Cognate aa-tRNAs. Both the purified and transcribed versions of each aa-tRNA display similar koff values, indicating that post-transcriptional modifications are not a critical contributor to aa-tRNA·A site stability for these aa-tRNAs. Values for cognate Val-tRNAVal(GAC) and Phe-tRNAPhe(GAA) are from Fahlman et al. 2004. tRNA2Ala was purified using a plasmid kindly provided by P. Schimmel (Scripps Institute) and the Nucleobond RNA/DNA Midi Kit (Clonetech). Purified tRNA3Thr was a generous gift from Margaret Saks (Northwestern University). (B) koff values of aminoacylated and misacylated tRNA(GAC)s. Aminoacyl-tRNA dissociation from the ribosomal A site was measured using a filter binding assay similar to that described by Fahlman and Uhlenbeck (2004). Tight-coupled E. coli 70S ribosomes were purified, stored, and activated as described by Fahlman et al. (2004). The ribosome binding (RB) buffer used in this work contained 50 mM HEPES (pH 7.0), 30 mM KCl, 70 mM NH4Cl, 1 mM DTT, and 100 μM EDTA. E. coli EF-Tu, purified as described by Fahlman and Uhlenbeck (2004) and stored in its GDP-bound form, was converted to its GTP-bound form immediately before use by incubating 5.2 μM EF-Tu, 20 μM GTP, 40 μg/mL pyruvate kinase, and 3 mM phosphoenolpyruvate in RB Buffer containing 15 mM MgCl2 (RB Buffer-15) for 30 min at 37°C. Ternary complex was formed by incubating 2.3 μM activated EF-Tu·GTP with 200 nM aa-[32P]-tRNA in RB Buffer-15 for 15 min on ice. Meanwhile, mRNA (5′-GGCAAGGAGGUAAAAAUGXXXGCACGU, where XXX represented the codon cognate to the given anticodon in the experiment) and deacyl-tRNAfMet were loaded onto E. coli ribosomes by incubating 2 μM ribosomes with 10 μM mRNA and 10 μM tRNAfMet in RB Buffer-15 for at least 15 min at room temperature. Aminoacyl-tRNAs were enzymatically loaded into the A site by mixing 20 nM EF-Tu·GTP·aa-[32P]-tRNA with 100 nM ribosome·mRNA·tRNAfMet for 2 min in RB Buffer-15 at room temperature. To prevent enzymatic reassociation of aa-tRNA during the subsequent chase reaction, EF-Tu·GTP and EF-Tu·GDP not bound to the ribosome were removed using an S-300 HR column (Amersham) equilibrated in RB Buffer-15. Dissociation from the A site was then initiated by mixing 5 μL of the column flow-through with 250 μL chase solution containing RB Buffer containing 10 mM MgCl2 (RB Buffer-10) and 30 nM unlabeled appropriate tRNA. The final concentration of ribosome·aa-tRNA complex was 0.4 nM. Aliquots were removed at desired times and filtered over a dual-membrane system as described by Fahlman and Uhlenbeck (2004), and koff values were determined from a semilogarithmic plot of fraction bound versus time. Control experiments verified that ribosomes remained active for aa-tRNA binding throughout the course of the experiments (data not shown).
Dissociation rate constants (koffs) of each aa-tRNA from the A site were measured using a filter binding assay (Fahlman and Uhlenbeck 2004). P site binding was prohibited by adding a saturating concentration of tRNAf Met to E. coli ribosomes programmed with an mRNA containing a Shine–Dalgarno sequence, an AUG start codon in the P site, and the appropriate codon in the A site. After each 3′-[32P]-labeled aa-tRNA was enzymatically loaded into the A site using EF-Tu, its dissociation was initiated by simultaneously diluting and adding unlabeled chase aa-tRNA. The free and ribosomally bound aa-tRNAs were subsequently separated at varying times using a double membrane filter. Control experiments showed that the same koff was observed whether the aa-tRNA was loaded into the A site enzymatically using EF-Tu·GTP or nonenzymatically by simply incubating the ribosomes and the aa-tRNA together. Additional control experiments confirmed that the koff values exhibited the weak Mg2+ dependence characteristic of A site, rather than P site, binding (data not shown).
Before performing experiments with misacylated tRNAs, it was important to evaluate whether or not the mutations made in the anticodon and the acceptor stem affected the ribosomal binding properties of the four cognate aa-tRNAs. In addition, we wanted to confirm that the effect of post-transcriptional modifications were small for these tRNA sequences. As shown in Figure 2A, the koff of each unmodified aa-tRNA with its cognate anticodon and normal acceptor stem is similar (twofold or less) to the koff of the corresponding fully modified aa-tRNA with the same sequence. This indicates that removal of post-transcriptional modifications has relatively little effect on the A site stabilities of this set of aa-tRNAs, which is in agreement with our previous work (Fahlman et al. 2004). Also, when unmodified Ala-tRNAAla, Thr-tRNAThr, and Val-tRNAVal with their cognate anticodons were compared to the same tRNA bodies with the GAC anticodon (Fig. 2), each pair of koff values was within error of each other. For example, the koff for Ala-tRNAAla(GGC) was 16 ± 3 × 10−3 min−1, while the koff for Ala-tRNAAla(GAC) was 19 ± 3 × 10−3 min−1. These controls indicate that these nucleotide substitutions had no effect on koff for these tRNAs. In the case of Phe-tRNAPhe, the introduction of the alanine identity elements in the acceptor stem and the GAC anticodon slowed the koff value by 2.5-fold, decreasing the koff to a value more similar to the purified Phe-tRNAPhe than the corresponding unmodified aa-tRNA. Finally, four of the misacylated tRNAs (Thr-tRNAVal, Phe-tRNAVal, Val-tRNAThr, and Phe-tRNAThr) were measured with and without the alanine identity elements, and again the koff was only marginally affected (data not shown). Taken together, these controls establish that neither the absence of post-transcriptional modifications nor the mutations made to permit misacylation appear to greatly alter the stabilities of these four aa-tRNAs in the A site.
Strikingly, the A site stability of the tRNAs was also unaffected by misacylation or by varying the tRNA body. As shown in Figure 2B, all 16 of the tested tRNAs have similar koff values (4.2 to 19 × 10−3 min−1), and these values are within the same range observed for the koff values of the fully modified aa-tRNAs with the normal anticodon (5.5 to 16 × 10−3 min−1; Fig. 2A). Furthermore, the range of koff values for misacylated tRNAs is also similar to the range that Fahlman et al. (2004) observed for a larger set of purified, fully modified tRNAs acylated with their cognate amino acid (2.7 to 6.4 × 10−3 min−1). These results indicate that this set of misacylated, anti-codon-substituted aa-tRNAs interacts with the ribosome just as well as the aa-tRNAs utilized by the ribosome in vivo.
Uniform binding of the misacylated tRNAs was unexpected based on the data that suggests the ribosome displays specificity for different amino acids. For example, Fahlman et al. (2004) observed a 10-fold difference in binding between Phe-tRNAPhe and deacyl-tRNAPhe, while no difference was seen between Val-tRNAVal and deacyl-tRNAVal. These results suggested that Phe adds significant stability of binding to the A site but Val does not. This preference of Phe over Val is consistent with the observation that the apparent Km for CpA-Phe was eightfold lower than the Kmapp of dinucleotides esterified with leucine or alanine in a ribosome-catalyzed dipeptide synthesis assay (Bhuta et al. 1981), indicating that aromatic side chains bind better than small aliphatic side chains. In addition, a p-methylphenyl derivative of puromycin has been found to be a 730-fold more effective inhibitor of in vitro translation than the analogous methyl puromycin derivative (Starck et al. 2003). Finally, the idea that Phe binds the A site better than Val is consistent with the available structural data, since it is easy to imagine that the large, aromatic Phe side chain will be stabilized in the somewhat hydrophobic A site crevice better than a small side chain, such as Val (Nissen et al. 2000; Schmeing et al. 2002; Hansen et al. 2003). Given this, one would expect a given tRNA body esterified with Phe to be significantly more stable in the A site than the same tRNA esterified with Val. Instead, all four tRNA bodies esterified with Phe exhibit virtually the same koff values as when they are esterified with Val (Fig. 2B).
Why weren’t differences in stability observed among the misacylated tRNAs? One explanation is that the ribosomal A site actually lacks specificity for either the amino acid side chain or the tRNA body, and the conclusions from the earlier biochemical experiments were incorrect because fragments of aa-tRNAs, rather than intact substrates, were used. It could be that each fragment binds the A site in a slightly different way, because the rest of the aa-tRNA is not present to aid in proper positioning. Although the binding of both deacylated tRNAs and CpA-aa derivatives to the A site is consistent with the chemical footprinting data acquired using intact aa-tRNAs (Nissen et al. 2000; Noller et al. 2001), these fragments could bind in position that was only 1 or 2 A ° different from the whole aa-tRNA. Such small differences in positioning could be sufficient to lead to different binding affinities for the fragments and would lead to the erroneous conclusion that specificity was present. Dramatically different biochemical properties of an intact aa-tRNA and a fragment were observed by Youngman et al. (2004), who found that mutations in the 23S ribosomal RNA (rRNA) in the peptidyl transferase center had a large effect on peptide bond formation when puromycin was used as a substrate, but had nearly no effect when an intact aa-tRNA was used.
Another explanation for why no difference in stability of the misacylated tRNAs was observed is that the ribosome is specific for different amino acid side chains and tRNA bodies, but this specificity is somehow masked in our experiments. It is possible that, unlike EF-Tu, the amino acid and tRNA body do not contribute independently to the binding affinity of aa-tRNAs for the A site. Instead, the presence of both the amino acid and the tRNA body could trigger the ribosome to undergo a conformational change to accept the aa-tRNA into the A site. Once such a ribosomal isomerization was induced, the dissociation of the aa-tRNA would be dependent on the reversal of that isomerization, and the uniform koff values observed in the current study would reflect the release of aa-tRNA by this rate-limiting ribosomal conformational change. In this view, the putative isomerization could not be induced without an intact aa-tRNA, and uniform binding would be predicted to be absent in experiments using aa-tRNA fragments, such as deacyl tRNAs (Fahlman et al. 2004), CpA-aa analogs (Bhuta et al. 1981), and puromycin derivatives (Starck et al. 2003). Biochemical and structural studies suggest that the ribosome undergoes a large conformational change upon a cognate anticodon·codon interaction during the decoding steps of translation (Pape et al. 2000; Ogle et al. 2002; Valle et al. 2003), and additional conformational changes occur when an aa-tRNA is accommodated into the A site (Pape et al. 1999; Blanchard et al. 2004; Gromadski and Rodnina 2004a,b). Thus, it may also be that aa-tRNA release from the A site measured in this work is limited by the rate of at least one ribosomal isomerization.
Regardless of the mechanism, all 12 misacylated tRNAs are just as stable in the A site as any cognate, purified aa-tRNA measured thus far (Fahlman et al. 2004; this study). An important implication of these results is that misacylated tRNAs are likely to function equally well in translation after they reach the ribosomal A site. It remains possible, however, that discrimination of misacylated tRNAs occurs during the earlier GTPase activation and accommodation steps of translation (Pape et al. 1998), which involve release of aa-tRNA from EF-Tu. Thus, the ability of EF-Tu to discriminate against misacylated tRNAs may be used twice, first in the initial formation of the ternary complex and then during the decoding steps on the ribosome. This view suggests that once a tRNA is fully bound to the A site of the ribosome, it requires only the presence of an amino acid in order to be a functional substrate.
Acknowledgments
This work was funded by National Institutes of Health Grant GM37552 to O.C.U.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2130505.
REFERENCES
- Asahara, H. and Uhlenbeck, O.C. 2002. The tRNA specificity of Thermus thermophilus EF-Tu. Proc. Natl. Acad. Sci. 99: 3499–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2005. Predicting the binding affinities of misacylated tRNAs for Thermus thermophilus EF-Tu·GTP. Biochemistry 44: 11254– 11261. [DOI] [PubMed] [Google Scholar]
- Battle, D.J. and Doudna, J.A. 2002. Specificity of RNA-RNA helix recognition. Proc. Natl. Acad. Sci. 99: 11676–11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuta, A., Quiggle, K., Ott, T., Ringer, D., and Chladek, S. 1981. Stereochemical control of ribosomal peptidyltransferase reaction. Role of amino acid side-chain orientation of acceptor substrate. Biochemistry 20: 8–15. [DOI] [PubMed] [Google Scholar]
- Blanchard, S.C., Gonzalez, R.L., Kim, H.D., Chu, S., and Puglisi, J.D. 2004. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 11: 1008–1014. [DOI] [PubMed] [Google Scholar]
- Cochella, L. and Green, R. 2005. An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308: 1178–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, J.F. and Yarus, M. 1989. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J. Mol. Biol. 209: 65–77. [DOI] [PubMed] [Google Scholar]
- Dale, T., Sanderson, L.E., and Uhlenbeck, O.C. 2004. The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry 43: 6159–6166. [DOI] [PubMed] [Google Scholar]
- Döring, V., Mootz, H.D., Nangle, L.A., Hendrickson, T.L., de Crécy- Lagard, V., Schimmel, P., and Marliere, P. 2001. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science 292: 501–504. [DOI] [PubMed] [Google Scholar]
- Fahlman, R.P. and Uhlenbeck, O.C. 2004. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry 43: 7575–7583. [DOI] [PubMed] [Google Scholar]
- Fahlman, R.P., Dale, T., and Uhlenbeck, O.C. 2004. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell 16: 799–805. [DOI] [PubMed] [Google Scholar]
- Giegé, R., Sissler, M., and Florentz, C. 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26: 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski, K.B. and Rodnina, M.V. 2004a. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell 13: 191–200. [DOI] [PubMed] [Google Scholar]
- ———. 2004b. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat. Struct. Mol. Biol. 11: 316–322. [DOI] [PubMed] [Google Scholar]
- Grosjean, H. and Fiers, W. 1982. Preferential codon usage in prokaryotic genes: The optimal codon–anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene 18: 199–209. [DOI] [PubMed] [Google Scholar]
- Grosjean, H.J., de Henau, S., and Crothers, D.M. 1978. On the physical basis for ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. 75: 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J.L., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2002. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. 99: 11670–11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J.L., Moore, P.B., and Steitz, T.A. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330: 1061–1075. [DOI] [PubMed] [Google Scholar]
- Janiak, F., Dell, V.A., Abrahamson, J.K., Watson, B.S., Miller, D.L., and Johnson, A.E. 1990. Fluorescence characterization of the interaction of various transfer RNA species with elongation factor Tu.GTP: Evidence for a new functional role for elongation factor Tu in protein biosynthesis. Biochemistry 29: 4268–4277. [DOI] [PubMed] [Google Scholar]
- Komine, Y., Adachi, T., Inokuchi, H., and Ozeki, H. 1990. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J. Mol. Biol. 212: 579–598. [DOI] [PubMed] [Google Scholar]
- LaRiviere, F.J., Wolfson, A.D., and Uhlenbeck, O.C. 2001. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294: 165–168. [DOI] [PubMed] [Google Scholar]
- Louie, A., Ribeiro, N.S., Reid, B.R., and Jurnak, F. 1984. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 259: 5010–5016. [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Noller, H.F., Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Lancaster, L., Dallas, A., Fredrick, K., Earnest, T.N., and Cate, J.H. 2001. Structure of the ribosome at 5.5 A° resolution and its interactions with functional ligands. Cold Spring Harbor Symp. Quant. Biol. 66: 57–66. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., and Ramakrishnan, V. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732. [DOI] [PubMed] [Google Scholar]
- Ott, G., Schiesswohl, M., Kiesewetter, S., Forster, C., Arnold, L., Erdmann, V.A., and Sprinzl, M. 1990. Ternary complexes of Escherichia coli aminoacyl-tRNAs with the elongation factor Tu and GTP: Thermodynamic and structural studies. Biochim. Biophys. Acta 1050: 222–225. [DOI] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., and Rodnina, M.V. 1998. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17: 7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., and Rodnina, M.V. 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18: 3800–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 7: 104–107. [DOI] [PubMed] [Google Scholar]
- Pedersen, S. 1984. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 3: 2895–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, J.R. and Uhlenbeck, O.C. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 85: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing, T.M., Seila, A.C., Hansen, J.L., Freeborn, B., Soukup, J.K., Scaringe, S.A., Strobel, S.A., Moore, P.B., and Steitz, T.A. 2002. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9: 225–230. [DOI] [PubMed] [Google Scholar]
- Starck, S.R., Qi, X., Olsen, B.N., and Roberts, R.W. 2003. The puromycin route to assess stereo- and regiochemical constraints on peptide bond formation in eukaryotic ribosomes. J. Am. Chem. Soc. 125: 8090–8091. [DOI] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Li, W., Stagg, S.M., Sengupta, J., Nielsen, R.C., Nissen, P., Harvey, S.C., Ehrenberg, M., and Frank, J. 2003. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryoelectron microscopy. Nat. Struct. Biol. 10: 899–906. [DOI] [PubMed] [Google Scholar]
- Wolfson, A.D. and Uhlenbeck, O.C. 2002. Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc. Natl. Acad. Sci. 99: 5965–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson, A.D., Pleiss, J.A., and Uhlenbeck, O.C. 1998. A new assay for tRNA aminoacylation kinetics. RNA 4: 1019–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman, E.M., Brunelle, J.L., Kochaniak, A.B., and Green, R. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117: 589–599. [DOI] [PubMed] [Google Scholar]