Abstract
Protein L4 from Thermus thermophilus (TthL4) was heterologously overproduced in Escherichia coli cells. To study the implication of the extended loop of TthL4 in the exit-tunnel and peptidyltransferase functions, the highly conserved E56 was replaced by D or Q, while the semiconserved G55 was changed to E or S. Moreover, the sequence -G55E56- was inverted to -E55G56-. When we incorporated these mutants into E. coli ribosomes and investigated their impact on poly(Phe) synthesis, high variations in the synthetic activity and response to erythromycin of the resulting ribosomes were observed. In the absence of erythromycin, ribosomes harboring mutations G55E and E56D in TthL4 protein were characterized by low activity in synthesizing poly(Phe) and decreased capability in binding tRNA at the A site. On the other hand, ribosomes possessing mutations G55E, G55S, G55E-E56G, or E56Q in TthL4 protein were unexpectedly more sensitive to erythromycin. Evidence in support of these findings was drawn by in vivo experiments, assessing the erythromycin sensitivity of E. coli cells expressing wild-type or mutant TthL4 proteins. Our results emphasize the role of the extended loop of L4 ribosomal protein in the exit-tunnel and peptidyltransferase center functions.
Keywords: ribosomal protein L4, poly(Phe) synthesis, peptidyltransferase, erythromycin, exit tunnel
INTRODUCTION
The prokaryotic ribosome consists of two subunits designated 50S and 30S, containing more than 50 different proteins in complex with rRNA molecules. Recently, complete atomic structures for the large (Ban et al. 2000; Harms et al. 2001) and small ribosomal subunit (Wimberly et al. 2000) from bacteria were determined by X-ray crystallography, and it is shown that the ribosomal primary activities, the decoding process and peptide bond formation, occur in the interior ribosomal compartment, which is largely composed of rRNA (Ban et al. 2000; Nissen et al. 2000).
In the large ribosomal subunit, proteins are abundant everywhere on its surface, except for the catalytic site cleft and the flat surface that contacts the small subunit. Nevertheless, certain ribosomal proteins contain extended tails that penetrate deeply into the interior of the subunit, making numerous, idiosyncratic interactions with the surrounding rRNA. More specifically, ribosomal proteins L4 and L22 protrude into the exit-tunnel region, where their extended tips along with rRNA residues form what appears to be a gate opening (Jenni and Ban 2003). It has been suggested that the exit tunnel, whose existence has been confirmed by crystallographic data (Nissen et al. 2000), is the normal exit path for nascent polypeptides before they emerge from the ribosome (Tenson and Ehrenberg 2002). However, there is also evidence suggesting that some proteins may leave the peptidyltransferase (PTase) center through other routes (Odom et al. 1990; Komar et al. 1997; Gabashvili et al. 2001). Interestingly, even if a growing peptide has decided to enter the tunnel, several exit possibilities remain, since the exit tunnel branches at the backside of the large subunit (Gabashvili et al. 2001).
The role of the exit-tunnel gate is still under investigation; it might function as a sensor, recognizing special features of the nascent peptide chain and transmitting messages to the PTase center through L22 and L4 proteins (Nissen et al. 2000), it might provide alternative exits for the nascent peptides (Komar et al. 1997; Gabashvili et al. 2001; Nakatogawa and Ito 2002), or it might regulate the protein elongation cycle by stopping or modulating the traffic (Gabashvili et al. 2001; Tenson and Ehrenberg 2002). Interestingly, several antibiotics, such as chloramphenicol, clindamycin, and several members of the macrolide family, inhibit protein elongation by binding to this gate (Schluenzen et al. 2001; Hansen et al. 2002, 2003). Their primary mode of action is modulated by the space available between the catalytic-site crevice and the bound drug molecule (Hansen et al. 2002; Tenson et al. 2003). Antibiotics that contain an extending arm, reaching the A site, inhibit directly the PTase reaction (Dinos and Kalpaxis 2000; Hansen et al. 2002), whereas antibiotics that lack such extensions, like erythromycin, inhibit protein synthesis by blocking the exit tunnel and inducing premature dissociation from the ribosome of short peptidyl-tRNAs with six, seven, or eight amino acid residues (Tenson et al. 2003; Lovmar et al. 2004).
Many reports have indicated that erythromycin does not inhibit poly(U)-directed poly(Phe) synthesis, although it is an effective inhibitor of other polypeptides, such as poly(Lys) (Odom et al. 1990). One possibility is that the highly hydrophobic nascent poly(Phe) chain may displace erythromycin from its binding site on 50S subunits. According to a second hypothesis, the typical movement through the exit tunnel does not take place during the synthesis of poly(Phe) and, therefore, the synthesis of this polypeptide is not inhibited by erythromycin (Odom et al. 1990). In any case, changes in the rate of poly(Phe) synthesis in response to erythromycin action could be interpreted on the basis of structural perturbations that would affect the local conformation of the exit tunnel.
In a previous study, Escherichia coli ribosomes harboring TthL4 protein mutated at positions 55 and 56 were used to investigate the implication of the extended loop region of this protein in puromycin reaction and binding of substrates (Leontiadou et al. 2003). In the present work, these and additional hybrid ribosomes were studied with respect to their ability to synthesize poly(Phe) in the presence and in the absence of erythromycin. In parallel, in vivo experiments were performed, assessing the erythromycin sensitivity of E. coli cells expressing wild-type and mutant TthL4 proteins. As shown, some of the mutations in TthL4 render poly(Phe) synthesis and growth of cells sensitive to erythromycin.
RESULTS AND DISCUSSION
Incorporation of wtTthL4 and its mutants into 70S ribosomes
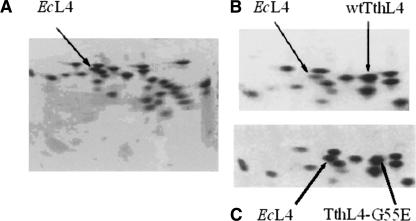
Figure 1 shows the results obtained by two-dimensional electrophoresis of TP-70 proteins from untreated E. coli cells (A) and from cells overproducing wtTthL4 or TthL4-G55E (B and C, respectively). In Figure 1, B and C, an extra spot is visible at the right of the native E. coli L4 (EcL4) position. As identified by electroblotting to PVDF membranes and automated Edmann degradation, this extra spot is attributed to the incorporation of the heterologous protein into a subpopulation of ribosomes. It is obvious from Figure 1, B and C, that the resolution of the electrophoretograms is excellent and that TthL4 species is well separated from other native proteins, adjacently moved. The relative intensity of EcL4 and TthL4 species was estimated by image analysis. The degree of incorporation (%) was calculated by the ratio A/(A + B)×100, where A represents the spot intensity of the heterologous protein and B is the spot intensity of EcL4 protein. The estimated value of incorporation for each mutant is given in Table 1.
FIGURE 1.
2D-urea PAGE electrophoresis of TP-70 proteins extracted from 70S ribosomes. TP-70 proteins were from ribosomes isolated from (A) wild-type E. coli cells and cells overproducing (B) wtTthL4 or (C) TthL4-G55E protein. Arrows indicate spots corresponding to endogenous protein L4 or spots corresponding to heterologous protein.
TABLE 1.
Degree of incorporation of different TthL4 proteins into E. coli ribosomes and ability of the resulting ribosomes to catalyze AcPhe-puromycin synthesis, bind AcPhe-tRNA at the P and A sites, and associate to 70S ribosomal complexes
| Assembled protein | Degree of incorporation (%) | kcata (min−1) | Ksa (μM) | P-site boundb AcPhe-tRNA per ribosome | A-site boundb AcPhe-tRNA per ribosome | 70S ribosomesc (%) |
| EcL4 | 100.0 | 1.250 ± 0.015 | 621 ± 80 | 0.230 ± 0.015 | 0.195 ± 0.015 | 87.0 ± 5.0 |
| WtTthL4 | 81.6 | 0.820 ± 0.040 | 615 ± 20 | 0.218 ± 0.015 | 0.185 ± 0.014 | 67.5 ± 4.5 |
| TthL4-E56D | 73.9 | 0.620 ± 0.035 | 625 ± 22 | 0.090 ± 0.006 | 0.088 ± 0.005 | 68.3 ± 5.0 |
| TthL4-E56Q | 92.7 | 1.290 ± 0.080 | 630 ± 80 | 0.505 ± 0.030 | 0.169 ± 0.014 | 83.7 ± 5.0 |
| TthL4-G55E-E56G | 81.9 | 1.010 ± 0.040 | 610 ± 25 | 0.183 ± 0.005 | 0.180 ± 0.015 | 65.5 ± 4.5 |
| TthL4-G55S | 87.5 | 0.820 ± 0.040 | 621 ± 8 | 0.200 ± 0.015 | 0.172 ± 0.014 | 79.1 ± 4.0 |
| TthL4-G55E | 61.6 | 0.800 ± 0.030 | 615 ± 15 | 0.232 ± 0.017 | 0.107 ± 0.005 | 42.3 ± 2.1 |
aThe values of kcat and Ks referring to the AcPhe-puromycin synthesis are expressed as means ± SD (n = 4, duplicate analysis in four ribosomal preparations) and have been corrected for the intervention of wild-type ribosomes in the mixed ribosomal population.
bBinding values are expressed as means ± SD (n = 4, duplicate analysis in four ribosomal preparations) and have been corrected for the intervention of wild-type ribosomes in the mixed ribosomal population.
cThe capability of ribosomal subunits to associate was checked by incubating five A260 units of each subunit under conditions that promote their association to 70S complexes. The percentages of the formed 70S ribosomal complexes are expressed as means ± SD (n = 3, duplicate analysis in three ribosomal preparations). One hundred percent represents the situation at which 100% of the added 50S subunits are associated with 30S subunits to form 70S ribosomes.
Although protein L4 has been shown to be an autogenous regulatory protein in E. coli (Zengel and Lindahl 1994), overproduction of the heterologous protein is not blocked, since, first, the plasmid-incorporated TthL4 gene is transcribed with phage T7 RNA polymerase under the control of the isopropyl-thio-α-D-galactoside-inducible lac UV5 promoter and, second, the plasmid-derived transcript has no leader sequence containing binding sites for the TthL4 protein. Nevertheless, overproduction of the heterologous protein may regulate transcription and translation of the S10 ribosomal protein operon in host cells, as shown previously for other heterologous L4 ribosomal proteins (Zengel et al. 1995). This hypothesis cannot be easily verified on the basis of the present results, since EcL4 is normally produced before induction and, therefore, it is included in the total product.
Undoubtedly, the properties of a chimeric E. coli ribosome reconstituted with a heterologously expressed L4 protein from Thermus thermophilus cannot be interpreted with the same rigor as studying a homologous construct. However, attempts to incorporate His-tagged EcL4 mutants into E. coli ribosomes showed that both the EcL4 as well as the His-tagged EcL4 comigrate and cannot be separately identified by two-dimensional urea PAGE. These restrictions do not apply to TthL4. Moreover, the N-terminal-domain residues of TthL4 and EcL4 are highly conserved. For these reasons, TthL4 (wild-type or mutant) was chosen to be heterologously overproduced in E. coli cells.
Influence of the wild-type and mutant TthL4s on AcPhe-tRNA binding, peptide bond formation, and the assembly and association of ribosomal subunits
In an attempt to examine the role of some amino acids of TthL4 on the function of the entrance of the exit tunnel, we mutated the highly conserved E56 and the semiconserved G55. Both amino acid residues are located within the extended loop of TthL4 (Schluenzen et al. 2001). Therefore, possible observed differences in poly(Phe) synthesis and its response against erythromycin could reveal an implication of those amino acids in the tunnel function.
It is already known that incorporation of TthL4-E56Q into E. coli cells causes improvement of the AcPhe-tRNA binding to the P site by >100% (Leontiadou et al. 2003). This was confirmed in the present study (Table 1). In addition, we observed that binding of AcPhe-tRNA to the P or A sites of ribosomes containing TthL4-E56D is impaired by >50%. A similar reduction was also observed in A-site binding for ribosomes containing TthL4-G55E. Nevertheless, the Ks values estimated by the puromycin reaction did not differ among the tested ribosomal species (Table 1), which means that TthL4 (wild-type or mutant) assembling into E. coli ribosomes does not affect the ribosomal affinity toward puromycin. In turn, this suggests that the observed differences in the binding of tRNA to ribosomes may be attributed to alterations in the interaction of 50S subunits with regions of tRNAs, other than their 3′-ends. With the exception of TthL4-E56Q, the incorporation of TthL4 species into E. coli ribosomes resulted in partial inactivation of PTase, especially incorporation of TthL4-E56D (Table 1).
The effects of TthL4 mutations on the binding of tRNA to ribosomes and on the activity of PTase suggest that the observed changes may be due to large-scale structural perturbations, provided that protein L4 belongs to the early assembled proteins (Rohl and Nierhaus 1982). To test this hypothesis, the sedimentation profile and the ability of ribosomes to associate were studied. No precursors of the 50S subunits were detected between 30S and 50S peaks. This is in agreement with recent studies demonstrating that the extended loops of L4 and L22 proteins are not required for ribosome assembly (Leontiadou et al. 2003; Zengel et al. 2003). However, replacement of EcL4 by wild-type or mutant TthL4 caused changes in the association capability of subunits, which in certain cases were dramatic. For instance, incorporation of TthL4-G55E into E. coli ribosomes resulted in about 50% reduction of association (Table 1). In fact, the degree of reduction might be >50%, provided that the degree of TthL4-G55E incorporation is only 61.6%.
Effects of wtTthL4 and its mutants on poly(Phe) synthesis
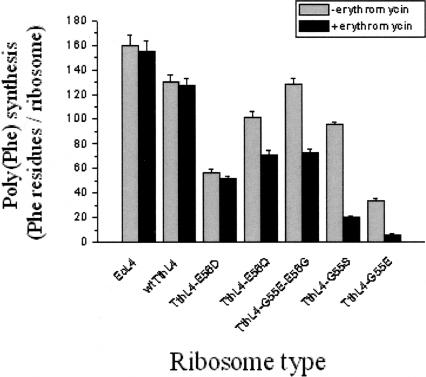
Poly(Phe) synthesis was carried out in the presence or in the absence of erythromycin. As shown in Figure 2, the level of poly(Phe) synthesis obtained after incubation for 60 min at 37°C depends on the type of hybrid ribosomes. This is clearly indicated for the mutants TthL4-E56Q and TthL4-G55E-E56G, whose effect on poly(Phe) synthesis and their degree of incorporation into ribosomes are inversely correlated. All the hybrid ribosomes exhibited moderate activity, compared with wild-type ribosomes; the lowest level of poly(Phe) synthesis was observed in ribosomes incorporating TthL4-G55E or TthL4-E56D, and the highest in ribosomes containing wtTthL4 or TthL4-G55E-E56G. The low level of poly(Phe) synthesis obtained with ribosomes incorporating TthL4-E56D can be attributed to their low PTase activity and their reduced capability in forming 70S complexes and binding tRNA at the P and A sites. It should be mentioned that 70S ribosomes constitute the functional subpopulation of ribosomal particles. Despite the extremely low activity of TthL4-G55E variant in synthesizing poly(Phe), it exhibited normal PTase activity. This behavior can be explained by the fact that the TthL4-G55E variant is characterized by a low content of 70S ribosomes and decreased efficiency for tRNA binding to the A site. It should be noted that the slowest step of the elongation cycle is the A-site occupation, which is much slower than the peptidyltransfer reaction (Schilling-Bartetzko et al. 1992). Consequently, a severe reduction of A-site occupation is expected to decrease more strongly the poly(Phe) synthesis than the puromycin reaction.
FIGURE 2.
Bar diagrams of poly(Phe) synthesis in hybrid ribosomes. The experiments were performed in the absence (gray bars) or in the presence (black bars) of 10 μM erythromycin. The level of poly(Phe) synthesis has been corrected for the intervention of wild-type ribosomes in the mixed ribosomal populations, and it is expressed in Phe residues incorporated per ribosome in 60 min. Abbreviations under the bars indicate L4 species assembled in each type of ribosome.
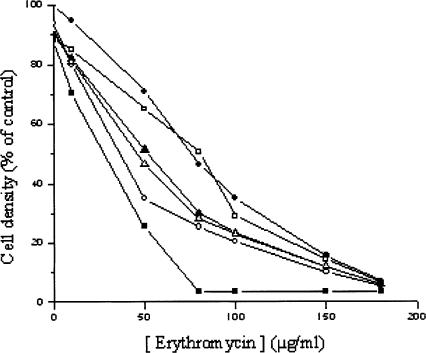
With the exception of untreated ribosomes and those incorporating wtThL4 or TthL4-E56D, all the rest of the species were clearly sensitive to erythromycin to different degrees (Fig. 2); the TthL4-G55E and TthL4-G55S variants exhibited the highest sensitivity, whereas the TthL4-E56Q and TthL4-G55E-E56G variants displayed a moderate behavior. Corroborative evidence was sought in experiments assessing the in vivo sensitivity of transformed E. coli cells against erythromycin. As shown in Figure 3, sensitivity against erythromycin was more evident in cells expressing TthL4-G55E and TthL4-G55S proteins. This means that the erythromycin sensitivity observed in vitro is physiologically relevant.
FIGURE 3.
Inhibition of growth of E. coli cells expressing wild-type or mutant TthL4 protein by erythromycin. Cells were grown at 37°C in LB medium containing 50 μg/mL ampicillin and erythromycin at the indicated concentrations until the optical density of the control culture (cells expressing wtTthL4 and grown in the absence of erythromycin) reached the value 1.000 at 600 nm. This optical density was taken as 100%. Overproduced protein: (•) wtTthL4, (○) TthL4-G55E, (▪) TthL4-G55S, (□) TthL4-E56D, (▴) TthL4-E56Q, and (▵) TthL4-G55E-E56G.
The efficiency of the mutated ribosomes for poly(Phe) synthesis was found to be related to the A-site binding. Therefore, it is reasonable to conclude that the conformation of the A site is targeted by remote modifications that are conveyed through the TthL4 backbone and 23S rRNA over a distance of 35–38 Å. On the other hand, the influence of TthL4 mutations on ribosome susceptibility to erythromycin seems to be related to a reorganization of the drug-binding region, again through a remote effect. A direct effect of mutations on erythromycin structure cannot be easily speculated; structural studies have denied any direct interaction of L4 with erythromycin (Schluenzen et al. 2001).
Interestingly, the effect of mutations at positions 55 and 56 of TthL4 on ribosome’s sensitivity to erythromycin is opposite to that previously reported for mutations concerning adjacent sites of the L4 extended loop (Chittum and Champney 1994; Gregory and Dahlberg 1999; O’Connor et al. 2004). In the latter case, all the tested mutations caused resistance rather than sensitivity to erythromycin. From the standpoint of pharmaceutical applications, our results could be used for the interpretation of the macrolide mechanism of action and design of more potent drugs.
MATERIALS AND METHODS
Materials
Restriction endonucleases and dNTPs were purchased from New England Biolabs. Expand Long Template PCR System and the Rapid Ligation Kit were from Boehringer Mannheim. Iso-propyl-thio-α-D-galactoside was from Appligene. Oligonucleotides used as primers in the PCRs were synthesized by TIB MOLBIOL Berlin.
PCR amplification, isolation, and cloning of TthL4 gene
Based on the known sequence of TthL4 gene (Pfeiffer et al. 1995), two oligonucleotides (5′-CCggAATTCATATgAAggAggTAgCggTgTACCA-3′ and 5′-CgCggATCCTCACgCCTCACCCCCTATg-3′) with suitable cloning sites at their 5′ ends (NdeI and BamHI, respectively) were utilized for the amplification of the TthL4 gene by PCR, using genomic DNA from T. thermophilus as template. The wild-type sequence of the amplified TthL4 was identified by nucleotide sequence and cloned into pET11a plasmid.
In vitro mutagenesis of TthL4
Six mutants of the TthL4 gene were produced, using a three-step PCR mutagenesis protocol (Picard et al. 1994). The codon for glutamic acid at position 56 was replaced by the codon for aspartic acid or glutamine, while the codon for glycine at the neighboring position 55 was replaced by serine or glutamic acid. Additionally, a double mutation was performed in which E56 was changed into glycine and G55 into glutamic acid. The template used was the recombinant plasmid pET11a/wtTthL4. The PCR products were purified, cloned into pET11a, and screened for the desired mutations by nucleotide sequencing.
Overexpression of wild-type or mutant TthL4 in E. coli BL21 (DE3) cells
E. coli BL21 (DE3) cells were transformed with the recombinant pET11a plasmids carrying either the wild-type TthL4 (wtTthL4) or the mutants. The transformed cells were grown at 37°C until absorption of 0.6–0.7 at 570 nm. The expression of the heterologous protein was induced by the addition of 1 mM iso-propyl-thio-α-D-galactoside to the growth medium. Cells harvested 3 h after induction were disrupted, and their protein content was analyzed by SDS-12% polyacrylamide gel electrophoresis. The proteins were visualized by staining with Coomassie Blue R-250. In addition, electrophorised proteins before staining were electro-transferred onto PVDF membranes (Immobilon-P, Millipore) and the overproduced protein was identified by a ProciseHT sequenator (Choli et al. 1989).
Ribosome preparation and evaluation of wild-type or mutant TthL4 incorporation
Ac[3H]Phe-tRNA was prepared from crude E. coli tRNA, as described previously (Kalpaxis et al. 1986). It was charged to 77%, 100% being 28 pmol of [3H]Phe per A260 unit (Sigma). Ac[3H]Phe-tRNA concentration in Table 1 is given as the fraction of crude tRNA active in charging Phe. Runoff ribosomes from E. coli BL21 (DE3) cells transformed with the appropriate recombinant plasmids were prepared according to Kalpaxis et al. (1986). The isolated ribosomes were further purified by two sequential centrifugations at 100,000g through a 0.5 M sucrose cushion made up in association buffer (100 mM Tris-HCl at pH 7.2, 100 mM NH4Cl, 6 mM MgCl2, 100 μM spermine, 6 mM 2-mercaptoethanol) to remove proteins artificially bound to native ribosomes. Ribosomal subunits were isolated by incubating runoff ribosomes in dissociation buffer (100 mM Tris-HCl at pH 7.2, 0.5 mM magnesium acetate, 150 mM NH4Cl, 6 mM 2-mercaptoethanol) for 30 min at 37°C. The incubation mixture was then separated into subunits by sucrose-gradient (10%–40%) centrifugation in dissociation buffer at 100,000g. To evaluate the ability of ribosomal subunits to associate, 5 A260 units of each subunit were incubated in association buffer for 30 min at 37°C. The mixture was then separated by sucrose-gradient (10%–30%) centrifugation in association buffer at 100,000g.
The incorporation of wild-type or mutant TthL4 into E. coli 70S ribosomes was examined by a 2D-urea PAGE electrophoresis, according to Geyl et al. (1981). Briefly, total proteins from 70S ribosomes (TP-70) extracted with two volumes of acetic acid (Hardy et al. 1969) were dialyzed against water, dried, and then subjected to electrophoresis followed by electroblotting onto PVDF membranes (Choli et al. 1989). After Coomassie Blue staining, the spots corresponding to TthL4 species were identified by N-terminal protein sequencing, as described above. The degree of incorporation was estimated by image analysis of the stained gels (Molecular Dynamics Densitometer and Image Quant 5.0 Software for scanning and quantitating wet gels and X-ray plates, University of Virginia, ITC-Academic Computing Health Sciences).
Binding of Ac[3H]Phe-tRNA to the ribosomal P and A sites
Binding was performed by incubating Ac[3H]Phe-tRNA with poly(U)-programmed 70S ribosomes prefilled (A-site occupation) or not prefilled (total binding) in their P sites by tRNAPhe (Leontiadou et al. 2003). The totally bound Ac[3H]Phe-tRNA was measured by nitrocellulose filtration. The P-site bound Ac[3H]Phe-tRNA was estimated from the total binding by titration with puromycin (2 mM, 10 min at 25°C). The estimated values were corrected, taking into account the intervention of wild-type ribosomes in the mixed ribosomal population (Leontiadou et al. 2003). Thus, the values shown in Table 1 correspond to hybrid ribosomes.
Puromycin reaction and poly(Phe) synthesis
The PTase activity was assessed by the puromycin reaction carried out at 25°C in the presence of 6 mM MgCl2, 100 mM NH4Cl, and puromycin in excess, as described elsewhere (Kalpaxis et al. 1986). The puromycin reaction was analyzed by pseudo-first-order kinetics, which allow the measure of the catalytic rate constant (kcat) of PTase and the dissociation constant (Ks) of the encounter complex between an initiation ribosomal complex (Ac[3H]Phe-tRNA·poly(U)·70S ribosome) and puromycin (Kalpaxis et al. 1986). The kcat and Ks values presented in Table 1 have been corrected for the intervention of wild-type ribosomes in the mixed ribosomal population (Leontiadou et al. 2003).
Poly(Phe) synthesis was performed with a mixed ribosomal population (wild-type and hybrid ribosomes), according to Rheinberger and Nierhaus (1990). As a control, native 70S ribosomes from E. coli BL21 (DE3) cells were used. The ionic conditions of this in vitro protein-synthesizing system were settled by the addition of 4.5 mM magnesium acetate, 150 mM ammonium acetate, 2 mM spermidine, and 0.05 mM spermine. After incubation for 60 min at 37°C, the protein-synthesizing mixture (30 μL) was precipitated by the addition of 2 mL 5% trichloroacetic acid, two drops of 1% bovine serum albumin, and by heating for 15 min at 90°C. Afterward, the samples were cooled down at 4°C for 20 min and then filtrated through glass filters (Schleicher & Schull). In another series of experiments, poly(Phe) synthesis was carried out in the presence of 10 μM erythromycin. The level of total poly(Phe) synthesis by the mixed population of ribosomes (wild-type and hybrids) was calculated by measuring the radioactivity trapped on the filters and expressing this radioactivity in Phe residues incorporated into the synthesized polypeptide chain per ribosome (Pt) in 60 min. From this level, the poly(Phe) synthesis managed by the hybrid ribosomes (Ph) was estimated by the formula
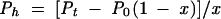 |
where P0 represents the poly(Phe) synthesis obtained by an equimolar amount of wild-type ribosomes and x is the fraction of hybrid ribosomes in the mixed ribosomal population.
Sensitivity to erythromycin of E. coli cells expressing wild-type or mutant TthL4 proteins
Cells (70 μL of a 0.900 OD600 preculture) were added in 7 mL of LB medium containing 50 μg/mL ampicillin and grown at 37°C in the presence or in the absence of erythromycin until the optical density of the control culture (E. coli cells expressing wtTthL4 protein and grown in the absence of erythromycin) reached the value 1.000 at 600 nm. The concentration of erythromycin ranged from 0 to 180 μg/mL. A 10 μg/mL concentration is equivalent to 13.62 μM erythromycin.
Statistics
One-way analysis of variance (ANOVA) was used to estimate the mean values and data variability. The Scheffé F test was used to determine significant differences between means. All statistical tests were performed using a SPSS program for MS Windows, Release 6.0.
Acknowledgments
We thank Dr. Francois Franceshi and Prof. Dr. Knund Nierhaus for their helpful discussions. Part of the biochemical work was carried out at Max-Planck Institute for Molecular Genetics in Berlin. This work was supported in part by Deutscher Akademischer Austauschdienst (DAAD) and by a grant (99ED-605) from the General Secretariat of Research and Technology, Ministry of Development of Greece, and the European Social Fund. M.A.X. and D.L.K. would like to thank the Operational Program for Educational and Vocational Training II (EPEAEKII), and particularly the Program IRAKLEITOS, for support.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2126205.
REFERENCES
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Chittum, H.S. and Champney, W.S. 1994. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J. Bacteriol. 176: 6192–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choli, T., Kapp, U., and Wittmann-Liebold, B. 1989. Blotting of proteins onto Immobilon membranes. In situ characterization and comparison with high performance liquid chromatography. J. Chromatogr. 476: 59–72. [DOI] [PubMed] [Google Scholar]
- Dinos, G.P. and Kalpaxis, D.L. 2000. Kinetic studies on the interaction between a ribosomal complex active in peptide bond formation and the macrolide antibiotics tylosin and erythromycin. Biochemistry 39: 11621–11628. [DOI] [PubMed] [Google Scholar]
- Gabashvili, I.S., Gregory, S.T., Valle, M., Grassucci, R., Worbs, M., Wahl, M.C., Dahlberg, A.E., and Frank, J. 2001. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell 8: 181–188. [DOI] [PubMed] [Google Scholar]
- Geyl, D., Bock, A., and Isono, K. 1981. An improved method for two-dimensional gel-electrophoresis: Analysis of mutationally altered ribosomal proteins of Escherichia coli. Mol. Gen. Genet. 181: 309–312. [DOI] [PubMed] [Google Scholar]
- Gregory, S.T. and Dahlberg, A.E. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23S ribosomal RNA. J. Mol. Biol. 289: 827–834. [DOI] [PubMed] [Google Scholar]
- Hansen, J.L., Ippolito, J.A., Ban, N., Nissen, P., Moore, P.B., and Steitz, T.A. 2002. The structure of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10: 117–128. [DOI] [PubMed] [Google Scholar]
- Hansen, J.L., Moore, P.B., and Steitz, T.A. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330: 1061–1075. [DOI] [PubMed] [Google Scholar]
- Hardy, S.J., Kurland, C.G., Voynow, P., and Mora, G. 1969. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry 7: 2897–2905. [DOI] [PubMed] [Google Scholar]
- Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688. [DOI] [PubMed] [Google Scholar]
- Jenni, S. and Ban, N. 2003. The chemistry of protein synthesis and voyage through the ribosomal tunnel. Curr. Opin. Struct. Biol. 13: 212–219. [DOI] [PubMed] [Google Scholar]
- Kalpaxis, D.L., Theocharis, A., and Coutsogeorgopoulos, C. 1986. Kinetic studies on ribosomal peptidyltransferase. The behaviour of the inhibitor blasticidin S. Eur. J. Biochem. 154: 267–271. [DOI] [PubMed] [Google Scholar]
- Komar, A.A., Kommer, A., Krasheninnikov, I.A., and Spirin, A.S. 1997. Contranslational folding of globin. J. Biol. Chem. 272: 10646–10651. [DOI] [PubMed] [Google Scholar]
- Leontiadou, F., Xaplanteri, M.A., Papadopoulos, G., Gerassimou, C., Kalpaxis, D.L., and Choli-Papadopoulou, T. 2003. On the structural and functional importance of the highly conserved Glu56 of Thermus thermophilus L4 ribosomal protein. J. Mol. Biol. 332: 73–84. [DOI] [PubMed] [Google Scholar]
- Lovmar, M., Tenson, T., and Ehrenberg, M. 2004. Kinetics of macrolide action: The josamycin and erythromycin cases. J. Biol. Chem. 279: 53506–53515. [DOI] [PubMed] [Google Scholar]
- Nakatogawa, H. and Ito, K. 2002. The ribosomal exit tunnel functions as discriminating gate. Cell 108: 629–636. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- O’Connor, M., Gregory, S.T., and Dahlberg, A.E. 2004. Multiple defects in translation associated with altered ribosomal protein L4. Nucleic Acids Res. 32: 5750–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom, O.W., Picking, W.D., Tsalkova, T., and Hardesty, B. 1990. The synthesis of polyphenylalanine on ribosomes to which erythromycin is bound. Eur. J. Biochem. 198: 713–722. [DOI] [PubMed] [Google Scholar]
- Pfeiffer, T., Jorcke, D., Feltens, R., and Hartmann, R.K. 1995. Direct linkage of str-, S10- and spc-related gene clusters in Thermus thermophilus HB8 and sequences of ribosomal proteins L4 and S10. Gene 167: 141–145. [DOI] [PubMed] [Google Scholar]
- Picard, V., Ersdal-Badju, E., Lu, A., and Bock, S.C. 1994. A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 22: 2587–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinberger, H.J. and Nierhaus, K.H. 1990. Partial release of AcPhe-Phe-tRNA fromribosomes during poly(U)-dependent poly(Phe) synthesis and the effects of chloramphenicol. Eur. J. Biochem. 193: 643–650. [DOI] [PubMed] [Google Scholar]
- Rohl, R. and Nierhaus, K.H. 1982. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl. Acad. Sci. 79: 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling-Bartetzko, S., Bartetzko, A., andNierhaus, K.H. 1992. Kinetic and thermodynamic parameters for transfer RNA binding to the ribosome and for translocation reaction. J. Biol. Chem. 267: 4703–4712. [PubMed] [Google Scholar]
- Schluenzen, F., Zarivach, R., Harms, J., Bashan, A., Tocilj, A., Albrecht, A., Yonath, A., and Franceschi, F. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase center in eubacteria. Nature 413: 814–821. [DOI] [PubMed] [Google Scholar]
- Tenson, T. and Ehrenberg, M. 2002. Regulatory nascent peptides in the ribosomal tunnel. Cell 108: 591–594. [DOI] [PubMed] [Google Scholar]
- Tenson, T., Lovmar, M., and Ehrenberg, M. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330: 1005–1014. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons Jr., W.M., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Zengel, J.M. and Lindahl, L. 1994. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acids Res. Mol. Biol. 47: 331–370. [DOI] [PubMed] [Google Scholar]
- Zengel, J.M., Vororheikina, D., Li, X., and Lindahl, L. 1995. Regulation of the Escherichia coli S10 ribosomal protein operon by heterologous L4 ribosomal proteins. Biochem. Cell Biol. 73: 1105–1112. [DOI] [PubMed] [Google Scholar]
- Zengel, J.M., Jerauld, A., Walker, A., Wahl, M.C., and Lindahl, L. 2003. The extended loops of ribosomal proteins L4 and L22 are not required for ribosomal assembly or L4-mediated autogenous control. RNA 9: 1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]