Abstract
MicroRNAs are sequence-specific regulators of post-transcriptional gene expression in many eukaryotes. They are believed to control the expression of thousands of target mRNAs, with each mRNA believed to be targeted by multiple microRNAs. Recent studies have uncovered various mechanisms by which microRNAs down-regulate their target mRNAs and have linked a well-known subcellular structure, the cytoplasmic processing bodies (PBs) to the microRNA pathway. The finding that microRNAs are misexpressed in cancers has reinforced the idea that their regulatory roles are very important.
Keywords: microRNAs, miRNPs, translational repression, P-bodies, Argonaute
INTRODUCTION
MicroRNAs (miRNAs) have come a long way from being an oddity of worms when they were first discovered over 10 years ago (Lee et al. 1993) to be recognized now as novel agents exercising post-transcriptional control over most eukaryotic genomes. They are a family of 21–25-nucleotides (nt)-long RNAs expressed in a wide variety of organisms ranging from plants to worms and humans. Many miRNAs are highly conserved across species (Bartel 2004), and components of the miRNA machinery are even found in archaea and eubacteria, revealing their very ancient ancestry. With the latest human miRNA count swelling to over 800, miRNAs easily account for >3% of all human genes (Bentwich et al. 2005). In worms, they were initially found to regulate developmental timing (Wightman et al. 1993), but now we know that they are involved in a wide variety of developmental processes and might have a role in networking and fine-tuning gene expression in the cell (Ambros 2003; Bartel 2004). With the number of predicted miRNA targets running into the thousands, miRNAs might very well form another layer of the regulatory circuitry that exists in the cell (Bartel and Chen 2004). Thus, any mis-regulation of the miRNAs could lead to great regulatory upheavals in the cell, perhaps leading to cancerous phenotypes. In fact, it was shown that the miRNA profiles are changed in a large number of cancers (Lu et al. 2005) and that the forced overexpression of miRNAs can lead to the development of tumors (He et al. 2005).
Another pathway that uses small RNAs as sequence-specific regulators is the RNA interference (RNAi) pathway, which is an evolutionarily conserved response to the presence of double-stranded RNA (dsRNA) in the cell (Meister and Tuschl 2004; Filipowicz 2005). The dsRNAs are cleaved into ~20-base pair (bp) duplexes of small-interfering RNAs (siRNAs) by Dicer. These small RNAs get assembled into multiprotein effector complexes called RNA-induced silencing complexes (RISCs). The siRNAs then guide the cleavage of target mRNAs with perfect complementarity. The finding that some aspects of biogenesis, protein complexes, and function are shared between siRNAs and miRNAs has greatly accelerated the study of miRNAs. Recent findings that improve our understanding of how miRNAs function are the subject of this review.
Biogenesis of miRNAs
MiRNAs are transcribed from different genomic locations as long primary transcripts (pri-miRNA) by RNA polymerase II (Kim 2005). They are embedded in either independent noncoding RNAs or the introns of protein-coding genes. Additionally, to allow coordinated expression, some miRNAs are clustered in polycistronic transcripts. Several miRNAs are expressed in a tissue-specific and developmental stage-specific manner. The expression of the intron-encoded miRNAs might be linked to the transcriptional regulation of their host gene promoters (Bartel 2004). A role for transcription factors like SRF (serum response factor), MyoD, and Mef2 in specifying cardiac tissue-specific expression of miR-1 (Zhao et al. 2005) and Myc in regulating a specific miRNA cluster (O’Donnell et al. 2005) was recently demonstrated. Although we know of only transcriptional regulation of miRNA genes, regulation at the level of miRNA processing remains a possibility.
Subsequent to transcription, the pri-miRNAs are processed by the successive action of two members of the RNase-III family of enzymes, Drosha and Dicer. In performing their tasks, these proteins are aided by their companion double-stranded RNA-binding domain (dsRBD)-containing proteins: Pasha functioning with Drosha, R2D2 with Drosophila Dicer-2 (Kim 2005), Loqs with Drosophila Dicer-1 (Forstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005), and TRBP with human Dicer (Chendrimada et al. 2005; Haase et al. 2005). An ~70-nt precursor called the pre-miRNA, which can be folded into a stem-loop structure containing multiple bulges and mismatches, is excised out from the pri-miRNA by the nuclear Drosha. The pre-miRNA is exported out to the cytoplasm by Exportin-5, in a Ran-GTP-dependent manner. In the cytoplasm, the pre-miRNA is cleaved by Dicer to generate an ~20-bp duplex intermediate (Sontheimer 2005). In keeping with the thermodynamic asymmetry rule, only one strand of the duplex accumulates as the mature miRNA. The asymmetry rule dictates that the 5′ end of the mature miRNA lies at the end of the duplex which has lower thermodynamic energy (Tomari and Zamore 2005). The mature miRNA then gets assembled into the effector complexes called miRNPs (miRNA-containing ribonucleo-protein particles) that share a lot of similarity to the RISC. Functional RISCs and miRNPs vary in size, ranging from the “minimal” RISC of ~160 kDa up to the holo RISC that fractionates at ~80S; human miRNAs were found in complexes of ~15S. Biochemical characterization of these different complexes has led to the identification of a number of protein components, among them Gemin3, Gemin4, vasa intronic gene (VIG), Fragile X mental retardation protein (FMRP), and a staphylococcal nuclease, Tudor-SN (Sontheimer 2005). The function of these proteins in the RNAi and miRNA pathways is not clear. However, the only protein consistently found in the RISC and miRNP complex is the highly conserved Argonaute (Ago) (Carmell et al. 2002). The general consensus is that the minimal effector complex contains a single-stranded small RNA associated with an Ago protein.
Once the miRNP is assembled, the miRNA guides the complex to its target by base-pairing with the target mRNA. In plants, miRNAs bind to a single, generally perfectly complementary site in either the coding or 3′-untranslated regions (UTRs) of the target mRNA. In contrast, most investigated animal miRNAs bind to multiple, partially complementary sites in the 3′-UTRs. However, the target sequences inserted into either coding or 5′-UTR sequences are also functional (Kloosterman et al. 2004). The complementarity is usually restricted to the nucleotides 2–8 in the 5′ end of the miRNA. These nucleotides form the “seed” sequence, implying that they nucleate binding between the miRNA and the target mRNA (Tomari and Zamore 2005). The fate of the target mRNA is decided by the extent of base-pairing to the miRNA. A miRNA will direct destruction of the target mRNA if it has perfect or near-perfect complementarity to the target (Hutvagner and Zamore 2002). On the other hand, the presence of multiple, partially complementary sites in the target mRNA will direct the inhibition of protein accumulation without strongly affecting mRNA levels (Bartel 2004). New findings discussed in this review suggest, however, that even in the latter situation, miRNAs can lead to the strong decrease in mRNA levels.
Modes of miRNA action
Human Ago2 as the mediator of mRNA cleavage in the RISC/miRNP
Both the siRNAs and miRNAs, when perfectly base-paired to their target mRNA, direct cleavage of a single phosphodiester bond in the target mRNA. The severed bond lies between the residues paired to siRNA nucleotides 10 and 11 (counting from the siRNA 5′ end) (Elbashir et al. 2001). This cleavage is a result of the “Slicer” activity in the RISC. Argonautes form the core of the RISC and are highly conserved ~100-kDa proteins with members present even in archaea and eubacteria. They contain the signature PAZ and PIWI domains (Carmell et al. 2002). Insight into the function of Ago proteins was obtained in structural studies revealing that the Argonaute PAZ domain has an oligonucleotide-binding fold that anchors the single-stranded 3′ end of small RNAs (Lingel and Sattler 2005). Most importantly, the PIWI domain of two archaeal proteins was shown to have a fold similar to that of RNase H (Parker et al. 2004; Song et al. 2004), an enzyme that cleaves the RNA strand in DNA–RNA hybrids. This immediately suggested that the Ago protein could harbor the mRNA-cleaving “Slicer” activity. Further information regarding recognition of the small RNAs came from co-crystal structures of an archaeal Ago protein with siRNA mimics, which revealed that the PIWI domain has a conserved binding pocket for the 5′ phosphate of small RNAs (Parker et al. 2005; Ma et al. 2005). These studies suggest a model in which the small RNA is wedged between the PAZ and PIWI domains of the Ago protein, positioning the target mRNA scissile bond in close proximity to the catalytic center.
Although all human Ago proteins bind both miRNAs and siRNAs, only Ago2-containing complexes support mRNA cleavage (Liu et al. 2004; Meister et al. 2004). Indeed, mutation analysis of Ago2 confirmed the catalytic role of a set of three DDH amino acids in the PIWI domain that are related to the DDE amino acids of RNase H involved in catalysis (Liu et al. 2004; Rivas et al. 2005). The finding that bacterially expressed Ago2 protein, when complexed with a single-stranded siRNA, can direct mRNA cleavage has proven conclusively that in humans Ago2 is the only protein needed for this activity (Rivas et al. 2005). The mammalian miR196 has a near-perfect complementary sequence in the 3′-UTR of the Hoxb8 mRNA and leads to direct mRNA cleavage and degradation of the target mRNA (Yekta et al. 2004). All the other mammalian miRNAs are believed to base-pair to their targets at partially complementary sites and inhibit protein accumulation.
Inhibition of protein synthesis
Early studies on miRNA-mediated repression of protein synthesis were performed in Caenorhabditis elegans with the miRNA lin-4 (Lee et al. 1993) and its target lin-14. Lin-4 base-pairs with the lin-14 mRNA with incomplete complementarity at multiple locations in the 3′-UTR, and inhibits protein synthesis (Wightman et al. 1993). A similar effect on protein accumulation was also demonstrated for other miRNAs in worms, flies, and humans (Reinhart et al. 2000; Brennecke et al. 2003; Zeng et al. 2003). Reporter constructs having multiple, partially complementary binding sites for transfected siRNAs were also shown to be repressed at the protein level (Doench et al. 2003; Zeng et al. 2003), reinforcing the idea that complementarity is the key factor that decides the fate of the target message. In all of the studies described above, the effect on protein accumulation is not accompanied by a major change in mRNA levels.
Artificially tethering human Ago proteins in a miRNA-independent manner to the 3′-UTR of a reporter mRNA inhibited protein synthesis in HeLa cells, without affecting reporter mRNA levels (Pillai et al. 2004). Tethering of the Drosophila Ago1 (dAgo1) protein to a reporter mRNA also inhibited the expression of the reporter (Rehwinkel et al. 2005). It not known whether the tethered Ago protein needs to be complexed with the miRNA. Since the immunoprecipitated, tagged human Ago2 used in the tethering assay was capable of supporting let-7-directed cleavage of a substrate RNA (Pillai et al. 2004), it is possible that tethering brings the whole miRNP complex to the reporter mRNA. These experiments demonstrated that miRNAs act as guides for their associated protein factors, and that the default effect of having the miRNP complex on reporter mRNAs is inhibition of protein synthesis.
Studies in worms on the mechanism of lin-4 action revealed that the repressed lin-14 mRNA was associated with polysomes (Olsen and Ambros 1999). A similar conclusion was reached in the analysis of lin-28 mRNA, another target of lin-4 (Seggerson et al. 2002). It was proposed that translation may be blocked at a step after initiation, for example, by the stalling of ribosomes on the message or by inducing proteolysis of the nascent polypeptides as soon as they exited the ribosome. Both models were not proved experimentally. Later, mammalian miRNAs and miRNP components were shown to be associated with polyribosomes (Krichevsky et al. 2003; Kim et al. 2004; Nelson et al. 2004). It was suggested that mammalian mi-RNAs also regulate translation of their target mRNAs by a mechanism similar to that suggested for lin-4 in worms. In these studies, however, the majority of the miRNAs and miRNP proteins were found in the top of the gradient, where the mRNPs sediment.
Targeted delivery of a protein product to the endoplasmic reticulum (ER) lumen should make it inaccessible to proteolysis by excluding it from the cytosol. However, experiments in HeLa cells revealed that ER targeting did not relieve the repression caused by both endogenous let-7 miRNA and tethered human Ago2. This suggested that translation per se might be the target of inhibition. In fact, polysome analysis revealed that let-7 miRNA inhibited translation initiation of a reporter mRNA in mammalian cells. A similar result was obtained with an independent method of repression mediated by tethered Ago proteins (Pillai et al. 2005). Experiments done to pinpoint the step of translation initiation that is affected by let-7 led to the conclusion that only cap-dependent translation is subject to repression. Translation initiated in a cap-independent manner by viral internal ribosome entry site (IRES) elements or by tethered initiation factors eIF4E and eIF4G (De Gregorio et al. 2001) was immune to repression by let-7 miRNA (Pillai et al. 2005). This suggested that the point of intervention lay upstream of the eIF4G recruitment via eIF4E. The molecular target of let-7 miRNP could be the m 7G cap structure itself, and the miRNP acts probably by interfering with its recognition by eIF4E. The immediate question that needs to be addressed is how let-7 miRNP can recognize the cap structure. A precedent for a 3′-UTR-bound factor affecting cap function exists in Drosophila (Cho et al. 2005 and refs. therein.). Additionally, the finding that in vitro synthesized reporter mRNAs, lacking any “nuclear experience,” were also repressed efficiently when transfected into HeLa cells demonstrated that unlike other translation-dependent post-transcriptional regulatory mechanisms like nonsense-mediated decay (NMD), nuclear history is not important for miRNA-mediated translation repression.
Thus, the translational fate of the target mRNA appears to be different in worms and humans. Perhaps this could be because of the different miRNAs studied or because miRNA-mediated inhibition of protein synthesis in worms and humans takes different routes. However, the presence of miRNAs on polysomes suggests that perhaps in mammals too, different mechanisms might be in operation. There is a report of the endogenous human lin-28 mRNA that is targeted by let-7 being associated with polysomes (Nelson et al. 2004). To gain a better understanding, the polysome analysis must be extended to other endogenous messages.
Role of PBs in the sequestration of miRNA target mRNAs
A major part of bulk mRNA degradation occurs via deadenylation followed by decapping and 5′–3′ exonuclease digestion. Proteins required for this pathway such as decapping enzymes, Dcp1/2, and the 5′–3′ exonuclease Xrn1 are concentrated in specialized cytoplasmic processing bodies (PBs) (Bashkirov et al. 1997; Parker and Song 2004). These structures are also known as GW bodies because of the presence of the RNA-binding protein GW182 (Eystathioy et al. 2003). A number of post-transcriptional regulatory pathways have been linked to the PBs (Parker and Song 2004; Andrei et al. 2005). Recently, new information linked the PBs to the miRNA pathway by showing that all four human Ago proteins were found to be enriched in discrete foci that colocalized with PBs (Liu et al. 2005; Pillai et al. 2005; Sen and Blau 2005). This colocalization is consistent with the observed interaction of Ago protein with components of the PBs. Additionally, both miRNAs and target reporter mRNAs were also shown to accumulate in PBs. Reporter mRNAs targeted by transfected miRNA mimics were also shown to accumulate in PBs (Liu et al. 2005; Pillai et al. 2005). This raised the possibility that relocalization of the target mRNA could be a means to achieve repression as PBs lack ribosomes and initiation factors (Teixeira et al. 2005).
Recently, a protein AIN-1 (Ding et al. 2005), which is homologous to the mammalian GW182, was shown to be in a complex with Dicer, Ago protein ALG-1, and a subset of miRNAs. Like its mammalian homolog, AIN-1 is also localized to cytoplasmic foci, colocalizing with worm decapping enzymes, suggesting that these might be worm PBs. Interestingly, when expressed alone, ALG-1 remains in the cytosol, but coexpression with AIN-1 resulted in the Ago protein accumulating in discrete foci that could be PBs. Mammalian GW182 was also shown to bind a subset of mRNAs, and it was proposed that GW182 participates in the post-transcriptional regulation of these mRNAs by sequestering them (Eystathioy et al. 2002). Knock-down experiments in Drosophila cell lines demonstrated that depletion of this protein leads to derepression of miRNA-mediated gene silencing (Rehwinkel 2005). A similar effect of depletion of GW182 was also observed for repression mediated by the miRNA-independent tethering of dAgo1. The derepression observed with depletion of GW182 was lower than that observed with depletion of dAgo1 protein, suggesting an effect on the robustness of the repression process, assuming that both proteins were depleted to a similar extent.
What could be the role of GW182 in the miRNA pathway? Experiments with AIN-1 suggest a role for this protein in transporting miRNPs containing ALG-1 to worm PBs, by direct interaction with ALG-1 (Ding et al. 2005). Interestingly, GW182 is one of the few PB components that are required for PB integrity (Yang et al. 2004). It is also known that PBs require a constant flow of mRNA into these structures to maintain themselves, as drugs that freeze mRNAs on ribosomes abolish PB formation (Teixeira et al. 2005). Thus one could imagine GW182 as having a general transport role in the larger context of PB formation. Studies in yeast have linked defects in translation initiation to the formation of PBs, suggesting that these are dynamic structures that are formed out of a need to warehouse repressed mRNAs (Teixeira et al. 2005). The need for a transport function assumes that the PBs are static structures to which the repressed mRNPs must be delivered. On the contrary, PBs are very dynamic (Kedersha et al. 2005), and it is possible that the rapid and random dance of the PBs is enough to pick up the repressed mRNAs in their path, without the need for any specific transport event. It is believed that aggregation of individual mRNP complexes leads to the formation of visible, large PBs (Coller and Parker 2004). Thus, one can also imagine GW182 in promoting such an aggregation event. It remains to be seen whether the factors that control such movement might also regulate the repression process. Thus, is relocalization the mechanism for repression or is it a consequence of translational initiation block (Pillai et al. 2005)? The movement of repressed mRNAs to the PBs might contribute to the robustness of the translational repression, by maintaining the repressed state. Are PBs sites of miRNA function? Is there miRNA-mediated repression outside of PBs?
Degradation of target mRNA
Recently the long-held view that animal miRNAs that base-pair to their targets with incomplete complementarity do not affect mRNA levels has been questioned. Microarray analysis has revealed that mammalian miRNAs can affect the mRNA levels of a large number of transcripts in the cell (Lim et al. 2005). Many of the down-regulated mammalian miRNA targets had complementarity to the miRNAs only in the “seed” sequence. Thus, it is clear that the effect on mRNA level is unlikely to be mediated by the RISC cleavage pathway, as for cleavage to occur, an A-form helix must be formed between the miRNA and the target mRNA (Chiu and Rana 2003; Haley and Zamore 2004). The absence of any repression by let-7 miRNA on mRNAs with IRES-mediated translation argues against direct destabilization of the target message upon miRNA binding (Pillai et al. 2005). Another study concludes that in C. elegans, lin-4 and let-7 miRNAs act by affecting target mRNA stability (Bagga et al. 2005). Those investigators reported that the miRNAs induced degradation of their target mRNAs when recognizing them with incomplete complementarity. The lin-14 and lin-28 mRNAs that are targeted by lin-4 miRNA were previously shown to be repressed at the translational level (Olsen and Ambros 1999; Seggerson et al. 2002).
How to explain the changes in the steady-state level of target mRNAs? One possibility is that the effect on mRNA levels is a consequence of translational repression and subsequent relocalization to PBs. The finding that degradation products accumulated in response to knock-down of predicted worm 5′–3 ′ exonucleases suggested that these exo-nucleases were perhaps recruited to the target mRNA (Bagga et al. 2005). It has not gone unnoticed that the homologs of these exonucleases are enriched in mammalian PBs, already shown to be sites containing reporter miRNA targets. Additionally, an increased accumulation of what appeared to be full-length target mRNA in these knockdown strains suggested that they might be degraded by decapping and subsequent exonucleolytic digestion. Mapping of the cleavage sites on the target mRNA indicated cleavages to either side of the miRNA-binding region, indicating a possible protection afforded by bound miRNP complexes from advancing exonucleases. In addition, experiments in Drosophila cell lines suggest that depletion of the DCP1:DCP2 decapping complex leads to a significant derepression of a reporter mRNA undergoing miRNA-mediated inhibition. Again, the derepression was modest compared to that observed by depletion of dAgo1 protein, suggesting that the decapping complex has a secondary role in the repression process. It is not clear whether there was any effect on target mRNA levels in these studies (Rehwinkel et al. 2005). Interestingly, the reporter mRNA (Pillai et al. 2005) targeted by endogenous let-7 miRNA in HeLa cells also showed slightly reduced mRNA levels.
A screen in Drosophila for factors involved in the ARE(AU-rich element)-mediated mRNA decay pathway identified a role for members of the RNAi machinery like Argonautes and Dicer (Jing et al. 2005). That study revealed that miR-16 has complementarity to core ARE sequences and that this complementarity is required to maintain the negative effect of miR-16 on mRNA stability. Interestingly, this complementarity is in the central part of the miR-16, which goes against the current notion of the requirement for a “seed” region complementary to the 5′ end of the miRNA. What distinguishes the effect of miR-16 on ARE-containing mRNAs from the normal effect of miRNAs on mRNA stability is the requirement of an ARE-binding protein, tristetraprolin (TTP), for rapid turnover. TTP was shown to interact with both the ARE and Argonaute proteins. This would suggest that TTP is recruited to the ARE by miR-16. Does this mean that miRNAs can act as carrier platforms for specific proteins? There are other miRNAs that have complementarity to several classes of conserved sequences known to mediate negative post-transcriptional regulation (Lai 2002), and further investigation is needed to understand the role of miRNAs in these regulatory pathways.
Endogenous miRNAs acting on viruses
Earlier studies in plants and Drosophila demonstrated an anti-viral role for the RNAi pathway (Hamilton and Baulcombe 1999; Li et al. 2002), where the viral RNAs are used by the RNAi machinery to make small RNAs that in turn target the viral genome. A recent report demonstrated an anti-viral role for RNAi in vertebrates by showing that an endogenous microRNA, miR-32, can restrict the proliferation of a primate retrovirus, PFV-1 (primate foamy virus type 1), in human cells (Lecellier et al. 2005). The miRNA binds to the viral transcripts with imperfect complementarity at a single site and leads to translational repression. It is not clear which gene is being down-regulated, as the target sequence is found in the 3′-UTR common to all viral mRNAs. This raised the possibility that any fortuitous base-pairing of the different cellular miRNAs to any viral RNA could act to protect us from the invading viruses.
The hepatitis C virus (HCV) was shown to require binding of the liver-specific miRNA, miR-122 (Jopling et al. 2005), for efficient replication. The miRNA has a single conserved binding site in the 5′ noncoding region of the viral RNA, with the complementarity restricted to the seed sequence. Sequestration of miR-122 or mutations in the binding site for miR-122 on the viral RNA reduced viral RNA maintenance in cells. Direct binding of miR-122 to the HCV RNA was demonstrated by rescuing the viral mutants by expression of miR-122 with compensatory mutations that restored base-pairing. It is not clear how the virus benefits from this interaction; perhaps it aids in RNA folding or recruitment into replication complexes. Binding of miR-122 did not affect translation or mRNA stability of the RNA. Is this also a fortuitous base-pairing? If so, it is curious that HCV gets some help from a liver-specific miRNA and not any other “housekeeping” miRNA.
Translational repression, degradation, or both?
The findings described above raise a pertinent question as to the contribution of translational repression and mRNA degradation in the observed reduction in protein levels. Clearly, in the case of worm miRNAs lin-4 and let-7, target mRNA degradation can account for the in vivo reduction in protein output (Bagga et al. 2005). However, in vitro experiments using a fragment of C. elegans lin-41 mRNA containing the let-7 complementary sites demonstrated that they are not cleaved by human let-7-programmed RISC activity (Hutvagner and Zamore 2002). Also, there are many documented examples where either target mRNA levels were not affected or the decrease in mRNA level alone could not account for the observed reduction in protein accumulation. There are also examples of endogenous mammalian mRNAs that are regulated by miRNAs at the level of translation without change in target mRNA levels (Poy et al. 2004; Cimmino et al. 2005). In this context, one can view translational repression as the primary event, with any reduction in mRNA levels being a consequence of translational repression and subsequent relocalization to PBs.
The PBs were initially shown to be sites of mRNA decay based on the finding that mRNA degradation intermediates accumulated in these structures (Sheth and Parker 2003). Further evidence linking the decapping activity in the cell to the size and number of PBs strengthened such a conclusion (Parker and Song 2004). This raises the question of whether the PBs are also sites of degradation of miRNA targets. Messages targeted for degradation by siRNAs are endonucleolytically cleaved by the “Slicer” activity of Ago protein, and the 5′ and 3′ cleavage fragments thus generated are degraded by the exosome and Xrn1, respectively (Orban and Izaurralde 2005). Such a pathway might also be true for mRNA targets with perfect or near-perfect complementarity to miRNAs. Although the cleavage fragments from such a reaction could be degraded in the PBs, only Xrn1 is known to be a component of PBs. However, a reporter mRNA undergoing miRNA-mediated cleavage was not detected in the PBs (Pillai et al. 2005), suggesting that such an activity of miRNAs may not be occurring in the PBs. Finally, the presence in PBs of human Ago proteins lacking cleavage activity and reporter mRNAs targeted for translational repression hints at another role for these structures in the miRNA pathway.
The finding that in Saccharomyces cerevisiae mRNAs that are repressed at the translation initiation step are sequestered in PBs and can exit the PBs to re-enter translation suggests that apart from their established role in mRNA decay, these structures can also act as sites for storage of repressed mRNAs (Brengues et al. 2005). It is important to note that release of repression and re-entry into translation from PBs has never been demonstrated for a miRNA target. Also, the very fact that miRNA-repressed mRNAs are detectable in the PBs suggests that these are not being actively degraded, as visualization of mRNA undergoing degradation in PBs required the depletion of essential proteins in the decay pathway to stabilize the decay intermediates (Sheth and Parker 2003). The finding that foci containing repressed mRNA and miRNAs do not always perfectly overlap with PBs but are sometimes found adjacent to them (Pillai et al. 2005) allows one to speculate that there might be some sort of compartmentalization within these structures for their roles in storage and mRNA decay.
Possible models of miRNA function
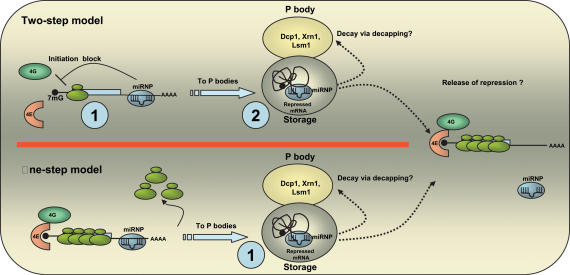
In the context of PBs, one can visualize two possible models to explain the mechanism of miRNA action. The one-step model (Fig. 1) envisages a scenario where the binding of the miRNP to the target mRNA would create a targeting signal that directs the mRNP:miRNP complex to the PBs. This could be viewed as an act of condemning them for either degradation (Bagga et al. 2005) or translational repression, as PBs lack ribosomes and translation factors (Teixeira et al. 2005). Prior to entry into the PBs the mRNAs must be cleared of the ribosomes and other translation factors. The alternative two-step model views the localization to PBs as a consequence of translational repression. It regards translational initiation block by miRNA as a primary event, with the relocalization to the PBs coming later as a means to store the mRNP. It is believed that although the translational repression machinery inhibits protein synthesis, the subsequent sequestration in PBs is what might further enforce this repression due to its translation-incompetent environment. It is difficult to fit into this model the data on the presence of miRNAs on polysomes, as any mechanism that retains messages on polysomes goes against the idea of PB formation. It remains a possibility that the presence of miRNAs/mRNAs on polysomes might reflect a state where mRNAs that are being actively translated are in the process of being engaged by these miRNAs for repression. Alternatively, these miRNAs might be in large aggregates that sediment in the range of polysomes. However, in the absence of data supporting any of these possibilities, an effect on other steps of translation or proteolysis of nascent polypeptides should be entertained in some situations.
FIGURE 1.
Models of miRNA function. The two-step model: Upon binding to the 3′-UTR of a target mRNA, the miRNP complex inhibits translation initiation, presumably by interfering with the 7mG cap function in recruiting eIF4E. Next, the repressed mRNA:miRNP complex is moved to the P-bodies (PBs) for storage. It is speculated that there are separate compartments for storage of the repressed mRNA and for the core PB components such as Dcp1, Xrn1, etc. It is possible that a part of the stored mRNA is degraded via decapping or that once repression is released, they exit the PBs and re-enter translation. One-step model: Binding of the miRNP to the target mRNA directs the mRNA to the PBs. Disassembly of ribosomes on the message occurs prior to entry into the PBs. The mRNA can then be either degraded or stored for later reuse. 7mG, 7-methyl guanine; AAAA, polyadenosine tail; 4E, eIF4E; 4G, eIF4G.
Although both of the models of miRNA function need to be tested, available data might support the two-step model. The finding that Ago2 with mutations in the PAZ domain and thus lacking miRNA-binding activity did not localize to the PBs (Liu et al. 2005) suggests that Ago proteins do not localize to PBs on their own but might do so in the context of a complex with miRNAs and perhaps with associated repressed mRNA. Additionally, the experiments with dicis-tronic constructs where the first cistron was repressed by let-7 while the second cistron was still being translated would suggest that miRNA-mediated translational repression was indeed occurring outside the PBs (Pillai et al. 2005). Both models allow for the hypothetical possibility that the repressed mRNAs can exit PBs and re-enter translation, once the repression is reversed. Alternatively, if these RNAs are deemed unnecessary, they can be cleared from the cell by channeling them into the decay pathway. This is an attractive possibility, as after translational repression by miRNAs and subsequent relocalization to the PBs, a part of the sequestered mRNAs might be degraded by the resident decapping and 5′–3′ exonuclease activities. This might explain the observed reduction in target mRNA levels. What decides whether the sequestered mRNA is to be degraded or stored for later reuse? Perhaps the incarceration in PBs tips the balance in favor of degradation, although it is possible that a message with a long enough half-life might escape back into the cytosol for translation, once the repression is reversed.
Conclusions
Recent studies have shed light on the mechanisms of animal miRNA function. The long-held view that animal miRNAs do not affect mRNA stability when base-paired to their targets with partial complementarity has been overturned. MicroRNAs have been shown to affect translation, both prior to and after the initiation step. The linking of cytoplasmic PBs to the miRNA pathway suggests a pathway where translational initiation arrest is the primary event and views the effect on mRNA stability as a consequence of relocalization of repressed mRNAs to the PBs. A cellular miRNA has been shown to affect replication of a virus without affecting stability or translational output of its target RNA. All of these findings raise the question of whether we can box these different observations into one unified model to explain miRNA function. Do we need a model? Perhaps there are multiple mechanisms in operation. Ultimately, the fate of a message is decided by the sum of all forces acting on it, and this can be quite different for each mRNA, with the miRNAs being only one of these forces.
Acknowledgments
I thank Witold Filipowicz in particular for his support, discussions, and critical reading of the manuscript. I also thank Suvendra Bhattacharyya, Daniela Schmitter, Fabrice Kolb, and Petr Svoboda for very useful comments on the manuscript, and Elisa Izaurralde for making her work available prior to publication. I apologize to many authors whose work could not be referenced due to space limitations. The Friedrich Miescher Institute for Biomedical Research is a part of the Novartis Research Foundation.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2248605.
EDITOR’S NOTEThe foregoing invited review is the first in an annual series to be published by the winner of the RNA Society/Scaringe Award. This award is given each year in recognition of exceptional contributions to RNA research by a graduate student or postdoctoral fellow. Dr. Pillai received the 2005 award for his work on microRNA-mediated repression of gene expression.
REFERENCES
- Ambros, V. 2003. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 113: 673–676. [DOI] [PubMed] [Google Scholar]
- Andrei, M.A., Ingelfinger, D., Heintzmann, R., Achsel, T., Rivera-Pomar, R., and Luhrmann, R. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., and Pasquinelli, A.E. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. and Chen, C.Z. 2004. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5: 396–400. [DOI] [PubMed] [Google Scholar]
- Bashkirov, V.I., Scherthan, H., Solinger, J.A., Buerstedde, J.M., and Heyer, W.D. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37: 766–770. [DOI] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., and Parker, R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. 2002. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 16: 2733–2742. [DOI] [PubMed] [Google Scholar]
- Chendrimada, T.P., Gregory, R.I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K., and Shiekhattar, R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, Y.L. and Rana, T.M. 2003. siRNA function in RNAi: A chemical modification analysis. RNA 9: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, P.F., Poulin, F., Cho-Park, Y.A., Cho-Park, I.B., Chicoine, J.D., Lasko, P., and Sonenberg, N. 2005. A new paradigm for translational control: Inhibition via 5′-3′ mRNA tethering by bicoid and the eIF4E cognate 4EHP. Cell 121: 411–423. [DOI] [PubMed] [Google Scholar]
- Cimmino, A., Calin, G.A., Fabbri, M., Iorio, M.V., Ferracin, M., Shimizu, M., Wojcik, S.E., Aqeilan, R.I., Zupo, S., Dono, M., et al. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. 39: 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J. and Parker, R. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73: 861–890. [DOI] [PubMed] [Google Scholar]
- De Gregorio, E., Baron, J., Preiss, T., and Hentze, M.W. 2001. Tethered-function analysis reveals that elF4E can recruit ribosomes independent of its binding to the cap structure. RNA 7: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., Spencer, A., Morita, K., and Han, M. 2005. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell 19: 437–447. [DOI] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20: 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy, T., Chan, E.K., Tenenbaum, S.A., Keene, J.D., Griffith, K., and Fritzler, M.J. 2002. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 13: 1338–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy, T., Jakymiw, A., Chan, E.K., Seraphin, B., Cougot, N., and Fritzler, M.J. 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz, W. 2005. RNAi: The nuts and bolts of the RISC machine. Cell 122: 17–20. [DOI] [PubMed] [Google Scholar]
- Forstemann, K., Tomari, Y., Du, T., Vagin, V.V., Denli, A.M., Bratu, D.P., Klattenhoff, C., Theurkauf, W.E., and Zamore, P.D. 2005. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, A.D., Jaskiewicz, L., Zhang, H., Laine, S., Sack, R., Gatignol, A., and Filipowicz, W. 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6: 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley, B. and Zamore, P.D. 2004. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 11: 599–606. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J.and Baulcombe, D.C.1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- He, L., Thomson, J.M., Hemann, M.T., Hernando-Monge, E., Mu, D., Goodson, S., Powers, S., Cordon-Cardo, C., Lowe, S.W., Hannon, G.J., et al. 2005. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. [DOI] [PubMed] [Google Scholar]
- Jiang, F., Ye, X., Liu, X., Fincher, L., McKearin, D., and Liu, Q. 2005. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes & Dev. 19: 1674–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, Q., Huang, S., Guth, S., Zarubin, T., Motoyama, A., Chen, J., Di Padova, F., Lin, S.C., Gram, H., and Han, J. 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120: 623–634. [DOI] [PubMed] [Google Scholar]
- Jopling, C.L., Yi, M., Lancaster, A.M., Lemon, S.M., and Sarnow, P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309: 1577–1581. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fitzler, M.J., Scheuner, D., Kaufman, R.J., Golan, D.E., and Anderson, P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, V.N. 2005. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell. Biol. 6: 376–385. [DOI] [PubMed] [Google Scholar]
- Kim, J., Krichevsky, A., Grad, Y., Hayes, G.D., Kosik, K.S., Church, G.M., and Ruvkun, G. 2004. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. 101: 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman, W.P., Wienholds, E., Ketting, R.F., and Plasterk, R.H. 2004. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 32: 6284–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., and Kosik, K.S. 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E.C. 2002. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30: 363–364. [DOI] [PubMed] [Google Scholar]
- Lecellier, C.H., Dunoyer, P., Arar, K., Lehmann-Che, J., Eyquem, S., Himber, C., Saib, A., and Voinnet, O. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308: 557–560. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Li, H., Li, W.X., and Ding, S.W. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., and Johnson, J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773. [DOI] [PubMed] [Google Scholar]
- Lingel, A. and Sattler, M. 2005. Novel modes of protein-RNA recognition in the RNAi pathway. Curr. Opin. Struct. Biol. 15: 107–115. [DOI] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., and Parker, R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7: 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., Getz, G., Miska, E.A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B.L., Mak, R.H., Ferrando, A.A., et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838. [DOI] [PubMed] [Google Scholar]
- Ma, J.B., Yuan, Y.R., Meister, G., Pei, Y., Tuschl, T., and Patel, D.J. 2005. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434: 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G. and Tuschl, T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15: 185–197. [DOI] [PubMed] [Google Scholar]
- Nelson, P.T., Hatzigeorgiou, A.G., and Mourelatos, Z. 2004. miRNP: mRNA association in polyribosomes in a human neuronal cell line. RNA 10: 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell, K.A., Wentzel, E.A., Zeller, K.I., Dang, C.V., and Mendell, J.T. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843. [DOI] [PubMed] [Google Scholar]
- Olsen, P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN- 14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671–680. [DOI] [PubMed] [Google Scholar]
- Orban, T.I. and Izaurralde, E. 2005. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R. and Song, H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11: 121–127. [DOI] [PubMed] [Google Scholar]
- Parker, J.S., Roe, S.M., and Barford, D. 2004. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 23: 4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2005. Structural insights into mRNA recognition from a PIWI domain–siRNA guide complex. Nature 434: 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Artus, C.G., and Filipowicz, W. 2004. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10: 1518–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Artus, C.G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., and Filipowicz, W. 2005. Inhibition of translational initiation by let-7 mcroRNA in human cells. Science 309: 1573–1576. [DOI] [PubMed] [Google Scholar]
- Poy, M.N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P.E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., et al. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230. [DOI] [PubMed] [Google Scholar]
- Rehwinkel, J., Behm-Ansmant, I., Gatfield, D., and Izaurralde, E. 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11: 1640–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Rivas, F.V., Tolia, N.H., Song, J.J., Aragon, J.P., Liu, J., Hannon, G.J., and Joshua-Tor, L. 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12: 340–349. [DOI] [PubMed] [Google Scholar]
- Saito, K., Ishizuka, A., Siomi, H., and Siomi, M.C. 2005. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 3: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggerson, K., Tang, L., and Moss, E.G. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243: 215–225. [DOI] [PubMed] [Google Scholar]
- Sen, G.L. and Blau, H.M. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7: 633–636. [DOI] [PubMed] [Google Scholar]
- Sheth, U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.J., Smith, S.K., Hannon, G.J., and Joshua-Tor, L. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J. 2005. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell. Biol. 6: 127–138. [DOI] [PubMed] [Google Scholar]
- Teixeira, D., Sheth, U., Valencia-Sanchez, M.A., Brengues, M., and Parker, R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari, Y. and Zamore, P.D. 2005. Perspective: Machines for RNAi. Genes & Dev. 19: 517–529. [DOI] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Jakymiw, A., Wood, M.R., Eystathioy, T., Rubin, R.L., Fritzler, M.J., and Chan, E.K. 2004. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 117: 5567–5578. [DOI] [PubMed] [Google Scholar]
- Yekta, S., Shih, I.H., and Bartel, D.P. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–596. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Yi, R., and Cullen, B.R. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. 100: 9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Samal, E., and Srivastava, D. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436: 214–220. [DOI] [PubMed] [Google Scholar]