Abstract
This commentary discusses the recent reports in RNA by Yan and colleagues and Westman and colleagues of the apparent failure of ribavirin to bind to recombinant eIF4E and inhibit 7-methyl guanosine cap-dependent exogenous mRNA translation of cell extracts in vitro. Measuring binding by using affinity chromatography of matrix-immobilized proteins and by using protein emission fluorescence spectroscopy in the presence of nucleotide ligands, as well as limitations of using cell extracts for the assessment of mechanisms of mRNA translation are discussed. Possible reasons for the discordant findings of Yan and colleagues and Westman and colleagues are suggested, and direct observation of the specific binding of ribavirin to eIF4E by using mass spectrometry is presented.
Keywords: ribavirin, eIF4E, m7-guanosine cap, physical mimic
The mechanism of action of ribavirin (Virazole), a triazole carboxamide ribonucleoside, has remained enigmatic since its discovery in the early 1970s (Smith et al. 1984). Much of the confusion stemmed from its apparent activity against a wide variety of seemingly unrelated viruses, as well as pleiotropic cellular effects dependent on concentration. Based on the similarities in the arrangement of hydrogen-bonding groups of ribavirin and guanosine, ribavirin was postulated to be a guanosine analog. This notion is consistent with ribavirin’s effects at millimolar concentrations on guanylyl transferases, inosine monophosphate dehydrogenase, and RNA-dependent RNA polymerases (Hong and Cameron 2002). In spite of these findings, definition of the cytostatic effects of ribavirin at low micromolar concentrations has remained elusive.
Our recent study sought to identify the nature of these specific effects, and thereby help to define the specific mechanism of action of ribavirin (Kentsis et al. 2004). Using ab initio quantum mechanics, we characterized the physical properties of a variety of guanosine analogs, noting a striking similarity in the electronic structure of ribavirin and 7-methyl guanosine. Using tryptophan emission fluorescence spectroscopy and nucleotide affinity chromatography, we measured the dissociation and inhibition constants of ribavirin and the 7-methyl guanosine (m7G) binding protein eIF4E to be in the low micromolar range. Using fluorescence microscopy, cell fractionation, Northern and Western analyses, and quantitative PCR, we observed ribavirin’s disruption of subcellular eIF4E localization, disruption of nuclear and cytoplasmic eIF4E:mRNA binding, inhibition of nucleocytoplasmic eIF4E-sensitive mRNA transport, and inhibition of eIF4E-sensitive mRNA translation in living cells, all at similar low micromolar concentrations. Using flow cytometry, colony formation, and tumor growth assays, we observed cytostatic and tumor-suppressive effects of ribavirin in models of eIF4E-dependent cancers in vitro and in vivo, as well as using eIF4E-over-expressing leukemic blasts isolated from human patients, also at low micromolar concentrations. While ribavirin exhibited physical properties similar to those of 7-methyl guanosine, its circularly permuted chemical analog Rib4C did not, failing to bind eIF4E, to inhibit its functions in mRNA transport, translation, and tumorigenesis. This led us to conclude that ribavirin is a physical mimic of the 7-methyl guanosine mRNA cap (Kentsis et al. 2004).
In recent issues of RNA, Yan et al. (2005) and Westman et al. (2005) present findings that dispute this conclusion. Their results are twofold: that ribavirin does not bind to recombinant eIF4E in vitro, and that ribavirin does not inhibit cap-dependent translation of exogenous mRNAs in extracts prepared from cells. We are concerned that these experiments failed to elicit ribavirin’s effects and, similarly to the authors, would like to discuss possible reasons for this.
First, it is well established that the binding of the m7G cap to eIF4E is highly dependent on solution conditions. Variations of several orders of magnitude (nanomolar to micromolar) can occur as a result of changes in ionic strength, pH, and temperature (Carberry et al. 1989; Kentsis et al. 2001; Niedzwiecka et al. 2002). This likely depends on the physical properties and exact geometries of the cap-binding site in the apo form of eIF4E under these conditions (L. Volpon, M. Osborne, and K.L.B. Borden, in prep.), which themselves are dependent on the relative populations of various structural substates that apo-eIF4E is known to adopt in solution (Matsuo et al. 1997; Kentsis et al. 2001, 2002, 2004). Given that ribavirin binds the cap-binding site of eIF4E, its apparent affinity for eIF4E would also be condition dependent. Furthermore, given that ribavirin’s triazole carboxamide likely makes fewer atomic contacts with eIF4E as compared to 7-methyl guanosine, ribavirin’s high-affinity binding to eIF4E would be expected to occur in a narrower range of solution conditions.
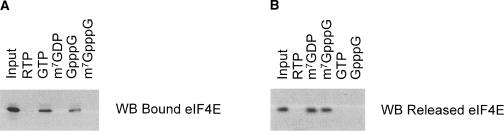
With this in mind, we reproduced our original affinity chromatography experiment using an independent operator and new reagents side by side with the experiment of Yan et al. (2005). We reproduced experimental conditions as published by us (0.3 M NaCl, 0.1 M sodium phosphate, 0.1% Nonidet P-40, 10 μM BSA at pH 7.5, room temperature), and those described by Yan et al. (2005) (0.1 M KCl, 10 mM HEPES-KOH, 0.2 mM EDTA at pH 8.0, presumed 4°C). In agreement with our reported findings (Kentsis et al. 2004), micromolar concentrations of ribavirin triphosphate (RTP) compete with the binding of eIF4E:m7G, similarly to that of m7GTP itself (Fig. 1A). In contrast, using the protocol of Yan et al. (2005) leads to an apparent failure of RTP to compete with m7G binding (Fig. 1B). Thermodynamic meta-stability of eIF4E under various solution conditions is well described in the literature (Matsuo et al. 1997; McGuire et al. 1998; Kentsis et al. 2001, 2004), leading to aggregation and linkage effects that can confound the apparent binding of ligands (Fletcher and Wagner 1998; Cohen et al. 2001), particularly when using matrix-immobilized proteins, as in the experiments of Yan et al. (2005). In addition, the structure of apo-eIF4E is sensitive to differences in pH between 7.5 and 8, as assessed by NMR chemical shift perturbation (L. Volpon, M. Osborne, and K.L.B. Borden, in prep.). Thus, the reported failure of ribavirin to bind eIF4E in vitro by Yan et al. (2005) appears to be due, at least in part, to the use of different solution conditions.
FIGURE 1.
Apparent binding of ribavirin to recombinant eIF4E in vitro is method and condition dependent. (A) As published previously, 20 μL of m 7GTP-Sepharose (Amersham) was mixed with 1 μg of eIF4E in Buffer B (0.3 M NaCl, 0.1 M sodium phosphate at pH 7.5, 10 μM protease-free BSA [UBS], 0.1% NP-40) with 0.1 mM GTP for 30 min at room temperature. Washed beads (three times with 75 bed volumes) were incubated with 50 μM of compounds as indicated for 30 min at room temperature. Beads were washed (three times with 75 bed volumes) to remove dissociated eIF4E, and eIF4E remaining bound to beads was resolved using SDS-PAGE, and visualized using Western blotting and chemiluminescence. Please note that here the buffer contained 0.1 mM GTP in order to emphasize the specificity of ribavirin’s competition of m7G:eIF4E binding. Also, here we used a fusion of mouse eIF4E with the B1 domain of protein G (G4E), which was a kind gift of Gerhard Wagner (Harvard Medical School, Boston, MA), as described in Zhou et al. (2001) and Kentsis et al. (2004). (B) As described by Yan et al. (2005), 1 μg of eIF4E was mixed with 20 μL of m 7GTP-Sepharose (Amersham) in 50 bed volumes of LCB buffer (10 mM HEPES at pH 8.0, 100 mM KCl, 0.2 mM EDTA at pH 8.0), supplemented with 10 μM protease-free BSA (UBS), 0.1% NP-40, and 0.1 mM GTP, for 20 min at presumed 4 °C, as the experimental temperature was not described (Yan et al. 2005). Washed beads (five times with 50 bed volumes of LCB buffer) were incubated with 5 bed volumes of 50 μM of compounds as indicated for 20 min at 4 °C. Then 20 μL of the supernatant containing dissociated eIF4E was transferred to a new tube, cleared of trace beads containing bound eIF4E, and resolved using SDS-PAGE, and visualized using Western blotting and chemiluminescence.
In contrast to Yan et al. (2005), who fail to observe ribavirin’s binding to eIF4E altogether, Westman et al. (2005) observe that ribavirin binds to recombinant eIF4E in vitro, but does so with an affinity two to four orders of magnitude lower than that measured by us (Kentsis et al. 2004; Westman et al. 2005). Measurements of ligand binding using quenching of fluorescence emission often require corrections for the intrinsic fluorescence of added ligand and its inner filter effect (Lakowicz 1999), neither of which appears to be considered in our reading of Westman et al. (Niedzwiecka et al. 2002; Westman et al. 2005). While fluorescence quantum yields of nucleotides are lower than those of amino acids, they can be significant at concentrations used in the above studies, potentially compensating for quenching of protein fluorescence upon binding, especially when ribavirin’s quenching efficiency is twofold lower than that of 7-methyl guanosine because of differences of the two compounds (Kentsis et al. 2001, 2004). In addition to this, titration of nucleotides leads to absorption of incident and/or emitted light, potentially reducing the apparent emission of fluorescence. The lower extinction coefficient of ribavirin as compared to 7-methyl guanosine (740 vs. 1600 M−1 cm−1 at 295 nm, respectively) may also contribute to the differences in apparent quenching (Kentsis et al. 2001, 2004). Unfortunately, a direct methodological comparison is precluded by our monitoring of fluorescence emission of tryptophans including those that directly bind 7-methyl guanosine (Kentsis et al. 2001, 2004), while Westman et al. measured emission by both tryptophans and tyrosines (Niedzwiecka et al. 2002; Westman et al. 2005), which may be preferentially quenched (excitation wavelengths of 295 nm and 280 nm, respectively). In addition, differences in solution conditions may also contribute to the observed differences in binding affinities. As suggested by Westman et al. (2005), these methodological differences may explain the apparent differences in the measured affinities.
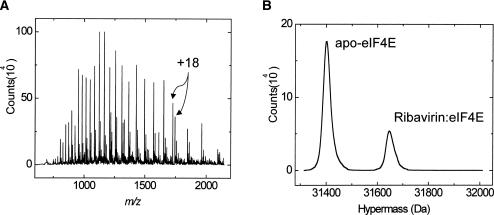
Nevertheless, in order to provide a decisive demonstration of binding of ribavirin to eIF4E, one that does not involve indirect or ambiguous probes of binding such as competition affinity chromatography and fluorescence quenching, we examined the binding of ribavirin and eIF4E by using electrospray mass spectrometry. A mixture of 20 μM purified recombinant eIF4E and fourfold excess of both ribavirin and GTP was electrosprayed directly, and its mass/ionization spectrum was measured. The recorded spectrum is shown in Figure 2A and contains two sets of multiply charged ions, one with a population-weighted mean molecular mass of 31,402 Da, corresponding to apo-G4E (Zhou et al. 2001; Kentsis et al. 2004), and another of 31,649 Da (Fig. 2B). This mass shift of 247 Da is due to specific binding of ribavirin (243 Da), and not of GTP (523 Da). In our published study, the specificity of ribavirin’s binding to eIF4E was established using mutation of the cap-binding site W56A, which disrupts binding of ribavirin, but not folding of the protein, similar to its disruption of binding of the 7-methyl guanosine cap (Kentsis et al. 2004). Ribavirin’s circularly permuted version Rib4C, which is chemically identical but is not positively charged, failed to bind eIF4E (Kentsis et al. 2004). And finally, ribavirin’s binding leads to a similar conformational rearrangement of eIF4E, as observed using NMR spectroscopy, as the one induced by the binding of 7-methyl guanosine cap, consistent with ribavirin’s binding of the cap-binding site (Kentsis et al. 2004). Additional specificity controls are described in Kentsis et al. (2004). Thus, eIF4E specifically binds to ribavirin using the cap-binding site, and experimental failure to observe this interaction may be due to challenges of the particular techniques used.
FIGURE 2.
Direct observation of specific binding of ribavirin to purified eIF4E in vitro. Mass spectra were recorded using the Agilent Technologies 1100 LC/MSD integrated liquid chromatograph single quadrupole electrospray mass spectrometer (ES-MS) operating in positive ion mode. A solution of 20 μM purified G4E (Zhou et al. 2001; Kentsis et al. 2004) was incubated with a mixture of 80 μM ribavirin (Calbiochem) and 80 μM GTP (Sigma) in 5% aqueous acetonitrile, 20 mM ammonium acetate (pH 6.5), for 1 min at room temperature. The solution was electrosprayed directly at 200 μL/min using nebulizer pressure of 20 psi, drying nitrogen gas at 200°C and 10 L/min, and capillary voltage of 4.5 kV. (A) ES-MS spectrum plotting ion abundance in 20 μL of the above mixture as a function of the mass/charge ratio is shown. An ion of ~1740 amu/z is labeled, corresponding to a +18 protonation state of apo-G4E (higher peak) and the complex of ribavirin with G4E (lower peak). (B) Hypermass reconstruction of the spectrum shown above was done according to standard methods (De Hoffmann and Stroobant 2001) and contains two species of population-weighted mean molecular masses of 31,402 and 31,649 Da, corresponding to apo-G4E and G4E bound to ribavirin (243 Da) with a molecular stoichiometry of 1:1, respectively. Please note that only a fraction of total ionized eIF4E appears to be bound to ribavirin because of the differences in ionization efficiencies of the apo- and ligand-bound species of eIF4E, wherein ligand binding occurs to the folded, more native-like, and therefore less ionizable, states (De Hoffmann and Stroobant 2001). Thus, obtaining affinities from mass spectrometry data is confounded by these differences in ionization. For comparison, we obtained Kd’s for eIF4E–ribavirin of 8.4 μM and for eIF4E–RTP, 0.13 μM, using fluorescence spectroscopy paralleling those differences previously observed for m7-guanosine and m7GTP (Kentsis et al. 2004).
Another question examined by Yan et al. (2005) and Westman et al. (2005) concerns the effect of ribavirin on eIF4E function. Both groups examined functional effects in vitro, whereas we were concerned with ribavirin’s effects in vivo (Kentsis et al. 2004). Cell extracts for translation of exogenous mRNAs in vitro are well known for their unique properties, having altered compositions, stoichiometries, and activities, as compared to those in living cells, where compartmentalization and molecular organization are maintained and are of paramount importance for a process as complex and regulated as mRNA translation. Although such extracts have been used with considerable success for the discovery of translation factors, their significance for the characterization of mechanisms of translation remains controversial. In this light, assessment of eIF4E activity by way of discrimination between efficiencies of translation of 7-methyl guanosine cap- and internal ribosome binding site (IRES)-driven transcripts is problematic for a number of reasons. The activity of each extract is optimized empirically in order to maximize the translational contribution of a particular feature of an exogenous mRNA, a process that in no way guarantees the overall mechanistic and functional fidelity that is absolutely required for the characterization of novel activities such as that of ribavirin.
For example, both Yan et al. (2005) and Westman et al. (2005) use cell extracts, albeit prepared from different cells and with different modifications, carefully optimized in order to maximize the translational synergy between the 5′-cap and 3′-poly(A) mRNA elements (Bergamini et al. 2000; Svitkin and Sonenberg 2004), a feature that depends neither solely nor specifically on the activity of eIF4E. This apparent synergy is due to the scaffolding activity of eIF4G, which concomitantly binds eIF4E, poly(A) binding proteins (PABPs), and the ribosome (Michel et al. 2000), thereby coupling the affinities of eIF4E and PABPs for the 5′-cap and 3′-poly(A) tail, respectively. Yet, the presence of the poly(A) tail alone can also stimulate translation from IRES in vitro (Svitkin et al. 2001), and eIF4E can recruit ribosomes in the absence of cap binding (De Gregorio et al. 2001).
Thus, in the experiments of Westman et al. (2005), although competition with m7GpppG and m7GTP inhibits cap-driven translation at analog concentrations of ~0.1 mM while RpppG does not, specificity of this difference and its mechanistic interpretation are indeterminate considering that the concentration of m7GpppG-capped transcript is ~1 nM (an excess of >100,000-fold) (Westman et al. 2005). The interpretation of the findings of Yan et al. (2005) is even more complicated by their use of a bicistronic construct containing both the 5′-cap and an IRES that minimizes relative differences in efficiency as a result of competition for rate-limiting translation factors (Yan et al. 2005). Thus, treatment with 1 mM m7GDP leads to a reduction of activity of cap-driven firefly luciferase from ~8 × to 2 × 105 light units: a fourfold effect, rather insignificant as compared to the molar excess of cap analog to mRNA of >1,000,000-fold (mRNA concentration of 5 μg/mL) (Yan et al. 2005). Considering that the concentration of eIF4E in similarly used cell extracts is estimated to be ~400 nM (Rau et al. 1996), the requirement of such high cap analog concentrations suggests that the examined process is not dependent strictly on eIF4E activity during mRNA translation.
While we do not dispute that ribavirin can be misincorporated into 5′-mRNA caps at millimolar concentrations, based on measurements of viral production by Yan et al. (2005) and careful analysis of cap structures by Westman et al. (2005), we question the specificity of the observed differences in translational efficiency between cap- and IRES-driven constructs in vitro, and their mechanistic interpretation with respect to the mechanism of action of ribavirin and our findings of its inhibition of eIF4E sensitive translation in vivo. In this context, although ribavirin failed to inhibit cap-dependent translation in vitro in the work of Yan et al. (2005) and Westman et al. (2005), this lack of an effect may have to do with the lack of sensitivity of current cell extracts to eIF4E activity.
In this regard, the distinction between cap- and eIF4E-sensitive translation may be of paramount significance. Although the interaction of the 5′ 7-methyl guanosine cap with eIF4E is required for the translation of cap-dependent mRNAs, up-regulation of eIF4E in cells does not increase levels of all proteins produced from cap-dependent transcripts, but only of a specific subset including cyclin D1 and VEGF, but not β-actin and GAPDH, for example (De Benedetti and Graff 2004). This effect occurs at the level of nucleocytoplasmic transport for some mRNAs, at the level of translation for others, and for some at both (Rousseau et al. 1996). Thus, just as eIF4E up-regulation does not globally increase cellular protein translation, ribavirin is not a global inhibitor. Such specificity of ribavirin’s effects on translation in cells was precisely observed in our measurements of polysomal loading of mRNAs of cyclin D1, GAPDH, VEGF, and ODC (Kentsis et al. 2004).
In summary, just like that of the 7-methyl guanosine cap, ribavirin’s binding to eIF4E is dependent on solution conditions, but nevertheless occurs robustly and specifically (Figs. 1, 2). To determine the physiological relevance of interactions assessed in vitro, it is important to assess their functionality in vivo. Thus, we assessed the physiological relevance of ribavirin’s binding to eIF4E in cells, in animal models, as well as in tissues isolated from human patients (Kentsis et al. 2004). In all of these systems, ribavirin antagonized eIF4E functions in transport and translation of eIF4E-sensitive mRNAs at low micromolar concentrations, similar to those at which it dissociates from purified eIF4E in vitro. We hope that future collaborative work will continue to define the specific mechanism and cellular effects of this rather simple chemically, but biologically complex, drug.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2238705.
REFERENCES
- Bergamini, G., Preiss, T., and Hentze, M.W. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6: 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carberry, S.E., Rhoads, R.E., and Goss, D.J. 1989. A spectroscopic study of the binding of m7GTP and m7GpppG to human protein synthesis initiation factor 4E. Biochemistry 28: 8078–8083. [DOI] [PubMed] [Google Scholar]
- Cohen, N., Sharma, M., Kentsis, A., Perez, J.M., Strudwick, S., and Borden, K.L. 2001. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 20: 4547–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti, A. and Graff, J.R. 2004. eIF-4E expression and its role in malignancies and metastases. Oncogene 23: 3189–3199. [DOI] [PubMed] [Google Scholar]
- De Gregorio, E., Baron, J., Preiss, T., and Hentze, M.W. 2001. Tethered-function analysis reveals that elF4E can recruit ribosomes independent of its binding to the cap structure. RNA 7: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoffmann, E. and Stroobant, V. 2001. Mass spectrometry: Principles and applications. John Wiley and Sons, New York.
- Fletcher, C.M. and Wagner, G. 1998. The interaction of eIF4E with 4E-BP1 is an induced fit to a completely disordered protein. Protein Sci. 7: 1639–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z. and Cameron, C.E. 2002. Pleiotropic mechanisms of ribavirin antiviral activities. Prog. Drug Res. 59: 41–69. [DOI] [PubMed] [Google Scholar]
- Kentsis, A., Dwyer, E.C., Perez, J.M., Sharma, M., Chen, A., Pan, Z.Q., and Borden, K.L. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312: 609–623. [DOI] [PubMed] [Google Scholar]
- Kentsis, A., Gordon, R.E., and Borden, K.L. 2002. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl. Acad. Sci. 99: 15404–15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis, A., Topisirovic, I., Culjkovic, B., Shao, L., and Borden, K.L. 2004. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc. Natl. Acad. Sci. 101: 18105–18110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz, J.R. 1999. Principles of fluorescence spectroscopy. Kluwer Academic/Plenum Publishers, New York.
- Matsuo, H., Li, H., McGuire, A.M., Fletcher, C.M., Gingras, A.C., Sonenberg, N., and Wagner, G. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4: 717–724. [DOI] [PubMed] [Google Scholar]
- McGuire, A.M., Matsuo, H., and Wagner, G. 1998. Internal and overall motions of the translation factor eIF4E: Cap binding and insertion in a CHAPS detergent micelle. J. Biomol. NMR 12: 73–88. [DOI] [PubMed] [Google Scholar]
- Michel, Y.M., Poncet, D., Piron, M., Kean, K.M., and Borman, A.M. 2000. Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 275: 32268–32276. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka, A., Marcotrigiano, J., Stepinski, J., Jankowska-Anyszka, M., Wyslouch-Cieszynska, A., Dadlez, M., Gingras, A.C., Mak, P., Darzynkiewicz, E., Sonenberg, N., et al. 2002. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319: 615–635. [DOI] [PubMed] [Google Scholar]
- Rau, M., Ohlmann, T., Morley, S.J., and Pain, V.M. 1996. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J. Biol. Chem. 271: 8983–8990. [DOI] [PubMed] [Google Scholar]
- Rousseau, D., Kaspar, R., Rosenwald, I., Gehrke, L., and Sonenberg, N. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. 93: 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R.A., Knight, V., and Smith, J.A.D. 1984. Clinical applications of ribavirin. Academic Press, Orlando, FL.
- Svitkin, Y.V. and Sonenberg, N. 2004. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 257: 155–170. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Imataka, H., Khaleghpour, K., Kahvejian, A., Liebig, H.D., and Sonenberg, N. 2001. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA 7: 1743–1752. [PMC free article] [PubMed] [Google Scholar]
- Westman, B., Beeren, L., Grudzien, E., Stepinski, J., Worch, R., Zuberek, J., Jemielity, J., Stolarski, R., Darzynkiewicz, E., Rhoads, R.E., et al. 2005. The antiviral drug ribavirin does not mimic the 7-methylguanosine moiety of the mRNA cap structure in vitro. RNA (in press). [DOI] [PMC free article] [PubMed]
- Yan, Y., Svitkin, Y., Lee, J.M., Bisaillon, M., and Pelletier, J. 2005. Ribavirin is not a functional mimic of the 7-methyl guanosine mRNA cap. RNA 11: 1238–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Lugovskoy, A.A., and Wagner, G. 2001. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J. Biomol. NMR 20: 11–14. [DOI] [PubMed] [Google Scholar]