Abstract
RNA-directed recombination can be used to catalyze a disproportionation reaction among small RNA substrates to create new combinations of sequences. But the accommodation of secondary and tertiary structural constraints in the substrates by recombinase ribozymes has not been explored. Here, we show that the Azoarcus group I intron can recombine oligoribonucleotides to construct class I ligase ribozymes, which are catalytically active upon synthesis. The substrate oligonucleotides, ranging in size from 58 to 104 nucleotides (nt), along with the 152-nt ligase ribozymes they reconstitute, can contain significant amounts of secondary structure. However, substrate recognition by the Azoarcus ribozyme depends on the existence of a single accessible CAU triplet for effective recombination. A biphasic temperature reaction profile was designed such that the sequential recombination/ligation events could take place in a thermocycler without human intervention. A temperature-dependent pH shift of the reaction buffer contributes to the success of the net reaction. When the substrate for the ligase ribozyme is introduced into the reaction mixture, as much as 11% can be observed being converted to product by the recombined ligase in the same reaction vessel. Recombination followed by ligation can also occur under isothermal conditions at 37°C. Tertiary structure formation of the ligase upon construction can provide some protection from cleavage by the Azoarcus ribozyme when compared to the constituent substrates. These data suggest that RNA-directed recombination can, in fact, articulate complex ribozymes, and that there are logical rules that can guide the optimal placement of the CAU recognition sequence.
Keywords: catalytic RNA, ribozymes, ligase, Azoarcus, recombination
INTRODUCTION
Group I and group II ribozymes can catalyze the energy-neutral disproportionation of RNA oligonucleotide substrates in vitro to produce new combinations of sequences (Zaug and Cech 1986; Mörl and Schmelzer 1990; Riley and Lehman 2003b). These recombination reactions proceed by the general formula A·B + C·D ↔ C·B + A·D, where A and C are the 5′ “head” portions of RNA substrate oligonucleotides, while B and D are the 3′ “tail” portions. Recombination is accomplished through a two-step mechanism. In the first step, A·B substrates are bound via the internal guide sequence (IGS) of the ribozyme to a complementary sequence in the substrate, and the 3′ portion B is transferred to the 3′ end of the ribozyme in a transesterification reaction while the A head is released. In the second step, either a freed C head or a C·D substrate binds to the B-tailed ribozyme and a second transesterification reaction results to produce the recombinant C·B molecule. Previously we have shown that a modified form of the Azoarcus group I intron is especially adept at performing recombination on short RNA oligonucleotides, and can transiently convert as much as 75% of a C·D input oligo into C·B products in <30 min (Riley and Lehman 2003b). The Azoarcus ribozyme is highly reactive in vitro at a temperature of 60°C, allowing relatively rapid binding and release of substrates or products, and hence efficient RNA recombinase activity. In one case we were able to recombine substrate oligos to produce a C·B product that itself was a ribozyme, a 33-nt hammerhead, which could bind and catalyze the cleavage of its own substrate in the same reaction vessel in which it was articulated (Riley and Lehman 2003b).
RNA-directed recombination of other ribozymes has great potential to illuminate the means by which RNAs can form networks of reactions. Class I ligase ribozymes promote the attack of a 3′-OH on an anhydride bond within a 5′ triphosphate moiety (Bartel and Szostak 1993). This is the correct chemistry needed for RNA polymerization, and hence ligases may model RNA reproduction during the putative RNA World. In fact, through a combination of ribozyme engineering and in vitro evolution, variants of the original class I ligase molecule have been discovered that can catalyze template-directed nucleotide triphosphate addition to a 3′-OH terminus (Johnston et al. 2001; McGinness and Joyce 2002), with a processivity up to at least 14 nt (Johnston et al. 2001).
Because of its potential as a primordial RNA replicase, the class I ligase represents an obvious goal for RNA-directed recombination from smaller constituent RNA pieces. However, these ligase molecules present two significant challenges in this regard. First, they are moderately sized molecules of 100–160 nt in length, significantly greater than that of the hammerhead motif. Second, they are structurally complex. The original b1–207 construct and its more kinetically proficient E100–3 and B16–19 variants all possess double nested pseudoknotted structures that can assume multiple folded conformations (Ekland and Bartel 1995; Ekland et al. 1995; Wright and Joyce 1997; Schmitt and Lehman 1999), unlike the relatively simple hammerhead motif with three independent stem-loop components. Previous experiments with the Tetrahymena group I intron have demonstrated its ability to bind, cleave, and exchange longer RNA substrates of 500 or more nucleotides (nt) in length (Woodson and Cech 1989; Sullenger and Cech 1994; Bell et al. 2002), albeit those without significant secondary structure.
With the expectation that the activity of the Tetrahymena group I intron on longer RNA substrates could be realized with the Azoarcus ribozyme—and perhaps improved—we designed a recombination scheme that would lead to the single-vessel construction of variants of the B16–19 ligase ribozyme from two constituent RNA oligonucleotides. We targeted mutants of the B16–19 ligase that could be spliced together as a result of their possession of a single 5′-CAU-3′ sequence, which is the complement of the IGS for the Azoarcus ribozyme. Guided by a recent model of the class I ligase (Bergman et al. 2004), we opted to place this CAU sequence in various locations, including an exterior region of the molecule that would also be distant from the active site (Fig. 1). We designed this strategy with the intent that the Azoarcus ribozyme could sequentially bind two longer (50–110 nt) oligonucleotides, and recombine them into a functional ligase ribozyme despite severe steric constraints imposed by secondary structure in both the oligonucleotides and the intact ribozyme (Fig. 2). Here, we show the viability of this strategy, the ability of the recombined ribozyme to then immediately perform ligation on its own substrate, and means to partition the various elements of this multistep reaction pathway even within a single homogeneous reaction mixture. In doing so we also provide insight into the influence that the CAU placement site has on the efficacy of recombination.
FIGURE 1.
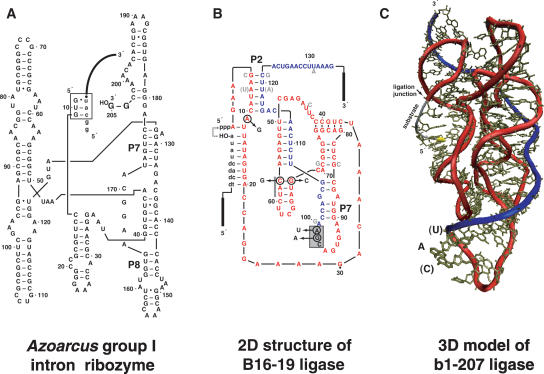
Ribozymes used in this study. (A) Azoracus group I intron designed with two 3′ guanosines such that it can perform recombination on exogenous RNA oligonucleotide substrates (Riley and Lehman 2003b). Substrate (lowercase letters) is in the form 5′-GGCAU·tail–3′ and is bound to the ribozyme through the internal guide sequence GUG (boxed). (B) B16–19 class I ligase ribozyme, which catalyzes the ligation of its substrate (lowercase letters) to its 5′ end (dashed arrow). Mutations added to make the B16–19RC construct are indicated by circles. The nine light gray nucleotides indicate differences for E100–3, although the A7:U119 pair was reverted to the B16–19 configuration for both the E100–3RC and E100–3nRC constructs. The boxed area denotes the insertion site for the IGS complement sequence CAU for both B16–19RC and E100–3RC; the gray bar denotes the insertion site for CAU in E100–3nRC. The red (C) and blue (B) portions of the molecule denote the two halves that derive from C·D and A·B constituent oligonucleotides, respectively, by recombination. (C) Three-dimensional model of the b1–207 class I ligase ribozyme (Bergman et al. 2004). The labeled nucleotides indicate the insertion location of the Azoarcus IGS complement sequence CAU for B16–19RC and E100–3RC; in the b1–207 model shown, the C is an adenine and the U is a cytidine. Backbone colors indicate source oligonucleotides for recombination as in panel B.
FIGURE 2.
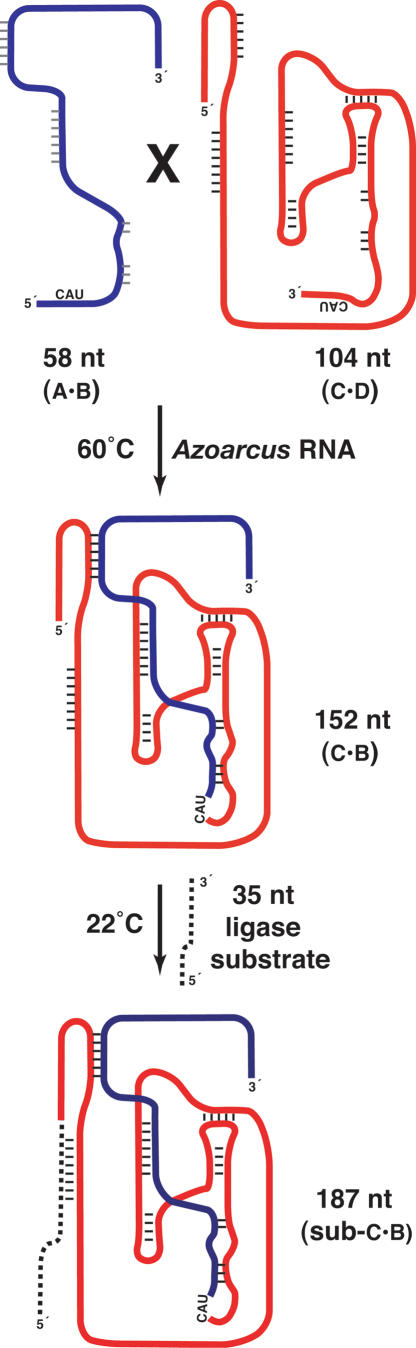
Schematic of the recombination of a class I ligase ribo-zyme from constituent halves and its subsequent catalysis of a ligation reaction to an exogenous substrate. All of these reactions are designed to take place in a single homogeneous aqueous milieu. In the recombination step at 60°C, the Azoarcus ribozyme first binds one RNA substrate (e.g., the 58-nt SL-5, blue = A·B) and performs a transesterification reaction to transfer the 3′ portion (following the CAU sequence) to its own 3′ end. Then the 5′ portion of the second substrate (e.g., the 104-nt SL-6, red = C·D) binds and a second trans-esterification reaction generates the recombined RNA (Riley and Lehman 2003b), in this case the 152-nt ligase ribozyme. Note that the converse reaction also occurs and produces a 10-nt RNA (GGCAU·AAAUA = A·D), not shown. In the ligation step at 22°C, the recombined ligase ribozyme binds its own substrate (e.g., S-163, dashed) and catalyzes a phosphoester bond formation to the 5′ end of the ligase.
RESULTS AND DISCUSSION
Ligase ribozyme design
We first selected suitable splice sites for the intact ligase ribozyme. We opted to place the IGS complement sequence CAU in a region of the ribozyme that should be solvent accessible when found in both of the component RNA oligonucleotides A·B and C·D and not involved in base-pairing (Fig. 1B). Ideally, this site would become buried and solvent inaccessible in the full-length reconstructed ribo-zyme, but this was not a requisite. The three-dimensional (3D) model that exists for the b1–207 ligase (Fig. 1C; Bergman et al. 2004) suggests that the 9-nt loop spanning nt 91–100 in the B16–19 ligase would possess these characteristics, provided the two ligases are homologous. Homology is reasonable because the B16–19 was derived via evolution in vitro from E100–3 (Schmitt and Lehman 1999), which in turn was derived from b1–207 (Wright and Joyce 1997). The b1–207 model indicates that the nucleotides in this loop are neither protected from hydroxyl-radical cleavage nor implicated in participating in the catalytic core (Bergman et al. 2004). We engineered two mutations, G98 → A and A99 → U, which would create a CAU triplet at positions 97–99. However, we also needed to remove two other CAU triplets within the B16–19 sequence. To do so we made three additional mutations, one by an A11 → G that eliminates the CAU at positions 10–12, and two others by a transposition of the C57:G68 base pair that eliminates the CAU at positions 57–59. The combination of these five mutations would be the molecule B16–19RC (Recombination Competent), but their effects on ligase activity were not known prior to the recombination experiments. We also designed a second recombination competent ligase ribozyme, E100–3RC, based on the E100–3 sequence. In addition to the A11 → G mutation and the C57:G68 base pair swap, this molecule required an A7:U119 base pair swap in comparison to the E100–3 sequence (Fig. 1B). These mutations would generate another single CAU-containing ribozyme, but one that was intermediate between E100–3 and B16–19 in the P2 stem. Third, we attempted to remove only the CAU spanning positions 10–12 in the E100–3 ribozyme and recombine using the other, intrinsic CAU at positions 57–59. This would create a third CAU-containing ribozyme, E100–3nRC, in which this sequence was buried within a relatively strong stem-loop structure that should be resistant to thermal melting.
Recombination of ligase ribozymes by the Azoarcus ribozyme
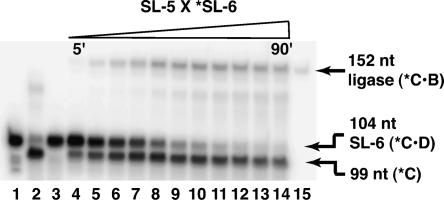
We then demonstrated that the Azoarcus ribozyme can perform recombination of A·B and C·D substrates to generate a full-length ligase ribozyme, C·B. In Figure 3, the 53-mer SL-5 is recombined with the 5′-labeled 104-mer SL-6 to generate the 152-mer B16–19RC. The production of the full-length ribozyme can be seen clearly after a 10-min incubation time and still increasing at 90 min, at which time 17% of the SL-6 RNA (C·D) has been recombined into B16–19RC (C·B). This peak recombination frequency (r.f.) is not as high as the 75% that can be seen with short, structureless RNA oligonucleotides but is comparable to the values seen for the recombination of the structurally simpler hammerhead ribozyme (Riley and Lehman 2003b). Of interest is the observation that the r.f. is not diminishing at time points as long as an hour or more. This is attributable in part to the relative stability of the C·B product compared to the C·D starting material and even to the free 5′ C cleavage products. One would expect the C·B product to be ephemeral, because it possesses the IGS complement sequence CAU of the Azoarcus ribozyme and thus is itself a substrate for Azoarcus RNA mediated cleavage. We suggest that the formation of the intact 152-nt ligase ribozyme partially buries the CAU inside the 3° structure of the ligase ribozyme once recombination has taken place, thereby protecting the construct from subsequent splicing. Specifically, the formation of the 9-nt loop that closes stem P7 (Fig. 1B) may contribute to making the CAU sequence less accessible to the Azoarcus ribozyme than it would be in the short, structureless head of C·D. As evidence, when we compared the relative abilities of SL-6 and the full-length B16–19RC RNA molecules to serve as substrates for the Azoarcus ribozyme, the latter is cleaved less than half as much after 30 min at 60°C (31% cleavage of B16–19RC into C and B vs. 71% cleavage of SL-6 into C and D).
FIGURE 3.
Time course of Azoarcus-ribozyme-directed recombination of the B16–19RC ribozyme (152 nt) from the constituent oligonucleotides SL-5 and SL-6. In all lanes, only the SL-6 RNA oligonucleotide (Table 1) initially has 5′ radiolabel (*). (Lane 1) Unincubated *SL-6. (Lane 2) Negative recombination control in the absence of SL-5: 0.2 μM *SL-6 and 1.5 μM Azoarcus ribozyme incubated for 30 min. (Lane 3) Negative recombination control in the absence of Azoarcus ribozyme: 0.2 μM *SL-6 and 0.2 μM SL-5 incubated for 30 min. (Lanes 4–14) Recombination time course: 0.2 μM *SL-6, 0.2 μM SL-5, and 1.5 μM Azoarcus ribozyme. At 90 min, the recombination frequency (r.f.) has reached 17%. (Lane 15) 5′-end-labeled B16–19 RNA as a 152-nt size control. All reactions performed at 60°C in 10 mM MgCl2, 5 mM CaCl2, and 30 mM EPPS, nominal pH 7.6.
In Figure 3, recombination was achieved using excess Azoarcus ribozyme and equimolar inputs of SL-5 and SL-6. Thus unlike previous experiments (Riley and Lehman 2003b), the recombination is not driven by an excess of one substrate oligonucleotide. With shorter substrates, we had found that an excess of the A·B substrate would improve the r.f. However, scout experiments with varying amounts of SL-5 and SL-6 indicated that the most favorable stoichiometry for recombination of B16–19RC was 1:7.5:1 < SL-5:Azoarcus:SL-6, and the most favorable conditions were 10 mM MgCl2, 5 mM CaCl2, pH 7–8, and 60°C (data not shown). The addition of CaCl2 to the reaction buffer and the near-neutral pH both appear to retard premature base-catalyzed hydrolysis of 5′-ribozyme·B-3′ intermediates.
The recombination is clearly dependent on Azoarcus RNA, because no 152-nt RNA can be seen when A·B and C·D are incubated in the absence of the ribozyme (Fig. 3, lane 3). Compared to the Tetrahymena version, the Azoarcus ribozyme is a more efficient recombinase (Riley and Lehman 2003b). For the Tetrahymena ribozyme, product release can be rate limiting (Herschlag and Cech 1990), as the 6-bp interaction between the IGS and its complement 5′-CCCUCU-3′ is relatively stable once formed. The overall intrinsic equilibrium favors forward splicing, consistent with the ribozyme’s biological function (Kumar and Joyce 2003), as opposed to the reverse splicing reaction that is the first step in recombination. The Azoarcus ribozyme is also unlike the Tetrahymena version in that Azoarcus RNA unfolds somewhat upon binding of substrate (i.e., substrate binding is not required for the molecule to assume an active three-dimensional conformation), implying that 2° structure constraints in the substrates are not as inhibitory, especially at 60°C (Kuo et al. 2004).
Recombination of active ligases
Next we tested whether the recombined ligases were active in the same milieu in which they were made. Because of their complex structures, we were not sure that class I ligases could fold properly once the 1° sequence was spliced together without some sort of denaturation and renaturation procedure. Consequently we designed a biphasic recombination/ligation protocol in which the favorable conditions for recombination were followed by conditions favorable to ligation (Fig. 2). Data from Figure 3 indicated that recombination at 60°C for 45 min is sufficient to engender r.f. values close to maximal. By contrast, ligation activities of the known ribozymes E100–3 and B16–19 are favored near room temperature and should be complete in <60 min, barring kinetic traps related to folding. Ligases should also be active in the buffer optimal for recombination containing 10 mM MgCl2 and 5 mM CaCl2 (Wright and Joyce 1997; Schmitt and Lehman 1999; Riley and Lehman 2003a). Our initial RNA network simulations were thus single 20-μL reaction vessels placed in a thermocycler containing Azoarcus RNA, A·B and C·D substrates, the 5′-γ32P-labeled ligase substrate, and buffer, to be incubated for 45 min at 60°C and then immediately for 60 min at 22°C.
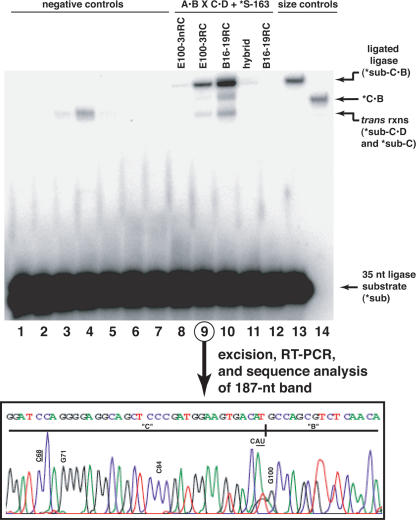
RNA-directed recombination does indeed produce nascent active ligase ribozymes. In Figure 4, we labeled only the S-163 substrate, meaning that only recombined ribo-zymes that can successfully perform ligation by covalent bond formation will themselves become visible by denaturing gel electrophoresis. In this fashion, we tested the ability of four ligase molecules to recombine and react: E100-nRC, E100–3RC, B16-RC, and a hybrid between the last two that contains a C-C mismatch in the first position of the P2 stem. The B16–19RC molecule exhibits the greatest amount of ligation ability, as measured by the relative intensity of the band at 187 nt, which corresponds to the full-length ligase ribozyme following ligation of S-163. The net amount of ligation under these conditions is modest: 0.87% of S-163 becomes covalently bound as a 187-mer. However, the extent of net reaction with B16–19RC is threefold greater than that with E100–3RC, which in turn is far greater than those of E100–3nRC and the hybrid molecule, which both exhibit only trace activities of <0.02%. Because the 187-nt band is the result of a sequence of reactions, it is difficult to pinpoint the advantage that B16–19RC has. The parent molecule B16–19 is not kinetically more proficient than E100–3, but it misfolds less often (Schmitt and Lehman 1999). When we analyzed the apparent pseudo-first-order rate constants of both parent molecules and both RC molecules (Table 2), it became clear that this trend holds for the RC molecules assayed in the divalent salt conditions used in recombination. Both parent molecules exhibit similar rate constants, at a value roughly 100-fold greater than the RC molecules. However, the B16–19RC molecule is more active when forced to fold after being recombined (Fig. 4). Taken together, we interpret these data as evidence that the B16–19RC molecule is more apt to fold into an active conformation following recombination, not that its sequence is more amenable to recombination or that its active form is a faster ligase. Additional support for this comes from the observation that the B16–19RC/E100–3RC hybrid recombinant (made by the SL-5 × SL-4 recombination), which should fold very poorly as a consequence of the mismatch in P2, shows almost no activity.
FIGURE 4.
Recombination and ligation in a single reaction mixture. The gel shows the separation of the recombined and reacted ligase ribozyme (187 nt) from the unreacted ligase substrate (35-mer). In lanes 1–13, only the ligase substrate S-163 (Table 1) initially has 5′ radiolabel (*). (Lanes 1–7) Negative controls (lane 1, Azoarcus ribozyme incubated only with *S-163; lanes 2–5, ligase halves incubated with *S-163 but in the absence of Azoarcus ribozyme: lane 2, SL-1 and SL-2; lane 3, SL-3 and SL-4; lane 4, SL-5 and SL-6; lane 5, SL-5 and SL-4; lane 6, SL-5 incubated with Azoarcus ribozyme in the absence of an SL-even RNA; lane 7, SL-6 incubated with Azoarcus ribozyme in the absence of an SL-odd RNA). (Lanes 8–12) Recombination then ligation, with expected ligase molecules indicated on gel (lane 8, SL-1 × SL-2; lane 9, SL-3 × SL-4; lane 10, SL-5 × SL-6; lane 11, SL-5 × SL-4; lane 12, SL-5 × SL-6 in 15 mM CaCl2 i.e., no Mg2+). (Lanes 13–14) Size controls (lane 12, *S-163 incubated with B16–19, which gives 187-nt RNA; lane 13, 5′-labeled B16–19, which is 152-nt RNA). In all lanes, the Azoarcus ribozyme, when present, is at 1.5 μM; the SL-oligonucleotides and S-163 substrate, when present, are at 0.2 μM. (Inset) Chromatogram of nucleotide sequence through the recombination junction of the E100–3RC ribozyme following recombination, excision of 187-nt band from the gel, and RT-PCR amplification. E100–3 specific mutations, as well as mutations required for E100–3RC (underlined), are indicated.
TABLE 2.
Kinetic characterization of class I ligase ribozymes in 10 mM MgCl2, 5 mM CaCl2, and 30 mM EPPS (pH 7.6) at 22°C
| Ribozyme | Origin | kcat (min−1) |
| E100–3 | Evolveda | 4.4 |
| B16–19 | Evolvedb | 3.6 |
| E100–3RC | Designed for recombination | 0.034 |
| B16–19RC | Designed for recombination | 0.033 |
aFrom Wright and Joyce (1997).
bFrom Schmitt and Lehman (1999).
The failure of E100-nRC to show activity can be attributed to the inability of the CAU sequence at positions 57–59 to serve as a viable site for access by the IGS of an Azoarcus ribozyme. The SL-1 and SL-2 substrates cannot be readily recombined into a full-length ribozyme, a consequence of the fact that the SL-1 oligonucleotide is almost immune to attack by Azoarcus RNA (data not shown). In SL-1, the CAU sequence is contained within the stem-loop formed from nt 57–68 that is believed to be a nucleation site for class I ligase folding (Bergman et al. 2004). This feature is one of the most likely 2° elements to be preserved at 60°C. Thus, while the ability of Azoarcus RNA to serve as a recombinase ribozyme is greatly bolstered by its reactivity at high temperatures, not all CAU placement sites in substrates are denatured and accessible at that temperature.
We performed controls to ensure that the net reaction was proceeding as outlined in Figure 2. No full-length ligation was detected when Azoarcus RNA or when one or both of the two A·B or C·D oligonucleotides were omitted from the reaction (Fig. 4, lanes 1–7). No reaction was detected when 15 mM CaCl2 was supplied as the sole divalent (Fig. 4, lane 12). Furthermore, when the 187-nt bands for E100–3RC and B16–19RC were excised from the gel and subjected to RT-PCR and sequence analysis, the expected nucleotide sequences through the splice sites were observed (e.g., Fig. 4, inset). However, the crosses of SL-3 × SL-4 and SL-5 × SL-6, leading to E100–3RC and B16–19RC, respectively, did display a small amount of shortened ligated product, with or without the presence of Azoarcus RNA. This product is most likely the result of a mild trans reaction, whereby the two halves of the ligase ribozyme can hybridize at 22°C without a covalent attachment and catalyze ligation. The product migrates in the 120–150-nt range, and, in fact, two different bands can be seen (Fig. 4, cf. lanes 4 and 10). The slower migrating band, seen in the absence of Azoarcus RNA, corresponds to the SL-4 oligonucleotide (104-mer) that has been ligated to S-163 (35-mer) to produce a 139-nt product, while the faster migrating band appears in the presence of Azoarcus RNA and would be the cleavage of the D tail from the 139-mer to give a 134-mer product. The intensities of the trans reactions are weak, being together about 10% of the covalent reaction in the presence of Azoarcus RNA. The trans reaction of class I ligases has been reported previously at a different location in the molecule; the b1–207t construct can promote ligation of an exogenous substrate to the 5′ portion of the ligase separated between the equivalents of positions 23 and 24 in Figure 1B (Bergman et al. 2000).
The Azoarcus RNA itself can apparently serve as a substrate for recombination. In searching for an explanation for the faint band migrating between 152 and 187 nt (e.g., Fig. 4, lane 10), we realized that the single CAU triplet that exists within the Azoarcus RNA sequence at positions 170–172 could serve to guide recombination between two molecules, a situation that would rarely, if ever, occur in vivo. If one Azoarcus RNA were able to recognize the CAU of another and catalyze a transesterification, then the first Azoarcus RNA would be extended by 33 nt on its 3′ end (205 − 172 = 33). Then, recombination with a reacted ligase molecule, which had been subsequently cleaved by an Azoarcus ribozyme (or produced and reacted via a trans event) to generate the 134-mer mentioned above, would generate a labeled 167-mer (134 + 33 = 167). This sequence of events is rare, as judged by the intensity of the band, underscoring the observation that CAU sequences buried in the core of a molecule would not present themselves easily for recombination. Positions 170–172 of Azoarcus RNA lie at the boundary between J7–8 and P7, and are in the ribozyme’s interior from a recent crystal structure (Adams et al. 2004).
It is important to note that the mutations added to make the ligase constructs recombination competent were deleterious for ligation activity (Table 2). The sole requirement for Azoarcus RNA mediated recombination of other catalytic RNAs is the possession of a single and (initially) accessible CAU sequence in the precursor oligonucleotides. The triplet CAU will occur on average once every 64 triplets by chance, and is present twice each in the B16–19 and E100–3 source ligase molecules. Yet the poor recombinability of the E100–3nRC ligase suggests that only a portion of CAUs can be used by the Azoarcus ribozyme. These observations led to two inferences. First, the recombination/ligation activity observed in Figure 4 occurred despite the ~100-fold dampened ligation rate of the RC constructs. Second, CAU sequences buried in strong base-pairing regions are inaccessible to the Azoarcus ribozyme, as expected. Thus, rational design of the recombination of a wide variety of RNA targets should be possible.
The effects of temperature, salt, and pH
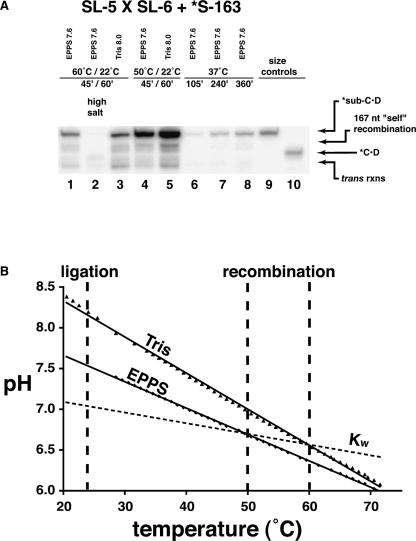
To optimize the combined recombination/ligation reaction pathway, we assayed the reactions under various temperature, salt, and pH conditions (Fig. 5A). While Azoarcus RNA mediated cleavage activity is known to be optimal at 60°C (Kuo et al. 1999), we found that the amount of S-163 converted to ligated product could be greatly enhanced by performing the recombination portion of the reaction at 50°C. In fact over a sixfold improvement can be seen, such that 6.0% net yield was observed (Fig. 5A, cf. lanes 1 and 4). On the other hand, increasing the ionic strength of the reaction buffer to 25 mM MgCl2 and 10 mM CaCl2 had the opposite effect, reducing the yield 25-fold down to a barely detectable 0.035%. The requirement that the recombinase ribozyme must retain its 3′ tail until another substrate can dock for recombination probably is the explanation for both of these effects. While higher temperatures and ionic strength favor the progress of Azoarcus RNA mediated RNA cleavage, to a certain extent, lowering these two parameters slows the site-specific hydrolysis of the tail long enough to maximize recombination. The trans and self-recombination reactions can also be observed at both temperatures, scaling roughly with the amount of 187-nt product.
FIGURE 5.
Effects of temperature, salt, and pH on single-vessel recombination/ligation. (A) The gel shows the higher molecular-weight products from recombination/ligation of B16–19RC. Reactions performed in the pH buffers indicated and in 10 mM MgCl2/5 mM CaCl2 (except lane 2) for the times and temperatures indicated. Lane 1 is the equivalent reaction as lane 10 in Figure 4. (B) The empirical response of pH to temperature of the buffers (Tris, nominal pH = 8.0; EPPS, nominal pH = 7.6) under simulated reaction conditions of cooling from 70°C to 20°C. The recombination and ligation temperatures employed in the gel experiment (60°C or 50°C and 22°C, respectively) are indicated by thick dashed lines.
The next parameter we investigated was that of pH. In Figure 4, the reaction was buffered with 30 mM EPPS (4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid) that had a nominal pH value of 7.6. But the pH of buffered solutions can be strongly influenced by temperature, usually becoming more acidic as the temperature rises. For EPPS, a reported ΔpH/ΔT is −0.0116 pH units/°C in 100 mM KCl (Fukada and Takahashi 1998). Because of the range of temperatures we were using for the combined assay, we were interested in how the pH would differ in the recombination and ligation phases. Recombination should be favored at lower pH values that promote retention of the ribozyme·B intermediates in the face of base hydrolysis. In contrast, ligation is favored at higher pH values, especially in Ca2+-containing buffers (Bergman et al. 2000; Riley and Lehman 2003a). Because the temperature change in the combined recombination/ligation experiment would produce a pH change in the favorable direction, we also tested the reaction in 30 mM Tris (tris(hydroxymethyl)aminomethane) buffer with a nominal pH of 8.0. Tris buffer has a much sharper reported ΔpH/ΔT of between −0.0280 and −0.0340 pH units/°C, and is, in fact, one of the least stable of the common buffers in this regard (Bates and Robinson 1973; Remette et al. 1977). Thus we anticipated that the steeper ΔpH/ΔT slope of Tris would lead to a greater overall amount of reacted ligase, and this correlates with what we observed (Fig. 5A). Compared to EPPS, in Tris there is ~150% of ligated ligase ribozyme after a 45-min recombination of B16–19C at 60°C followed by a 60-min ligation reaction. To demonstrate that this improvement is not simply a consequence of a single-temperature pH difference between EPPS and Tris, we empirically measured the pH variation of the two buffers as a function of temperature in a simulated recombination/ligation environment between 70°C and 20°C (Fig. 5B). Auspiciously, the slope of the Tris relationship is 137% that of EPPS, and, in fact, the Tris buffer rises 1.7 pH units during the transition from recombination at 60°C to ligation at 22°C. Both buffers actually have behaviors that augment the duality of environment within a single homogenous aqueous reaction vessel. This is a manifestation in part of the temperature shift and in part of the pH shift, which actually spans the acidic–basic boundary even though the neutral pH of water itself changes nearly 0.8 log units over this range. When temperature, salt, and pH conditions were all optimized (i.e., 50°C recombination followed by 22°C ligation in 10 mM MgCl2, 5 mM CaCl2, Tris nominal pH 8.0), we detected 11.2% conversion of ligase substrate into ligated B16–19RC ribozyme (Fig. 5A, lane 5), an order-of-magnitude improvement over the conditions used for Figure 4 (Fig. 5A, lane 1).
Isothermal reactions
Despite the expectation that Azoarcus-ribozyme-directed RNA cleavage should be almost 100-fold slower at 37°C than at 60°C (Kuo et al. 1999), we examined the ability of recombination and ligation to occur isothermally at 37°C. This would be a stringent test of the ability to perform recombination on substrates replete with 2° structure, given the observation that class I ligases retain high activity at this temperature (Wright and Joyce 1997; Schmitt and Lehman 1999; Riley and Lehman 2003a). Amazingly, isothermal recombination/ligation is possible at 37°C (Fig. 5A, lanes 6–8). The yield of ligated B16–19RC is not great, reaching only 0.58% after 6 h, but of interest is the fact that the yield continues to improve with time. We attribute this to the ability of the ligase to protect its CAU into 3° once recombined, thus precluding the reverse reaction via Azoarcus-RNA-directed cleavage. Furthermore, the trans-mediated ligation reaction appears to be hindered at 37°C, as would be expected if this temperature lies well above the melting temperature for the two ligase half molecules. Multistep RNA-catalyzed reactions are thus feasible in an isothermal environment, as would be in systems under homeostasis.
Conclusions
Our experiments demonstrate the plausibility of using recombination to construct complex ribozymes that possess significant structure, even pseudoknots. Disproportionation reactions resulting from ribozyme-catalyzed phosphoester-transfer events can generate new ribozyme functionalities that were previously weak or nonexistent. The Azoarcus self-splicing intron serves as an excellent model for these events because it can operate at high temperatures, allowing the ability both to recombine RNAs that contain significant 2° and 3° features and to perform multiple turnover for recombination. Exploitation of the ΔpH/ΔT behavior of certain buffers, as well as isothermal reactions, is also possible, giving rise to a “virtual compartmentalization” of an otherwise homogeneous aqueous milieu in order to better partition the components of a multistep reaction pathway of catalytic RNAs.
MATERIALS AND METHODS
Construction of RNAs and ribozymes
The source of the Azoarcus ribozyme (Reinhold-Hurek and Shub 1992) was a generous gift of Louis Kuo (Lewis and Clark College, Portland, OR) and consisted of the gene cloned into a pUC19 plasmid without 3′ guanosines at positions 204 and 205. These were restored in the molecule by PCR amplification using the primers TPM: 5′-TAATACGACTCACTATAG-3′ (which creates a promoter for T7 RNA polymerase) and T20a: 5′-CCGGTTTGTGTGAC TTTCGCC-3′ (which adds G204 and G205 via the underlined nucleotides). The source of the ligase ribozymes B16–19 and E100–3 was PCR DNA derived from plasmid preparations as described previously (Schmitt and Lehman 1999). The B16–19RC, E100–3RC, and E100–3nRC ligase ribozymes, along with the RNA oligonucleotides used to construct these ribozymes via RNA-directed recombination (Table 1), were transcribed from double-stranded DNA templates prepared by recursive PCR gene synthesis (Engels and Uhlmann 1988). Generally between four and eight partially overlapping single-stranded DNA oligonucleotides were incubated in equal molar ratios in 20-cycle PCR-like reactions using Vent DNA polymerase (New England Biolabs). The Azoarcus-RNA-directed recombination of ligase ribozymes was accomplished prior to the acquisition of DNA oligos and gene synthesis of the full-length ribozymes for kinetic analysis. This strategy insured that the RC ligase ribozymes could not exist prior to their construction via recombination. Transcriptions of ribozymes and long RNA oligos were carried out in reaction mixtures containing 1 × transcription buffer (15 mM MgCl2, 25 mM Tris at pH 7.5, 5 mM dithiothreitol, 2 mM spermidine), 2 mM rNTPs, 10 μL eightfold concentrated PCR DNA, and 1200 U of T7 RNA polymerase (Ambion) in a 200 μL volume for 4–12 h at 37°C. The resulting RNA was purified on 6% polyacrylamide/8 M urea gels. DNA oligonucleotides were purchased from Operon. The DNA/RNA chimeric ligase substrate S-163 (Table 1) was purchased from IDT.
TABLE 1.
Oligoribonucleotides used in this study
| RNA | Sequence | Length |
| SL-1 | GGCAU·CUUCGGAUGCAGGGGAGGCAGCUCCCGAUGGAAGUGACGAGCCAGCGUUCUCAACAGUAUUGACUGAACCUAAAAGCCAAUCGCAGGCUCAGC | 98 |
| SL-2 | AGAACAAUACGUUAUAGUGACCAGGAAAAGACAAAUCUGCCCUCAGAGCUUGAGAACAU·AAAUA | 64 |
| SL-3 | GGCAU·GCCAGCGUUCUCAACAGUAUUGACUGAACCUAAAAGCCAAUCGCAGGCUCAGC | 58 |
| SL-4 | AGAACAAUACGUUAUAGUGACCAGGAAAAGACAAAUCUGCCCUCAGAGCUUGAGAAGAUCUUCGGAUCCAGGGGAGGCAGCUCCCGAUGGAAGUGACAU·AAAUA | 104 |
| SL-5 | GGCAU·ACCAGCGUUCUCAACAGUAUUCACUGAACCUUAAAGCCAAUCGCAGGCUCAGC | 58 |
| SL-6 | AGAAGAAUACGUUAUAGUGACCAGGAAAAGACAAAUCUGCCCUUAGAGCUUGAGAAGAUCUUCGGAUCCACGGGAGGCAGCUCGCGAUGGAAGUGACAU·AAAUA | 104 |
| S-163 | d (CTTGACGTCAGCCTGGACTAATACGACTCA) UAUA | 35 |
Underlined region is sequence complementary to the IGS region of the Azoarcus group I ribozyme; dot indicates expected cleavage and transesterification site. All SL oligos are written 5′–head · tail–3′ (e.g., A·B). Ligase ribozymes produced upon recombination are as follows: SL-1 × SL-2 = E100-3nRC; SL-3 × SL-4 = E100-3RC; SL-5 × SL-6 = B16-19RC. The chimeric DNA/RNA oligonucleotide S-163 is the substrate for ligase ribozymes.
RNA-directed RNA recombination
Single reaction vessel recombination of A·B and C·D RNA oligonucleotides was mediated by the Azoarcus ribozyme at 60°C following the general protocols reported earlier (Riley and Lehman 2003b). For initial investigations of the efficacy of Azoarcus RNA to recombine large substrates into a 152-nt ligase molecule, the SL-3 × SL-4 and SL-5 × SL-6 crosses were performed to obtain the E100–3RC and B16–19RC molecules, respectively. Here, 15 pmol of Azoarcus RNA was incubated with 2 pmol each of A·B and C·D substrates in a 10-μL reaction volume. The odd-numbered substrates were envisioned as the A·B substrates, while the even-numbered substrates were envisioned as C·D to aid in representation of recombination as diagrammed in Figure 2. Prior to the addition of the substrate oligonucleotides, the Azoarcus RNA was first preheated in the presence of water to 80°C for 2 min. Immediately thereafter, 2 μL of a 5× buffer was added and the resulting solution was allowed to cool to room temperature. At 1× strength, all buffers used contained 30 mM of a pH buffer, either Tris or EPPS, and 0–50 mM divalent cation chloride. Magnesium, calcium, and mixtures of the two salts were used in various experiments. The pH value was variable (see below). To start the recombination reaction, the A·B and C·D substrates were added, in that order, and the reaction was brought to 60°C in a PTC-100 thermocycler (MJ Research) with a heated lid. After an appropriate incubation time, reactions were quenched with 10 μL of a solution containing 1 volume of 0.5 M EDTA and 9 volumes of 2× polyacrylamide gel loading dye containing formamide and urea. The resulting products were size partitioned by 8% polyacrylamide/8 M urea gel electrophoresis. Typically the C·D substrate would have been previously 5′-end-labeled by T4 polynucleotide kinase and [γ32P]·ATP, allowing for visualization of C, C·B, and C·D products by phosphorimaging.
Recombination with ligation
Recombination reactions with subsequent ligase-catalyzed phosphoester-bond formation were performed as follows in 20-μL final volumes in thin-walled 600-μL centrifuge tubes. Thirty picomoles of Azoarcus RNA were first heat denatured in water at 80°C for 2 min, followed by the immediate addition of 5× buffer that would be 10 mM MgCl2, 5 mM CaCl2, and 30 mM Tris or EPPS at 1× concentration. As in the recombination reactions decribed above, the solution was allowed to cool to room temperature. Then 4 pmol each of A·B, C·D, and the ligase substrate S-163 were added to the reaction, in that order. In these experiments, radiolabel was not present in either the A·B or the C·D oligonucleotides, only in the S163, which had been previously 5′-end-labeled by T4 polynucleotide kinase and [γ32P]·ATP, gel purified, and diluted 1:10 with gel-purified but unlabeled S163. Once all the components of the reaction had been added, the tubes were capped and placed into a thermocycler with the heated lid activated. The thermocycler was programmed to heat to 60°C for 45 min, then cool to 22°C for 60 min, and then cool to 4°C, at which time the reactions were removed and quenched. Isothermal reactions were incubated at 37°C without interruption for varying lengths of time. The migration of the S-163 was then assayed by electrophoresis on an 8% polyacrylamide/8 M urea gel followed by overnight exposure to a phosphor screen and phosphorimaging.
Genotypic analysis
To determine whether the recombination and ligation reactions had indeed proceeded as depicted in Figure 2, bands corresponding to the recombined/ligated molecule (187 nt) were excised from the gel following phosphorimaging. The RNA was eluted from the gel by osmosis, and purified and concentrated on a Nanosep 3K size-exclusion filter (Pall Gelman). One-half of the sample was then subject to RT-PCR using primers that would only amplify successfully ligated full-length ribozymes (primer 1 = primer for reverse transcription as well = 5′-GCTGAGCCTGCGATTGG-3′ primer 2 = 5′-CTTGACGTCAGCCTGGA-3′) (Wright and Joyce 1997; Schmitt and Lehman 1999). The resulting PCR DNA was purified on QIAquick spin columns (Qiagen) and subjected to cycle-sequence analysis by BigDye (v. 3) chemistry and capillary electrophoresis on a 3100 Avant automated sequencer (ABI).
Kinetic analysis
Formal kinetic analyses of the full-length ligase ribozymes were performed after the conclusion of the recombination experiments using RNAs transcribed from either plasmid DNA or recursively amplified PCR DNA as described above (i.e., not from recombined RNA). The kcat parameter was estimated for the E100–3, E100–3RC, B16–19, and B16–19RC ribozymes by the y-intercept of modified Eadie-Hofstee plots of the observed rate of reaction (kobs) as a function of kobs/[ribozyme]. The kobs values of the ligation reaction were determined under ribozyme excess conditions of 5 nM 5′-[γ32P]-labeled S163 substrate and 0.5–4.0 μM ligase ribozyme at 22°C. Six time points ranging between 5 sec and 60 min were taken when the ribozymes were incubated in buffer containing 10 mM MgCl2, 5 mM CaCl2, and 30 mM EPPS (pH 7.6). The extent of reaction was quantified as the fraction of total RNA in the ligated form after electrophoresis through 5% polyacrylamide/8 M urea gels. Single-turnover self-ligation kobs values were obtained for each ribozyme concentration by an exponential curve fit to the equation f = A(1 − exp(−kobs * t)), where f is the fraction of substrate reacted at time t, and A is the asymptote (projected maximum f). Only kobs values from individual time series where the half-time of the reaction was 10 sec or greater were utilized.
pH dependence
Because both the Azoarcus ribozyme transesterification activity (Kuo et al. 1999) and the ligase ribozyme phosphodiester bond formation activity (Bergman et al. 2000) are pH dependent, the initial recombination trials were performed using Tris and EPPS buffers at nominal (i.e., room temperature) pH values ranging from 7.0 to 8.5. To measure the empirical ΔpH/°C behaviors of these buffers, 30 mL solutions were prepared containing 30 mM of either buffer at selected pH values at 22°C (295 K), along with 10 mM MgCl2, 5 mM CaCl2, and a generic DNA oligonucleotide at a concentration that mimicked that of the total nucleic acids in recombination/ligation (2.3 μM). The solutions were covered with parafilm and heated gradually from 22°C to 75°C and cooled back down again while pH values were measured every 1–2°C. Corrections for electrode dependence on temperature can be made with the Nernst equation and were accomplished using pHact = pHobs ° (295/T), where pHact is the actual pH value, pHobs is the pH value indicated by the electrode, and T is the temperature in Kelvin of the electrode. Values of Kw (here, a symbol to represent the pH of neutral water) as a function of temperature were obtained from the CRC Handbook of Chemistry and Physics.
Acknowledgments
We thank K. Rusterholtz and W. Yu for technical support, L. Kuo and P. Unrau for advice and useful discussions, and NASA for funding (NNG04-GM20G).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2125305.
REFERENCES
- Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J., and Strobel, S.A. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. and Szostak, J.W. 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418. [DOI] [PubMed] [Google Scholar]
- Bates, R.G. and Robinson, R.A. 1973. Tris(hydroxymethyl)aminomethane—A useful secondary pH standard. Anal. Chem. 45: 420. [Google Scholar]
- Bell, M.A., Johnson, A.K., and Testa, S.M. 2002. Ribozyme-catalyzed excision of targeted sequences from within RNAs. Biochemistry 41: 15327–15333. [DOI] [PubMed] [Google Scholar]
- Bergman, N.H., Johnston, W.K., and Bartel, D.P. 2000. Kinetic framework for ligation by an efficient RNA ligase ribozyme. Biochemistry 39: 3115–3123. [DOI] [PubMed] [Google Scholar]
- Bergman, N.H., Lau, N.C., Lehnert, V., Westhof, E., and Bartel, D.P. 2004. The three-dimensional architecture of the class I ligase ribo-zyme. RNA 10: 176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland, E.H. and Bartel, D.P. 1995. The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucleic Acids Res. 23: 3231–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland, E.H., Szostak, J.W., and Bartel, D.P. 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269: 364–370. [DOI] [PubMed] [Google Scholar]
- Engels, J. and Uhlmann, E. 1988. Gene synthesis. Adv. Biochem. Eng. Biotechnol. 37: 73–127. [DOI] [PubMed] [Google Scholar]
- Fukada, H. and Takahashi, K. 1998. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Protein. Struct. Funct. Genet. 33: 159–166. [PubMed] [Google Scholar]
- Herschlag, D. and Cech, T.R. 1990. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry 29: 10159–10171. [DOI] [PubMed] [Google Scholar]
- Johnston, W.K., Unrau, P.J., Lawrence, M.S., Glasner, M.E., and Bartel, D.P. 2001. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292: 1319–1325. [DOI] [PubMed] [Google Scholar]
- Kumar, R.M. and Joyce, G.F. 2003. A modular, bifunctional RNA that integrates itself into a target RNA. Proc. Natl. Acad. Sci. 100: 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, L.Y., Davidson, L.A., and Pico, S. 1999. Characterization of the Azoarcus ribozyme: Tight binding to guanosine and substrate by an unusually small group I ribozyme. Biochim. Biophys. Acta 1489: 281–292. [DOI] [PubMed] [Google Scholar]
- Kuo, L., Perara, N., and Tarpo, S. 2004. Metal ion coordination to 2′ functionality of guanosine mediates substrate-guanosine coupling in group I ribozymes: Implications for conserved role of metal ions and for variability in RNA folding in ribozyme catalysis. Inorg. Chim. Acta 357: 3934–3942. [Google Scholar]
- McGinness, K.E. and Joyce, G.F. 2002. RNA-catalyzed RNA ligation on an external RNA template. Chem. Biol. 9: 297–307. [DOI] [PubMed] [Google Scholar]
- Mörl, M. and Schmelzer, C. 1990. Group II intron RNA-catalyzed recombination of RNA in vitro. Nucleic Acids Res. 18: 6545–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek, B. and Shub, D.A. 1992. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature 357: 173–176. [DOI] [PubMed] [Google Scholar]
- Remette, R.W., Culberson, C.H., and Bates, R.G. 1977. Acid–base properties of tris(hydroxymethyl)aminomethane (Tris) buffers in seawater from 5 to 40°C. Anal. Chem. 49: 867–870. [Google Scholar]
- Riley, C.A. and Lehman, N. 2003a. Expanded divalent metal-ion tolerance of evolved ligase ribozymes. Biochimie 85: 683–689. [DOI] [PubMed] [Google Scholar]
- ———. 2003b. Generalized RNA-directed recombination of RNA. Chem. Biol. 10: 1233–1243. [DOI] [PubMed] [Google Scholar]
- Schmitt, T. and Lehman, N. 1999. Non-unity molecular heritability demonstrated by continuous evolution in vitro. Chem. Biol. 6: 857–869. [DOI] [PubMed] [Google Scholar]
- Sullenger, B.A. and Cech, T.R. 1994. Ribozyme-mediated repair of defective mRNA by targeted, trans-splicing. Nature 371: 619–622. [DOI] [PubMed] [Google Scholar]
- Woodson, S.A. and Cech, T.R. 1989. Reverse self-splicing of the Tetrahymena group I intron: Implication for the directionality of splicing and for intron transposition. Cell 57: 335–345. [DOI] [PubMed] [Google Scholar]
- Wright, M.C. and Joyce, G.F. 1997. Continuous in vitro evolution of catalytic function. Science 276: 614–617. [DOI] [PubMed] [Google Scholar]
- Zaug, A.J. and Cech, T.R. 1986. The intervening sequence RNA of Tetrahymena is an enzyme. Science 231: 470–475. [DOI] [PubMed] [Google Scholar]