Abstract
In eukaryotic cells degradation of bulk mRNA in the 5′ to 3′ direction requires the consecutive action of the decapping complex (consisting of DCP1 and DCP2) and the 5′ to 3′ exonuclease XRN1. These enzymes are found in discrete cytoplasmic foci known as P-bodies or GW-bodies (because of the accumulation of the GW182 antigen). Proteins acting in other post-transcriptional processes have also been localized to P-bodies. These include SMG5, SMG7, and UPF1, which function in nonsense-mediated mRNA decay (NMD), and the Argonaute proteins that are essential for RNA interference (RNAi) and the micro-RNA (miRNA) pathway. In addition, XRN1 is required for degradation of mRNAs targeted by NMD and RNAi. To investigate a possible interplay between P-bodies and these post-transcriptional processes we depleted P-body or essential pathway components from Drosophila cells and analyzed the effects of these depletions on the expression of reporter constructs, allowing us to monitor specifically NMD, RNAi, or miRNA function. We show that the RNA-binding protein GW182 and the DCP1:DCP2 decapping complex are required for miRNA-mediated gene silencing, uncovering a crucial role for P-body components in the miRNA pathway. Our analysis also revealed that inhibition of one pathway by depletion of its key effectors does not prevent the functioning of the other pathways, suggesting a lack of interdependence in Drosophila.
Keywords: Argonaute, GW182, mRNA decay, NMD, RNAi, siRNAs, miRNAs, P-bodies
INTRODUCTION
In eukaryotic cells, bulk messenger RNA (mRNA) is degraded via two alternative pathways, each of which is initiated by the removal of the poly(A) tail by deadenylases (for review, see Parker and Song 2004). Following this first step, mRNAs can be degraded from their 3′ ends by the exosome, a multimeric complex of 3′ to 5′ exonucleases. Alternatively, after deadenylation, the cap structure is removed by the DCP1:DCP2 decapping complex, and the mRNA is degraded by the major cytoplasmic 5′ to 3′ exonuclease XRN1 (for review, see Parker and Song 2004).
Proteins required for 5′ to 3′ mRNA degradation (e.g., DCP1, DCP2, and XRN1) colocalize in specialized cytoplasmic bodies or mRNA decay foci, also known as mRNA processing bodies (P-bodies) or GW-bodies, because of the accumulation of the RNA binding protein GW182 in these bodies (Eystathioy et al. 2002, 2003; Ingelfinger et al. 2002; van Dijk et al. 2002; Sheth and Parker 2003). Additional components of P-bodies in yeast and/or human cells include the deadenylase Ccr4, the cap binding protein eIF4E and its binding partner eIF4E-transporter (eIF4E-T), auxiliary decay factors such as the LSm1–7 complex, Pat1p/Mtr1p, and the putative RNA helicase Dhh1/rck/p54 (Eystathioy et al. 2002, 2003; Ingelfinger et al. 2002; Lykke-Andersen 2002; van Dijk et al. 2002; Sheth and Parker 2003; Cougot et al. 2004; Andrei et al. 2005). Among these, human GW182, eIF4E-T, and Dhh1 are required for P-body formation, while the decapping enzymes and XRN1 are dispensable (Ingelfinger et al. 2002; Yang et al. 2004; Andrei et al. 2005). In addition, mRNA decay intermediates, microRNA (miRNA) targets, and miRNAs have been localized to P-bodies, suggesting that these bodies are sites where translationally silenced mRNAs are stored before undergoing decay (Sheth and Parker 2003; Cougot et al. 2004; Liu et al. 2005; Pillai et al. 2005; Teixeira et al. 2005).
Recently, proteins involved in other post-transcriptional processes have been localized to P-bodies in human cells. These include the proteins SMG5, SMG7, and UPF1 involved in the nonsense-mediated mRNA decay (NMD) pathway (Unterholzner and Izaurralde 2004; Fukuhara et al. 2005), and the Argonaute (AGO) proteins that play essential roles in RNA silencing (Liu et al. 2005; Pillai et al. 2005; Sen and Blau 2005). Moreover, XRN1 is recruited by both the NMD and the RNA interference (RNAi) machineries to degrade targeted mRNAs (Souret et al. 2004; Orban and Izaurralde 2005; for review, see Conti and Izaurralde 2005), suggesting a possible link between NMD, RNAi, and P-bodies.
NMD is an mRNA quality control (or surveillance) mechanism that degrades aberrant mRNAs having premature translation termination codons (PTCs), thereby preventing the synthesis of truncated and potentially harmful proteins (for review, see Conti and Izaurralde 2005). Core components of the NMD machinery include the proteins UPF1, UPF2, and UPF3, which form a complex whose function in NMD is conserved. The activity of UPF1 is regulated in multicellular organisms by additional proteins (i.e., SMG1, SMG5, SMG6, and SMG7) that are also required for NMD in all organisms in which orthologs have been characterized (for review, see Conti and Izaurralde 2005).
In yeast and human cells, a major decay pathway for NMD substrates involves decapping and 5′ to 3′ degradation by XRN1 (for review, see Conti and Izaurralde 2005). Although degradation of nonsense transcripts in Drosophila is initiated by endonucleolytic cleavage near the PTC, the resulting 3′ decay intermediate is also degraded by XRN1 (Gatfield and Izaurralde 2004). A molecular link between the NMD machinery and the decay enzymes localized in P-bodies is provided by SMG7 in human cells. Indeed, when overexpressed, human SMG7 localizes in P-bodies and recruits both UPF1 and SMG5 to these bodies (Unterholzner and Izaurralde 2004; Fukuhara et al. 2005), suggesting that NMD factors may reside at least transiently in P-bodies.
RNA silencing pathways are evolutionarily conserved mechanisms that elicit decay or translational repression of mRNAs selected on the basis of complementarity with small interfering RNAs (siRNAs) or miRNAs, respectively (for review, see Filipowicz 2005). siRNAs are fully complementary to their targets and elicit mRNA degradation via the RNAi pathway. Animal miRNAs are only partially complementary to their targets and do not generally elicit decay, but repress translation instead (for review, see Filipowicz 2005).
To perform their function, the siRNAs and miRNAs associate with the AGO proteins to form multimeric RNA-induced silencing complexes (RISC) (for review, see Filipowicz 2005). Drosophila AGO1 mediates miRNA function, while AGO2 catalyzes the endonucleoytic cleavage of siRNA targets within the region complementary to the siRNA. Following this initial cleavage, the resulting 5′ mRNA fragment is degraded by the exosome, while the 3′ fragment is degraded by XRN1 (Souret et al. 2004; Orban and Izaurralde 2005). The localization of AGO proteins in P-bodies in human cells provides a possible link between these bodies and silencing pathways (Liu et al. 2005; Pillai et al. 2005; Sen and Blau 2005).
The NMD, the siRNA, and the miRNA pathways are therefore interlinked by the use of common decay enzymes and/or the coexistence of components of these pathways in P-bodies, suggesting a possible interdependence between these post-transcriptional mechanisms. Evidence for a link between NMD and RNAi has been reported in Caenorhabditis elegans where UPF1, SMG5, and SMG6 are required for persistence of RNAi, though not to initiate silencing (Domeier et al. 2000; Kim et al. 2005). In contrast, UPF2, UPF3, and SMG1, which are also essential for NMD, are not required to maintain silencing, suggesting that UPF1, SMG5, and SMG6 may have evolved specialized functions in RNAi (Domeier et al. 2000).
In this study we sought to investigate the interplay between NMD, RNAi, and the miRNA pathway using the Drosophila Schneider cell line 2 (S2 cells) expressing reporters allowing us to monitor NMD, RNAi, or miRNA function. To this end, factors involved in NMD (UPF1, UPF2, UPF3, SMG1, SMG5, and SMG6), RNAi (AGO2), or the miRNA pathway (AGO1) were depleted and the effect on the expression of the reporters analyzed. These proteins showed a high degree of functional specificity. To determine the role of P-body components in these pathways we depleted the DCP1:DCP2 decapping complex, the decapping coactivators LSm1 and LSm3, the 5′ to 3′ exonuclease XRN1, GW182, and the Drosophila protein CG32016, which shares limited sequence homology with human eIF4E-T (Dostie et al. 2000). Our results uncovered a crucial role for GW182 and the DCP1:DCP2 decapping complex in the miRNA pathway.
RESULTS AND DISCUSSION
Components of the NMD, RNAi, and miRNA pathways exhibit functional specificity in Drosophila
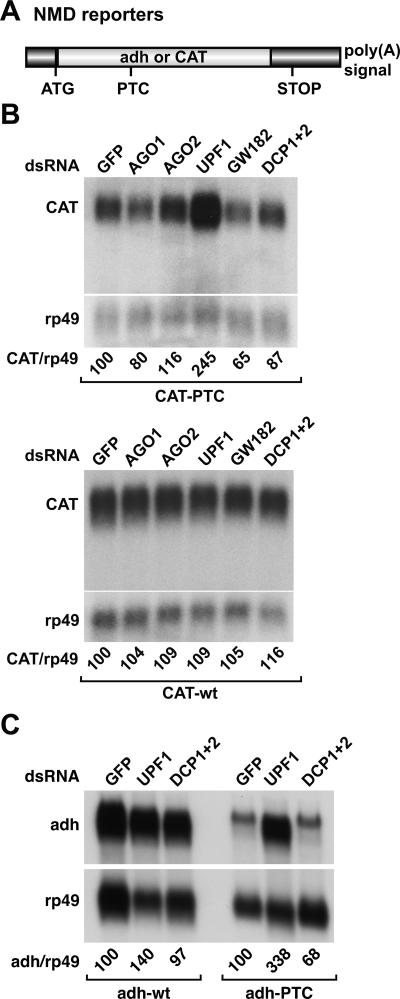
To investigate a potential role of components of RNA silencing pathways or of P-body components in NMD we made use of previously described cell lines expressing wild-type or PTC-containing reporter constructs in which the coding regions of the bacterial chloramphenicol acetyl transferase (CAT) or the Drosophila alcohol dehydrogenase (adh) genes are placed downstream of inducible or constitutive promoters (Gatfield et al. 2003; Gatfield and Izaurralde 2004). The PTCs are inserted at codon 72 and 83 of the CAT and adh open reading frames, respectively (Fig. 1A ▶; Gatfield et al. 2003). P-body components and proteins involved in NMD, RNAi, or the miRNA pathway were depleted by treating the cells with double-stranded RNAs (dsRNAs) specific for the different factors. A dsRNA that targets green fluorescent protein (GFP) served as a control. The steady-state levels of the wild-type and PTC-containing mRNAs were analyzed by Northern blot and normalized to those of the endogenous rp49 mRNA (encoding ribosomal protein L32).
FIGURE 1.
NMD, RNAi, and miRNA factors are functionally specific in Drosophila. (A) Schematic representation of the NMD reporters used in this study. (B,C) S2 cell-lines expressing wild-type (wt) or PTC-containing transcripts were treated with the dsRNAs indicated above the lanes. RNA samples were analyzed by Northern blot. The steady-state levels of wild-type and PTC mRNAs were quantitated and normalized to those of rp49 mRNA. These values were set to 100 in cells treated with GFP dsRNA.
The effects of the depletions on the expression of all reporters used in this study are shown in Figures 1 ▶–3 ▶ ▶ and summarized in Table 1 ▶. Only the results obtained for proteins inhibiting at least one pathway were considered as being meaningful. We also defined a change in the expression levels of the reporters of at least twofold as being significant (see Materials and Methods for an explanation of the twofold cutoff).
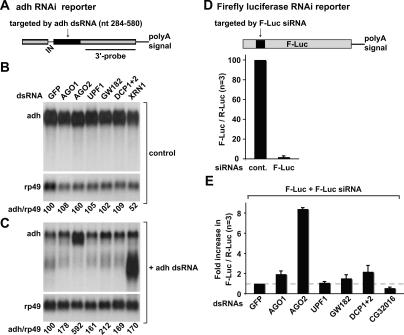
FIGURE 2.
siRNA-mediated gene silencing occurs independently of NMD or the miRNA pathway. (A) Schematic representation of the adh reporter as described by Orban and Izaurralde (2005). (Gray boxes) exons; (black box) sequences complementary to the dsRNAs; (IN) intron. The region of the transcript detected by the 3′ probe used in panels B and C is indicated. (B,C) S2 cells constitutively expressing adh mRNA were treated with the dsRNAs indicated above the lanes. In panel C, cells also were treated with adh dsRNA. The levels of the reporters were quantitated and normalized to those of rp49 mRNA. These values were set to 100 in cells treated with GFP dsRNA. (D) Schematic representation of the firefly luciferase RNAi reporter. The region complementary to F-Luc siRNA is shown in black. S2 cells were transfected with plasmids expressing firefly and Renilla luciferase (F-Luc and R-Luc, respectively). siRNAs targeting firefly luciferase (F-Luc siRNA) or a control siRNA (cont.) were cotransfected. Four days post-transfection, firefly and Renilla luciferase activities were measured in three independent experiments (n = 3). Firefly luciferase activity was normalized to that of Renilla and set to 100 in cells treated with control siRNA. Mean values ± standard deviations are shown. (E) S2 cells were treated with the indicated dsRNAs. Six days after the first knockdown, cells were transfected with plasmids expressing firefly and Renilla luciferase, and either F-Luc or control siRNAs. Firefly and Renilla luciferase activities were measured 4 d post-transfection. For each knockdown, firefly luciferase activity was normalized to that of the Renilla and set to 100 in cells transfected with control siRNA (not shown). The normalized luciferase activity in cells transfected with F-Luc siRNA and treated with GFP dsRNA was then set to 1. Mean values ± standard deviations are shown.
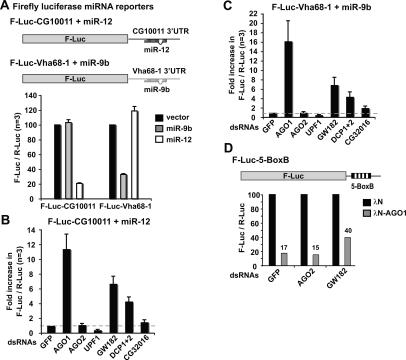
FIGURE 3.
A role for GW182 and the DCP1:DCP2 complex in the miRNA pathway. (A) Schematic representation of miRNA reporter constructs. F-Luc is flanked by the 3′ UTRs of CG10011 or Vha68–1 mRNAs, which contain binding sites for miR-12 or miR-9b, respectively. S2 cells were transfected with the reporters (F-Luc-CG10011 or F-Luc-Vha68–1) together with plasmids expressing miR-9b (gray bars), miR-12 (white bars), or the corresponding empty vector (black bars). A plasmid expressing Renilla luciferase (R-Luc) was included as a transfection control. Firefly luciferase activity was normalized to that of the Renilla in three independent experiments. Normalized firefly luciferase activities in cells transfected with the empty vector (black bars) were set to 100%. Mean values ± standard deviations are shown. (B,C) S2 cells were treated with the indicated dsRNAs. On day 6, cells were cotransfected with F-Luc-CG10011 reporter and a plasmid expressing miR-12 or with F-Luc-Vha68–1 reporter and a plasmid expressing miR-9b. A plasmid encoding Renilla luciferase was included in all transfection mixtures. Firefly and Renilla luciferase activity were measured 4 d post-transfection. Firefly luciferase activity was normalized to that of the Renilla and set to unity in cells treated with GFP dsRNA. Mean values of at least three independent experiments ± standard deviations are shown. (D) Schematic representation of the F-Luc-5-BoxB tethering reporter. (Black boxes) The five BoxB sequences that bind the λN-peptide. S2 cells were treated with the indicated dsRNAs. Six days after the first knockdown, cells were transfected with the F-Luc-5-BoxB reporter, a plasmid expressing Renilla luciferase, and vectors expressing the λN-peptide (black bars) or λN-AGO1 (gray bars). Firefly and Renilla luciferase activities were measured 3 d post-transfection. For each knockdown, firefly luciferase activity was normalized to that of the Renilla and set to 100 in cells expressing the λN-peptide alone.
TABLE 1.
Functional specificity of P-body components and of proteins involved in NMD, RNAi, or the miRNA pathway
| Depleted proteins | CG number | NMD | RNAi | miRNA pathway |
| NMD factors | ||||
| UPF1 | CG1559 | + | − | − |
| UPF2 | CG2253 | + | − | − |
| UPF3 | CG11184 | + | − | − |
| SMG1 | CG4549 | + | − | − |
| SMG5 | CG8954 | + | − | − |
| SMG6 | CG6369 | + | − | − |
| RNAi | ||||
| AGO2 | CG7439 | − | + | − |
| miRNA pathway | ||||
| AGO1 | CG6671 | − | − | + |
| P-body components | ||||
| GW182 | CG31992 | − | ± | + |
| CG32016 | CG32016 | − | − | ± |
| XRN1 (Pacman) | CG3291 | 3′ fragmenta | 3′ fragment | − |
| DCP1+DCP2 | − | ± | + | |
The effects of the depletions were scored as (−) when the changes in reporter levels were below twofold relative to the levels observed in cells treated with GFP dsRNA. (±) Indicates a twofold effect with at least one reporter assay; (+) indicates an increase above the twofold cutoff.
aXRN1 is required for degradation of the 3′ decay intermediates.
Relative to the expression levels of the wild-type mRNAs, the levels of the corresponding PTC-containing transcripts are reduced because these transcripts are rapidly degraded via the NMD pathway (Fig. 1B,C ▶; Gatfield et al. 2003). As reported already, depletion of UPF1 inhibits NMD, so the levels of the PTC-containing mRNAs are restored (Fig. 1B,C ▶; Gatfield et al. 2003). Depletion of AGO1 or AGO2, both singly and in combination, did not interfere with the NMD pathway (Fig. 1B ▶; Table 1 ▶; data not shown), although these depletions do inhibit siRNA- or miRNA-mediated gene silencing (Figs. 2 ▶, 3 ▶; Table 1 ▶). The levels of the CAT wild-type transcript were not affected by the depletions (Fig. 1B ▶, CAT-wt). Similar results were obtained with the NMD reporter based on the adh gene (data not shown). Together, these results indicate that inhibition of RNAi or of the miRNA pathway does not interfere with NMD.
XRN1 is the only P-body component known to be required for degradation of decay intermediates arising from mRNAs undergoing NMD in Drosophila (Gatfield and Izaurralde 2004). Nevertheless, in cells depleted of XRN1 the NMD pathway is not inhibited, and only the 3′ decay intermediate generated by endonucleolytic cleavage of the mRNA accumulates (Gatfield and Izaurralde 2004). In contrast to XRN1, none of the P-body components tested including GW182 and the DCP1:DCP2 decapping complex affected NMD or the accumulation of the 3′ decay intermediate (Fig. 1B ▶; Table 1 ▶). The lack of a significant effect of the depletion of the DCP1:DCP2 complex was confirmed using the adh reporter (Fig. 1C ▶). The decapping enzymes are certainly involved in NMD in yeast and human cells because the major decay pathway for NMD substrates is initiated by decapping in these organisms (for review, see Conti and Izaurralde 2005). Thus, it is possible that the requirement for P-body components and/or P-body integrity in NMD varies across species.
RNA interference functions independently of NMD or the miRNA pathway
We used two different approaches to investigate the RNAi pathway. In one approach, a cell line constitutively expressing the wild-type Drosophila adh gene was treated with a dsRNA complementary to a central region of ~300 nucleotides (nt) of adh mRNA (adh dsRNA) (Fig. 2A ▶, black box). In previous studies we have shown that this dsRNA elicits decay of the adh mRNA via the RNAi pathway (Orban and Izaurralde 2005). Cells were treated with dsRNAs targeting various factors in the presence or absence of adh dsRNA (Fig. 2, C and B ▶, respectively). The steady-state levels of the adh mRNA were analyzed by Northern blot and normalized to those of the rp49 mRNA.
In cells treated with GFP dsRNA, the normalized levels of the adh transcript were reduced to 4% after addition of adh dsRNA, relative to the levels detected in the absence of adh dsRNA. In cells depleted of AGO2, a sixfold increase of adh mRNA levels was observed despite the presence of adh dsRNA (Fig. 2C ▶). In contrast, when AGO1 was depleted, adh dsRNA could still trigger a reduction of adh mRNA levels, though a slight increase in transcript levels was observed (Fig. 2C ▶). Similarly, depletion of UPF1 did not prevent silencing of adh expression by adh dsRNA. These results indicate that UPF1 is not required for RNAi in Drosophila. We also tested additional NMD components (i.e., UPF2, UPF3, SMG1, SMG5, and SMG6; no SMG7 ortholog has been identified in Drosophila) (Gatfield et al. 2003). We could not observe a significant change in the efficacy of RNAi under the conditions in which NMD was inhibited (Table 1 ▶).
Similarly to the results reported for the NMD pathway, depletion of XRN1 leads to the accumulation of the 3′ decay intermediate generated by endonucleolytic cleavage by RISC (Fig. 2C ▶; Gatfield and Izaurralde 2004; Orban and Izaurralde 2005), while depletion of the DCP1:DCP2 decapping complex did not prevent RNAi or the degradation the 3′ decay intermediate (Fig. 2C ▶; Table 1 ▶). In contrast, depletion of GW182 led to a modest increase in the adh mRNA level in the presence of adh dsRNA, suggesting that this protein could influence the efficiency of RNAi (Fig. 2C ▶).
In a second approach, RNAi was triggered by an siRNA instead of a long dsRNA, to uncouple RISC activity from processing of dsRNAs. To this end, S2 cells were transiently transfected with a plasmid expressing firefly luciferase (F-Luc) and an siRNA targeting the luciferase coding sequence (F-Luc siRNA) or a control siRNA (cont., Fig. 2D ▶). A plasmid encoding Renilla luciferase (R-Luc) was included to normalize for transfection efficiencies. Cotransfection of the F-Luc reporter with the F-Luc siRNA led to a 50-fold inhibition of firefly luciferase activity relative to the activity measured when the control siRNA was cotransfected (Fig. 2D ▶), indicating that F-Luc siRNA effectively silences firefly luciferase expression.
The results obtained with the luciferase reporter correlate well with those obtained with adh mRNA, in spite of differences between the methods used to detect changes in reporter levels (RNA levels vs. protein levels), and the nature of the RNA trigger (long dsRNA vs. siRNA). Indeed, depletion of AGO2 impaired silencing of firefly luciferase expression by the F-Luc siRNA, leading to an eightfold increase in firefly luciferase activity relative to the activity of the Renilla control. Depletion of AGO1 led to a twofold increase of firefly luciferase activity (Fig. 2E ▶).
The observation that depletion of AGO2, but not AGO1, significantly inhibits RNAi is in agreement with previous reports showing that only AGO2-containing RISC is able to catalyze mRNA cleavage triggered by siRNAs (Liu et al. 2004; Meister et al. 2004; Okamura et al. 2004; Rand et al. 2004). Our results together with these observations indicate that Drosophila AGO1 and AGO2 are not redundant.
Depletion of GW182 or the DCP1:DCP2 complex led to a 1.5- to twofold increase of the firefly luciferase activity, although RNAi was not abolished (Fig. 2E ▶). These results together with those obtained with the adh reporter suggest that GW182 and the DCP1:DCP2 complex are not absolutely required for RNAi but may modulate siRNA function.
Finally, depletion of core NMD components did not inhibit the silencing of firefly luciferase expression by F-Luc siRNA (Fig. 2E ▶; Table 1 ▶). Our results are consistent with results from C. elegans showing that NMD per se is not required for the establishment of silencing (Domeier et al. 2000).
GW182 and the DCP1:DCP2 complex play a crucial role in the miRNA pathway
To investigate the miRNA pathway we generated firefly luciferase reporters in which the coding region of firefly luciferase is flanked by the 3′ UTRs of the Drosophila genes CG10011 or Vha68–1 (Fig. 3A ▶). These genes were identified as miRNA targets in a genome-wide analysis of mRNAs regulated by AGO1 (to be reported elsewhere). The 3′ UTR of CG10011 mRNA contains two binding sites for miR-12, while the 3′ UTR of Vha68–1 has two binding sites for miR-9b. Expression of the firefly luciferase construct fused to the 3′ UTR of CG10011 (F-Luc-CG10011) was strongly reduced by cotransfection of a plasmid expressing the primary (pri) miR-12 transcript, but not pri-miR-9 (Fig. 3A ▶). Conversely, expression of the firefly luciferase reporter fused to the 3′ UTR of Vha68–1 (F-Luc-Vha68–1) was inhibited by cotransfection of pri-miR-9b, but not of pri-miR-12 (Fig. 3A ▶).
Silencing of luciferase expression by the cognate miRNAs was prevented in cells depleted of AGO1 (Fig. 3B,C ▶). Indeed, despite the presence of the transfected miRNAs, in cells depleted of AGO1 we observed an 11-fold and a 16-fold increase of firefly luciferase expression from the F-Luc-CG10011 and F-Luc-Vha68–1 reporters, respectively. Notably, the firefly luciferase activity measured in AGO1-depleted cells in the presence of the transfected miRNAs was at least twofold higher than the activity measured in control cells in the absence of exogenously added miRNAs. Since endogenous miR-9b and miR-12 are expressed in S2 cells (data not shown), these results suggest that depletion of AGO1 also suppresses silencing mediated by the endogenous miRNAs. Depletion of AGO2 did not suppress the effect of coexpressing the reporters with the cognate miRNAs (Fig. 3B,C ▶; Table 1 ▶). These results provide additional evidence supporting the conclusion that the siRNA and miRNA pathways are not interdependent.
miRNA-mediated silencing of firefly luciferase expression was not affected by depletion of UPF1 or of the additional NMD factors (i.e., UPF2, UPF3, SMG1, SMG5, and SMG6) (Fig. 3B,C ▶; Table 1 ▶). Thus, the individual NMD factors and NMD per se are not required for miRNA function.
Unexpectedly, although the efficiency of NMD and RNAi was unaffected or only modestly affected in cells depleted of GW182 or the DCP1:DCP2 complex, miRNA-mediated silencing of firefly luciferase expression was effectively relieved in these cells (Fig. 3B,C ▶). In the presence of cognate miRNAs, depletion of GW182 resulted in a sixfold increase of firefly luciferase expression. Therefore, despite the presence of transfected miRNAs, firefly luciferase activity in GW182-depleted cells was similar to that measured in controls cells in the absence of transfected miRNAs. Codepletion of DCP1 and DCP2 led to a fourfold increase of firefly luciferase expression. Finally, depletion of CG32016 resulted in a twofold increase of firefy luciferase activity, but only for the F-Luc-Vha68–1 reporter, suggesting that this effect may not be significant.
To investigate whether depletion of GW182 affects RISC activity directly, as opposed to interfering with miRNA processing, we made use of a tethering assay described by Pillai et al. (2004). This assay involves the expression of a λN-fusion of AGO1 that binds with high affinity to five BoxB sites (5-BoxB) in the 3′ UTR of a firefly luciferase reporter mRNA (Fig. 3D ▶). When AGO1 is tethered to this reporter transcript, luciferase expression is inhibited relative to the activity measured in cells expressing the λN-peptide alone (Fig. 3D ▶). The inhibition was partially relieved in cells depleted of GW182 but not of AGO2.
We conclude that GW182 and the decapping DCP1: DCP2 complex play a critical role in the effector step of the miRNA pathway. These results are in agreement with the observation that Argonaute proteins localize to P-bodies and interact with DCP1 and DCP2 independently of RNA or of P-body integrity (Liu et al. 2005; Pillai et al. 2005; Sen and Blau 2005).
CONCLUSION
In this paper we show that despite convergence in P-bodies, NMD, RNAi, and the miRNA pathway are not interdependent in Drosophila. This conclusion is based on the observation that the inhibition of one pathway by depleting key effectors may slightly interfere with, but does not significantly inhibit, the functioning of the other pathways. The lack of interdependence between RNAi and the miRNA pathway is further supported by the observation that knockouts of AGO1 or AGO2 in Drosophila have different phenotypes (Okamura et al. 2004). Nevertheless, cross-talk between the RNAi and the miRNA pathways may still occur at the initiation step, as Dicer-1 plays a role in RISC assembly (Lee et al. 2004).
Biochemical and genetic approaches in several organisms have led to the identification of essential components of the miRNA pathway. These include AGO1 and the enzymes required for miRNA processing, such as Drosha and Dicer-1 and their respective cofactors, Pasha and Loqs (for review, see Filipowicz 2005). However, the mechanisms by which miRNAs inhibit protein expression without affecting mRNA levels are not completely understood. Recent evidence suggests that translation initiation is inhibited and that the targeted mRNAs are stored in P-bodies, where they are maintained in a silenced state either by associating with proteins that prevent translation or possibly by removal of the cap structure (Liu et al. 2005; Pillai et al. 2005; Sen and Blau 2005). In this study we have identified the P-body components GW182 and the DCP1:DCP2 decapping complex as proteins required for the miRNA pathway. The precise molecular mechanism by which these proteins participate in this pathway remains to be established. These proteins may have an indirect role in the miRNA pathway by affecting P-body integrity. Alternatively, these proteins may play a direct role in this pathway by escorting miRNA targets to P-bodies or facilitating mRNP remodeling steps required for the silencing of these targets. Consistent with a direct role for the DCP1:DCP2 decapping complex, and thus for the cap structure, in miRNA function is the observation that mRNAs translated via a cap-independent mechanism are not subject to miRNA-mediated silencing (Pillai et al. 2005). In conclusion, our results uncover an important role for the P-body components, GW182 and the DCP1:DCP2 complex, in miRNA-mediated gene silencing.
MATERIALS AND METHODS
Cell culture and RNA interference
RNAi was performed essentially as described by Gatfield et al. (2003). dsRNAs used in this study correspond to fragments encompassing ~700 nt of the coding sequences. Depletions described in this study were carried out by treating cells with the corresponding dsRNAs on day 0 and day 4. Thirty micrograms of dsRNA were used per six-well dish containing ~2 × 106 cells.
NMD and RNAi reporters
The NMD reporters and the adh reporter used in Figures 1 ▶ and 2 ▶ have been described (Gatfield et al. 2003; Orban and Izaurralde 2005). Firefly and Renilla luciferase were cloned between the KpnI and EcoRI or the EcoRI and XhoI sites of vector pAc5.1 (Invitrogen), respectively. The plasmid expressing Renilla luciferase (R-Luc) served as a transfection control. The transfection mixtures contained 0.05 μg of pAc5.1-F-Luc and 0.5 μg of pAc5.1-R-Luc plasmids. The following siRNAs were used: luciferase siRNA (5′-CGUACGCGGAAUACUUCGAdTdT), control siRNA (5′-GGA-CAGAUUCAAAUAACAAdTdT). The siRNAs were transfected at a final concentration of 26 nM. Transfections were performed in six-well dishes using Effectene transfection reagent (Qiagen).
RNA analysis and luciferase assays
Total RNA samples were analyzed by Northern blot as described by Gatfield et al. (2003). Body-labeled DNA probes were generated by unidirectional PCR or random priming according to standard protocols. The levels of the reporters and intermediates were quantitated and normalized to the levels of rp49 mRNA. Luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega).
Generation of luciferase reporters for the miRNA pathway
DNA fragments corresponding to the 3′ UTRs of Vha68–1 and CG10011 transcripts (including the polyadenylation signals) were amplified by PCR from a Drosophila S2 cell cDNA library and cloned downstream of the firefly luciferase coding region, between the XbaI and XhoI sites of plasmid pJ-Luc. Plasmid pJ-Luc derives from the GFP sensor described before (Brennecke et al. 2003), in which GFP is replaced by firefly luciferase. The firefly luciferase coding region is inserted between KpnI and NotI sites, downstream of the constitutive tubulin promoter, and includes an optimal Kozak consensus sequence. The pAc5.1 plasmid expressing Renilla luciferase described above served as a transfection control. For expression of miR-9b and miR-12, a fragment of ~200 nt encompassing the miRNA gene was amplified from genomic DNA and cloned in vector pAc5.1A. Transfections were performed as described above. The transfection mixtures contained 50 ng of firefly luciferase reporter plasmid, 0.3 μg of the transfection control plasmid, and 0.7 μg of plasmids expressing miRNAs. In an experiment in which dsRNAs targeting 1140 Drosophila genes were tested using the assay shown in Figure 3B ▶, the normalized firefly luciferase activity was set to an average value of 1. The standard deviation of all measurements was ±0.43. We therefore judged a change of at least twofold to be a meaningful cutoff.
Tethering assay
The F-Luc-5-BoxB plasmid was constructed by inserting the 5-Box sequences into the EcoRI–XhoI sites of the vector pAC5.1-F-Luc described above. AGO1 cDNA was amplified with primers containing appropriate restriction sites, using a (dT)15-primed S2 cDNA library as template. The amplified cDNA was cloned into a vector allowing the expression of λN-peptide fusions (pAc5.1-λN).
Acknowledgments
We are grateful to Tamas I. Orban for help with the reporter assays and David Thomas for comments on the manuscript. This study was supported by the European Molecular Biology Organization (EMBO) and the Human Frontier Science Program Organization (HFSPO).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2191905.
REFERENCES
- Andrei, M.A., Ingelfinger, D., Heintzmann, R., Achsel, T., Rivera-Pomar, R., and Luhrmann, R. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Conti, E. and Izaurralde, E. 2005. Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr. Opin. Cell. Biol. 17: 316–325. [DOI] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., and Seraphin, B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeier, M.E., Morse, D.P., Knight, S.W., Portereiko, M., Bass, B.L., and Mango, S.E. 2000. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289: 1928–1931. [DOI] [PubMed] [Google Scholar]
- Dostie, J., Ferraiuolo, M., Pause, A., Adam, S.A., and Sonenberg, N. 2000. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 19: 3142– 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy, T., Chan, E.K., Tenenbaum, S.A., Keene, J.D., Griffith, K., and Fritzler, M.J. 2002. A phosphorylated cytoplasmic auto-antigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 13: 1338–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy, T., Jakymiw, A., Chan, E.K., Seraphin, B., Cougot, N., and Fritzler, M.J. 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW-bodies. RNA 9: 1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz, W. 2005. RNAi: The nuts and bolts of the RISC machine. Cell 122: 17–20. [DOI] [PubMed] [Google Scholar]
- Fukuhara, N., Ebert, J., Unterholzner, L., Lindner, D., Izaurralde, E., and Conti, E. 2005. SMG7 is a 14–3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell 18: 537–547. [DOI] [PubMed] [Google Scholar]
- Gatfield, D. and Izaurralde, E. 2004. Nonsense-mediated mRNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578. [DOI] [PubMed] [Google Scholar]
- Gatfield, D., Unterholzner, L., Ciccarelli, F.D., Bork, P., and Izaurralde, E. 2003. Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO J. 22: 3960–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger, D., Arndt-Jovin, D.J., Lührmann, R., and Achsel, T. 2002. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Kim, J.K., Gabel, H.W., Kamath, R.S., Tewari, M., Pasquinelli, A., Rual, J.F., Kennedy, S., Dybbs, M., Bertin, N., Kaplan, J.M., et al. 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., and Parker, R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7: 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15: 185–197. [DOI] [PubMed] [Google Scholar]
- Okamura, K., Ishizuka, A., Siomi, H., and Siomi, M.C. 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 18: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban, T.I. and Izaurralde, E. 2005. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex and the exosome. RNA 11: 459– 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R. and Song, H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11: 121–127. [DOI] [PubMed] [Google Scholar]
- Pillai, R.S., Artus, C.G., and Filipowicz, W. 2004. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10: 1518–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Artus, C.G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., and Filipowicz, W. 2005. Inhibition of translational initiation by let-7 microRNA in human cells. Science http://www.sciencemag.org/cgi/content/abstract/1115079v1 [DOI] [PubMed]
- Rand, T.A., Ginalski, K., Grishin, N.V., and Wang, X. 2004. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. 101: 14385–14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, G.L. and Blau, H.M. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7: 633–636. [DOI] [PubMed] [Google Scholar]
- Sheth, U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret, F.F., Kastenmayer, J.P., and Green, P.J. 2004. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15: 173–183. [DOI] [PubMed] [Google Scholar]
- Teixeira, D., Sheth, U., Valencia-Sanchez, M.A., Brengues, M., and Parker, R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner, L. and Izaurralde, E. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16: 587–596. [DOI] [PubMed] [Google Scholar]
- van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., and Seraphin, B. 2002. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21: 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Jakymiw, A., Wood, M.R., Eystathioy, T., Rubin, R.L., Fritzler, M.J., and Chan, E.K. 2004. GW182 is critical for the stability of GW-bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 117: 5567–5578. [DOI] [PubMed] [Google Scholar]