Abstract
Short interfering RNA (siRNA) binding by Dicer is important for RNA interference in Drosophila, but human Dicer (hDcr) has been reported to lack siRNA binding activity. We used native gel electrophoresis to characterize the siRNA-binding activity of endogenous hDcr-containing complexes in extracts from human cells. We identified a complex (D) that contains hDcr, as demonstrated by antibody supershift. Complex D appears to contain double-stranded siRNAs, and requires structural features of authentic siRNAs. Glycerol gradient sedimentation indicates that Complex D is ~250 kDa, slightly larger than hDcr alone. In addition, we found that purified recombinant hDcr (rhDcr) alone has siRNA binding activity. Complex D migrates more slowly than the rhDcr/siRNA complex in a native gel, suggesting that it contains at least one additional factor. hDcr directly contacts siRNAs within Complex D, as indicated by crosslinking. The endogenous complex is significantly enhanced by ATP, unlike the siRNA-binding activity of purified rhDcr, suggesting the existence of additional factors that can enforce the ATP dependence of endogenous hDcr/siRNA interactions. Complex D could impinge upon the RISC assembly pathway in humans, similar to an analogous complex in Drosophila.
Keywords: Dicer, RNA-induced silencing complex (RISC), RNA interference (RNAi), short interfering RNA (siRNA)
INTRODUCTION
RNA interference (RNAi) is a sequence-specific response to double-stranded RNA (dsRNA) that down-regulates genes at the level of mRNA stability (Fire et al. 1998). A class III ribonuclease, Dicer, cleaves long dsRNA into 21–23-nucleotide (nt) duplexes. These short interfering RNAs (siRNAs) are subsequently unwound and incorporated into RISC, where they direct the site-specific cleavage of complementary mRNA (for reviews, see Meister and Tuschl 2004; Filipowicz 2005; Sontheimer 2005; Tomari and Zamore 2005).
Although the RNAi response was discovered nearly a decade ago, the pathway’s mechanism remains only partially characterized. Much of the initial characterization has been done in Drosophila, leading to the identification of numerous RISC components (for reviews, see Sontheimer 2005; Tomari and Zamore 2005). Dcr-2 is required to initiate RISC assembly on siRNAs (Lee et al. 2004; Pham et al. 2004). An additional protein, R2D2, is needed to stabilize Dcr-2 and to facilitate siRNA binding (Liu et al. 2003). The R2D2/Dcr-2/siRNA complex progresses to intermediate complexes that then assemble into holo-RISC (Pham et al. 2004; Tomari et al. 2004a).
Active RISC contains only one strand of the siRNA duplex, and strand selection is thought to occur in an asymmetric fashion. Studies suggest that the strand with the less thermodynamically stable 5′ end is favored for RISC assembly (Khvorova et al. 2003; Schwarz et al. 2003; Reynolds et al. 2004). Intriguingly, there is evidence to suggest that the R2D2/Dcr-2 heterodimer senses this asymmetry, with Dicer preferentially binding the 5′ end of the strand favored for RISC entry (the guide strand), and R2D2 preferentially binding the 5′ end of the other (passenger) strand (Tomari et al. 2004b).
In addition to the extensive body of work in Drosophila, there have also been important advances made in human RNAi biochemistry. A human RISC of ~160 kDa was partially purified (Martinez et al. 2002) and copurifies with the proteins Argonaute1 and Argonaute2 (Ago1 and Ago2). Human Ago2 has been shown to act as the endonuclease responsible for target cleavage (Liu et al. 2004; Meister et al. 2004; Song et al. 2004; Yuan et al. 2005). A functional human ortholog of R2D2 was only recently reported (Chendrimada et al. 2005; Haase et al. 2005). A single Dicer enzyme generates siRNAs and miRNAs in humans and other mammals, but conflicting data exist on whether mammalian Dicer participates in later stages of the RNAi pathway (Martinez et al. 2002; Doi et al. 2003; Chendrimada et al. 2005; Kanellopoulou et al. 2005; Kim et al. 2005; Rose et al. 2005; Siolas et al. 2005).
Despite our growing body of knowledge, much remains unknown about the mechanism of RNAi in higher eukaryotes. It was recently shown that hAgo2 can bind single-stranded siRNA (Liu et al. 2004; Rivas et al. 2005), but double-stranded siRNA-binding proteins in the human RISC assembly pathway are only beginning to be characterized (Chendrimada et al. 2005; Haase et al. 2005). Although hDcr binds long dsRNA (Provost et al. 2002; Zhang et al. 2002), two reports state that it does not possess double-stranded siRNA-binding activity (Provost et al. 2002; Chendrimada et al. 2005). This raises questions about the similarity of mammalian RNAi to the Drosophila system, in which siRNA binding by R2D2/Dcr-2 is required to initiate RISC assembly (Liu et al. 2003; Lee et al. 2004; Pham et al. 2004; Tomari et al. 2004a). In addition, the role of hDcr’s ATPase domain remains unknown, since purified rhDcr’s only biochemical activities reported to date (dsRNA substrate binding and processing, and pre-miRNA processing) are ATP-independent (Provost et al. 2002; Zhang et al. 2002). To address these and other issues, we set out to identify and characterize human siRNA-binding complexes. We identified one such complex in which hDcr interacts directly with the siRNA in an ATP-dependent fashion.
RESULTS AND DISCUSSION
siRNP complexes in human cell extracts
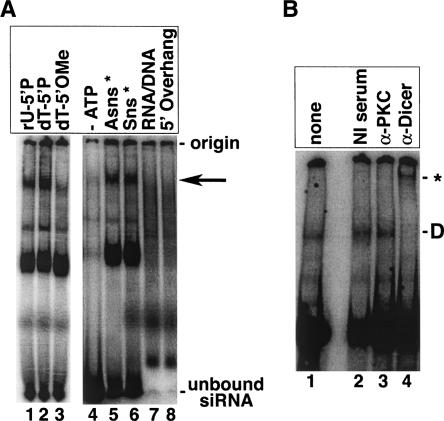
We used native gel electrophoresis to identify and characterize siRNA-containing complexes in human cell extracts and to determine whether hDcr was present in any of these complexes. After incubating HEK 293 S100 extract with a radiolabeled siRNA duplex, we observed several complexes on a native gel (Fig. 1A, lane 1). These complexes formed in HEK 293 and HeLa S100 extracts, both of which are competent for in vitro RNAi (data not shown). To assess the siRNA-binding specificity of the complexes, we altered the structure of the nucleic acid substrate. Only one of the complexes (denoted by an arrow in Fig. 1) was significantly reduced when the guide strand was modified with a 5′-terminal 2′-deoxy-5′-methoxythymidine residue, which blocks 5′ phosphorylation (Fig. 1A, lane 3). The same complex was unaffected when the guide strand contained a 5′-terminal 2′-deoxythymidine residue (Fig. 1A, lane 2). Because a 5′-phosphate group is important for siRNA function (Nykänen et al. 2001), this result suggests that the 5′-methoxy-sensitive complex alone has the binding specificity expected of a bona fide RNAi complex, and we characterized this complex further.
FIGURE 1.
An siRNA-binding activity from human cell extracts requires ATP and contains hDcr. (A) Radiolabeled siRNA was incubated with HEK 293 S100 extract and analyzed by native gel electrophoresis. The samples shown in lanes 1–3 and 5–8 contained ATP and an ATP-regenerating system. The sample in lane 4 was depleted for ATP. The siRNAs in lanes 1–3 were labeled at the 3′ end of the antisense (guide) strand; the passenger strand contained a 5′ hydroxyl. In lanes 4–8, the siRNAs were labeled at their 5′ end with [γ32P]-ATP, and the unlabeled strand contained a 5′ phosphate. In lanes 4 and 5, the guide strand was labeled, and in lanes 6–8 the passenger strand was labeled. “rU-P” denotes the unmodified siRNA with a uridine 5′-phosphate at its 5′ terminus. “dT-P” denotes an siRNA modified with a 2′-deoxythymidine 5′-phosphate at its 5′ terminus. “dT-OMe” denotes a 2′-deoxy-5′-methoxythymidine-modified siRNA. The asterisk (*) in lanes 5 and 6 denotes the labeled strand. “RNA/DNA” refers to an siRNA duplex in which the guide strand is DNA rather than RNA. “5′ Overhang” indicates a duplex with 2-nt 5′ overhangs. The 5′-methoxy-sensitive complex is denoted by an arrow. (B) Radiolabeled siRNA was incubated with extract and α-hDcr antibodies, and then analyzed by native gel electrophoresis. As negative controls, nonimmune serum (“NI serum”) and a nonspecific antibody (“α-PKC”) were also used. Lane 1 (“none”) indicates the mobility of the hDcr-containing species (Complex D) in the absence of any antibody. The supershifted complex is denoted by an asterisk (*).
The 5′-phosphate-dependent siRNA binding activity is greatly enhanced by ATP, because ATP depletion severely reduced complex levels (Fig. 1A, lane 4). The complex appeared equally when either strand of a reportedly asymmetric siRNA (Schwarz et al. 2003) was labeled, suggesting that it contains double-stranded RNA (Fig. 1A, cf. lanes 5 and 6). Moreover, no single-stranded RNA was observed in partially purified fractions containing this complex (data not shown). It did not form on a hybrid RNA/DNA duplex (Fig. 1A, lane 7) or on an siRNA containing a 2-nt 5′ overhang as opposed to the characteristic 2-nt 3′ overhang (Fig. 1A, lane 8). Furthermore, its formation is sequence-independent, as it formed on multiple, unrelated siRNAs (data not shown). The complex formed very early in time-course experiments (data not shown). Target cleavage activity did not cofractionate with the complex in a 10%–30% glycerol gradient, indicating that it is not a mature form of RISC.
The 5′-methoxy-sensitive complex contains hDcr
Some of the features of the 5′-methoxy-sensitive complex are reminiscent of an siRNA- and Dcr-2-containing complex (R1) that initiates RISC assembly in Drosophila (Pham et al. 2004). To address whether hDcr is present within the complex, we performed antibody supershift assays using our native gel system. In the presence of nonimmune serum or an unrelated antibody, the complex formed as usual (Fig. 1B, lanes 1–3). In contrast, incubation with antibodies raised against hDcr (Billy et al. 2001) resulted in the disappearance of the complex and the concomitant appearance of a more slowly migrating species (Fig. 1B, lane 4). We conclude that the complex contains the hDcr protein, and we now refer to it as Complex D.
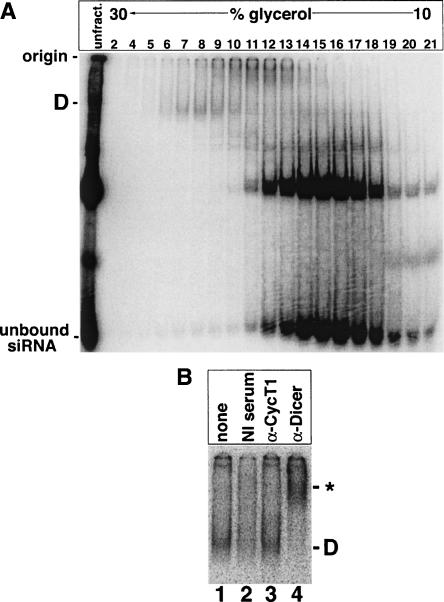
To further characterize Complex D, we set out to partially purify it. We incubated HEK 293 S100 extract with radiolabeled siRNA and subjected the reaction to 10%–30% glycerol gradient sedimentation. Fractions were analyzed by native gel, yielding a clear Complex D-containing peak (Fig. 2A). To confirm that this complex was truly the D complex, we repeated the antibody supershift assay. In the presence of the α-hDcr antibody, the partially purified complex shifted in a native gel (Fig. 2B, cf. lanes 1 and 4). This shift did not occur when the complex was incubated with nonimmune serum or a nonspecific antibody (Fig. 2B, lanes 1–3). Therefore, the siRNA-bound species observed in the glycerol gradient peak corresponds to hDcr-containing Complex D. We estimated the size of Complex D as 250–300 kDa based on comparison with the sedimentation behavior of the U1 snRNP, which has a known molecular weight of 240 kDa (Stark et al. 2001).
FIGURE 2.
Partially purified D complex contains hDcr. (A) HEK 293 S100 extract was incubated with radiolabeled siRNA and sedimented on a 10%–30% glycerol gradient. Fractions were collected from the bottom of the gradient and analyzed by native gel electrophoresis. Complex D peaked in fractions 6–10. (B) As in Figure 1B, except that the Complex D-containing fraction from A was substituted for S100 extract. Antibodies against cyclin T1 (“α-CycT1”) were used as a negative control.
hDcr directly contacts the siRNA within Complex D
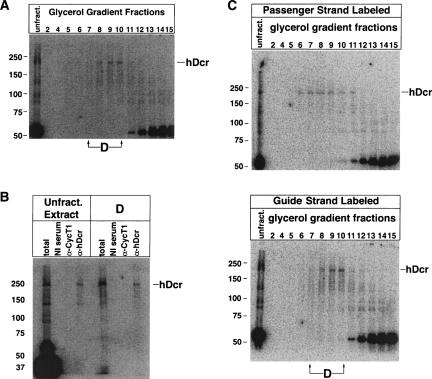
Since hDcr and the siRNA coexist within the D complex, we tested whether the siRNA directly contacts the hDcr protein by crosslinking with a radiolabeled siRNA containing a photo-activatable 5-iodouracil at its 3′ end. This siRNA was incubated with extract, exposed to 312-nm light, and fractionated by 10%–30% glycerol gradient sedimentation. We analyzed fractions by native gel and SDS-PAGE, and observed a radiolabeled protein of ~220 kDa that cofractionated with Complex D (Fig. 3A). This protein was specifically immunoprecipitated with an α-hDcr antibody both from irradiated crude extract and from irradiated Complex D-containing gradient fractions (Fig. 3B), indicating that it is hDcr. Interestingly, hDcr was photo-crosslinked with equal or slightly greater intensity when the guide strand contained the 5-iodouracil at its 3′ end, as opposed to when the passenger strand contained the 5-iodouracil at its 3′ end (Fig. 3C). This is contrary to results obtained in Drosophila (Tomari et al. 2004b), where Dcr-2 crosslinks much more efficiently to the 3′ end of the passenger strand. This suggests that siRNA asymmetry might not be sensed identically in humans and Drosophila.
FIGURE 3.
hDcr directly contacts the siRNA within Complex D. (A) Labeled siRNA containing a 5-iodouracil at position 20 was incubated in extract and irradiated with 312-nm light. The crosslinked mixture was sedimented on a 10%–30% glycerol gradient, and fractions were separated on a 6% polyacrylamide-SDS gel. (B) Crosslinked extract or Complex D-containing fractions from A were added to Protein-A sepharose beads bound with nonimmune serum (“NI serum”) or antibodies against cyclin T1 or hDcr. The immunoprecipitated samples were denatured and analyzed by electrophoresis on a 6% polyacrylamide-SDS gel. (C) Radiolabeled siRNA duplex containing a 5-iodouracil on either the guide or passenger strand was incubated with extract, crosslinked with 312-nm light, and sedimented on a 10%–30% glycerol gradient. Fractions were collected and analyzed by electrophoresis in 6% polyacrylamide-SDS gels. The labeled strand (containing the 5-iodouracil) is indicated at the top of each panel, and the Complex D-containing fractions are indicated below. The mobility of hDcr is given on the right.
Recombinant human Dicer binds siRNA
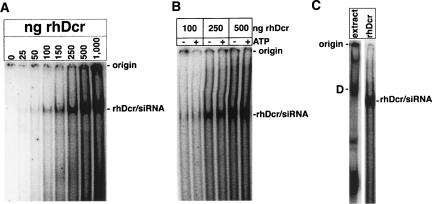
Purified recombinant hDcr (~220 kDa) sediments more slowly than endogenous Dicer from mouse extracts (Zhang et al. 2004), suggesting that most hDicer stably associates with one or more additional proteins (probably TRBP; Chendrimada et al. 2005; Haase et al. 2005). Our estimation that Complex D is ~250–300 kDa (see above) is consistent with this. To further test the possibility that the D complex contains additional proteins besides hDcr, we compared its electrophoretic mobility to that of siRNA-bound purified recombinant Dicer (rhDcr) in native gels. As shown in Figure 4A, we were able to detect rhDcr binding to the radiolabeled siRNA, contrary to earlier reports (Provost et al. 2002; Chendrimada et al. 2005). The siRNA-binding activity of rhDcr was ATP-independent (Fig. 4B), in contrast to the siRNA-binding activity of endogenous hDcr within Complex D (Fig. 1A). This rhDcr/siRNA complex migrated faster than Complex D on a native gel (Fig. 4C), consistent with the possibility that the endogenous complex contains one or more factors in addition to hDcr.
FIGURE 4.
Recombinant hDcr binds siRNA. (A) Increasing amounts of rhDcr protein were incubated with radiolabeled siRNA and analyzed by native gel electrophoresis. The amount of rhDcr is given at the top of each lane. (B) As in A, except that 0.5 mM ATP and an ATP-regenerating system were either omitted or included, as indicated at the top of each lane. (C) Radiolabeled siRNA was incubated with either HEK 293 S100 extract or 250 ng rhDcr and analyzed on a native gel.
Is Dicer involved in RISC assembly in mammals?
A significant question is whether Complex D is a true functional intermediate in RISC assembly in humans. This would usually be assessed by preforming the complex and challenging it with a large excess of competitor substrate, and then testing its ability to progress into a mature, functional complex upon the addition of other required components. However, despite exhaustive attempts, we have been unable to conduct such a “chase” experiment with Complex D, because control experiments indicate that the siRNA off-rate is too high to allow the complex to survive the challenge with a large amount of competitor siRNA.
Several published reports are consistent with roles for Dicer in RISC assembly in humans. Following dsRNA processing by rhDcr, the enzyme remains associated with some of the siRNA product, and product inhibition could account for rhDcr’s low catalytic efficiency under multiple turnover conditions (Zhang et al. 2002). Furthermore, HeLa cells depleted of either hDcr or its recently identified binding partner TRBP show a decreased RNAi response, even when triggered by siRNAs (Doi et al. 2003; Chendrimada et al. 2005; Haase et al. 2005). Three recent reports demonstrated that hDcr substrates are more efficient than siRNAs at triggering RISC activity in human cells (Kim et al. 2005; Rose et al. 2005; Siolas et al. 2005), consistent with the possibility that Dicer processing potentiates RISC assembly. Finally, Dcr-2 plays a role in RISC assembly in Drosophila (Lee et al. 2004; Pham et al. 2004; Tomari et al. 2004a), and a lack of involvement for hDcr in human RISC assembly would indicate a dramatic and unexpected difference between the otherwise well conserved RNAi pathways in these two organisms.
Although numerous reports suggest that Dicer may be involved in mammalian RISC assembly, several lines of evidence indicate that it is not strictly required. In contrast to in vivo results in human cells (Doi et al. 2003; Chendrimada et al. 2005; Haase et al. 2005), HeLa extract immuno-depleted of hDcr retained full siRNA-triggered RISC activity (Martinez et al. 2002). Additionally, Dicer-null mouse embryonic stem cells were capable of siRNA-mediated RISC activity (Kanellopoulou et al. 2005). Although these data indicate that RISC is capable of assembling in the absence of hDcr, they do not exclude the possibility that hDcr normally plays a role in enhancing RISC assembly and function.
In addition to identifying a Dicer/siRNA complex in human cell extract, we have presented evidence that purified rhDcr alone is capable of binding to siRNA. This binding activity is contrary to previous reports (Provost et al. 2002; Chendrimada et al. 2005). The rhDcr/siRNA complex migrates faster on a native gel than Complex D, suggesting that the endogenous complex contains one or more additional factors. It is likely that an additional factor is TRBP (Chendrimada et al. 2005; Haase et al. 2005), although TRBP is not absolutely required for Dicer/siRNA binding, as appears to be the case for Dicer-2 and R2D2 in Drosophila. It is possible that Complex D can function as a precursor in RISC assembly, analogous to a similar complex observed in Drosophila.
Most Dicers (including hDcr and Drosophila Dcr-2) contain an apparent N-terminal ATPase domain (Carmell and Hannon 2004), but the function of this domain has been enigmatic. Missense mutations in the Dcr-2 ATPase domain block dsRNA processing activity but have no effect on subsequent phases of RNAi (Lee et al. 2004). Furthermore, ATP is required for efficient dsRNA cleavage by Drosophila extract (Bernstein et al. 2001; Nykänen et al. 2001) and by recombinant Dcr-2 (Liu et al. 2003). In contrast, ATP depletion (Provost et al. 2002; Zhang et al. 2002) or a mutation predicted to block ATP binding (Zhang et al. 2002) has no effect on dsRNA processing by rhDcr. Thus, ATP appears to be required for dsRNA processing in insects but not mammals. In the case of siRNA binding activity, the situation is reversed: Drosophila Dcr-2/R2D2 does not require ATP for siRNA association (Liu et al. 2003; Pham et al. 2004), whereas endogenous hDcr/siRNA interaction within Complex D does require ATP (Fig. 1A). Although the D complex’s ATP requirement cannot yet be ascribed to ATP binding or hydrolysis by the hDcr N-terminal domain, these results suggest that this domain is likely to play different roles in different Dicer enzymes. Interestingly, the siRNA binding activity of purified rhDcr (Fig. 4B) indicates that hDcr/siRNA interactions are not intrinsically ATP-dependent, and suggests that additional factors present in human cell lysates may enforce Complex D’s ATP dependence. Uncovering the basis for this effect will illuminate the roles of Dicer and ATP in human RNAi.
MATERIALS AND METHODS
General methods
HEK 293 and HeLa S100 extracts were prepared as previously described (Dignam et al. 1983) except the final dialysis buffer contained 10% glycerol. The Pp-luc siRNA sequences used in this study were previously described in Nykänen et al. (2001). The siRNA was labeled either at the 3′ end of the antisense strand with an [α32P]-cordycepin 5′-triphosphate (Perkin Elmer, 5000 Ci/mmol) using poly(A) polymerase (Amersham), or on the 5′ end of either strand with [γ32P]-ATP (MP Biomedicals, 7000 Ci/mmol) using T4 polynucleotide kinase (New England Biolabs) according to the manufacturers’ instructions. Duplex siRNAs were annealed as described in Pham et al. (2004).
For all experiments outlined in this study, RNAi reactions contained 75% extract (vol/vol), 3 mM MgOAc, 2.5 mM DTT, 0.5 mM ATP, 12.5 mM creatine phosphate, 0.015 μg/mL creatine kinase (Calbiochem), 4.5 units RNAguard (Amersham), and 5000–10,000 cpm siRNA duplex. The reactions were incubated at 37°C for 1 h, unless otherwise specified. Standard RNAi reactions were 10 μL. For ATP depletion experiments, ATP, creatine phosphate, and creatine kinase were omitted and replaced with 10 mM glucose and 0.05 U/mL hexokinase (Calbiochem). The ATP depletion reactions were incubated at 37°C for 15 min before addition of siRNA. In the rhDcr experiments, the extract was removed and replaced with 1 μL purified rhDcr-HisC (Zhang et al. 2002) and 6.5 μL lysis buffer. For the rhDcr experiments lacking ATP, creatine phosphate, creatine kinase, and ATP were omitted from the reaction and replaced with water. RISC activity was assayed as described in Pham et al. (2004).
Native gel electrophoresis
Reactions to be run on a native gel were supplemented with 10% (vol/vol) glycerol before incubation at 37°C. The native gel system was as described in Pham et al. (2004). No heparin was added to the reactions. For the antibody supershift assay, RNAi reactions (or reactions with partially purified Complex D in place of extract) were incubated at 37°C for 30 min, after which nonimmune serum or antibodies against protein kinase C (PKC), cyclin T1 (CycT1), or hDcr (D347, as described in Billy et al. 2001) were added. The mixtures were incubated at 4°C for 5 h, then analyzed by native gel electrophoresis.
Glycerol gradient sedimentation
An RNAi reaction (400 μL, 400,000 cpm) was layered on a 10%–30% glycerol gradient in Buffer A (Zhang et al. 2004). The gradient was centrifuged at 36,000 rpm for 24 h at 4°C in a Beckman SW 41 Ti rotor. Fractions (~550 μL) were collected by dripping from the bottom of the tube.
Photo-crosslinking
Pp-luc siRNA containing a 5-iodouracil at position 20 of one strand was 5′-end-labeled and annealed as described above. This siRNA was used in an RNAi reaction, then placed on an inverted 96-well plate in ice, covered with a Petri dish lid to minimize exposure to shortwave UV light, and irradiated with a 312-nm lamp (15 W, 1000 μW/cm2) for 10 min. For the immunoprecipitations, Protein A-sepharose beads (24 mg) were rinsed with a wash buffer (20 mM Tris-HCl pH 7.5, 200 mM NaCl, 0.5% Igepal, 2.5 mM MgCl2, Protease Inhibitors-EDTA [Roche]), blocked in a 500 μL mixture of BSA, glycogen, and carrier tRNA (10 μg/mL each in wash buffer) for 15 min at 4°C, and washed three times. Nonimmune human serum or antibodies against either CycT1 or hDcr were added and the beads were gently rocked at room temperature for 1 h, then washed 3 × 500 μL with wash buffer. A crosslinking reaction or Complex D-containing fraction from a crosslinked gradient were added to the beads with 300 μL wash buffer, gently rocked for 4 h at 4°C, and washed 4 × 200 μL with wash buffer. The samples were eluted with 1× SDS-PAGE sample buffer, heated at 100°C for 10 min, then loaded onto a 6% SDS-PAGE gel.
Acknowledgments
We thank all the members of the Sontheimer lab and A. Haase for their advice and support, and J. Widom for sharing his ultracentrifuge rotor. This work was supported by an NIH Cellular and Molecular Basis of Disease Training Grant to J.L.P. and an American Cancer Society Research Scholar Grant to E.J.S.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2102805.
REFERENCES
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Billy, E., Brondani, V., Zhang, H., Muller, U., and Filipowicz, W. 2001. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. 98: 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell, M.A. and Hannon, G.J. 2004. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 11: 214–218. [DOI] [PubMed] [Google Scholar]
- Chendrimada, T.P., Gregory, R.I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K., and Shiekhattar, R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, N., Zenno, S., Ueda, R., Ohki-Hamazaki, H., Ui-Tei, K., and Saigo, K. 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 13: 41–46. [DOI] [PubMed] [Google Scholar]
- Filipowicz, W. 2005. RNAi: The nuts and bolts of the RISC Machine. Cell 122: 17–20. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Haase, A., Jaskiewicz, L., Zhang, H., Laine, S., Sack, R., Gatignol, A., and Filipowicz, W. 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. http://www.nature.com/embor/journal/vaop/ncurrent/full/7400509.html#abstract [DOI] [PMC free article] [PubMed]
- Kanellopoulou, C., Muljo, S.A., Kung, A.L., Ganesan, S., Drapkin, R., Jenuwein, T., Livingston, D.M., and Rajewsky, K. 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & Dev. 19: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., and Jayasena, S.D. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Behlke, M.A., Rose, S.D., Chang, M.S., Choi, S., and Rossi, J.J. 2005. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23: 222–226. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., and Wang, X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Meister, G. and Tuschl, T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15: 185–197. [DOI] [PubMed] [Google Scholar]
- Nykänen, A., Haley, B., and Zamore, P.D. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321. [DOI] [PubMed] [Google Scholar]
- Pham, J.W., Pellino, J.L., Lee, Y.S., Carthew, R.W., and Sontheimer, E.J. 2004. A Dicer-2-dependent 80S complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94. [DOI] [PubMed] [Google Scholar]
- Provost, P., Dishart, D., Doucet, J., Frendewey, D., Samuelsson, B., and Radmark, O. 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21: 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W.S., and Khvorova, A. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22: 326–330. [DOI] [PubMed] [Google Scholar]
- Rivas, F.V., Tolia, N.H., Song, J.J., Aragon, J.P., Liu, J., Hannon, G.J., and Joshua-Tor, L. 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12: 340–349. [DOI] [PubMed] [Google Scholar]
- Rose, S.D., Kim, D.H., Amarzguioui, M., Heidel, J.D., Collingwood, M.A., Davis, M.E., Rossi, J.J., and Behlke, M.A. 2005. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 33: 4140–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208. [DOI] [PubMed] [Google Scholar]
- Siolas, D., Lerner, C., Burchard, J., Ge, W., Linsley, P.S., Paddison, P.J., Hannon, G.J., and Cleary, M.A. 2005. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 23: 227–231. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Smith, S.K., Hannon, G.J., and Joshua-Tor, L. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J. 2005. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell. Biol. 6: 127–138. [DOI] [PubMed] [Google Scholar]
- Stark, H., Dube, P., Luhrmann, R., and Kastner, B. 2001. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409: 539–542. [DOI] [PubMed] [Google Scholar]
- Tomari, Y. and Zamore, P.D. 2005. Perspective: Machines for RNAi. Genes & Dev. 19: 517–529. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Du, T., Haley, B., Schwarz, D.S., Bennett, R., Cook, H.A., Koppetsch, B.S., Theurkauf, W.E., and Zamore, P.D. 2004a. RISCassembly defects in the Drosophila RNAimutant armitage. Cell 116: 831–841. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Matranga, C., Haley, B., Martinez, N., and Zamore, P.D. 2004b. A protein sensor for siRNA asymmetry. Science 306: 1377–1380. [DOI] [PubMed] [Google Scholar]
- Yuan, Y.-R., Pei, Y., Ma, J.-B., Kuryavyi, V., Zhadina, M., Meister, G., Chen, H.-Y., Dauter, Z., Tuschl, T., and Patel, D.J. 2005. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 19: 405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Kolb, F.A., Brondani, V., Billy, E., and Filipowicz, W. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21: 5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Kolb, F.A., Jaskiewicz, L., Westhof, E., and Filipowicz, W. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68. [DOI] [PubMed] [Google Scholar]