Abstract
The mRNA processing body (P-body) is a cellular structure that regulates gene expression by degrading cytoplasmic mRNA. The objective of this study was to identify and characterize novel components of the mammalian P-body. Approximately 5% of patients with the autoimmune disease primary biliary cirrhosis have antibodies directed against this structure. Serum from one of these patients was used to identify a cDNA encoding Ge-1, a 1401-amino-acid protein. Ge-1 contains an N-terminal WD40 motif and C-terminal domains characterized by a repeating ψ(X2–3) motif. Ge-1 co-localized with previously identified P-body components, including proteins involved in mRNA decapping (DCP1a and DCP2) and the autoantigen GW182. The Ge-1 C-terminal domain was necessary and sufficient to target the protein to P-bodies. Following exposure of cells to oxidative stress, Ge-1-containing P-bodies were found adjacent to TIA-containing stress granules. During the recovery period, TIA returned to the nucleus while Ge-1-containing P-bodies localized to the perinuclear region. siRNA-mediated knock-down of Ge-1 resulted in loss of P-bodies containing Ge-1, DCP1a, and DCP2. In contrast, Ge-1-containing P-bodies persisted despite knock-down of DCP2. Taken together, the results of this study show that Ge-1 is a central component of P-bodies and suggest that Ge-1 may act prior to the 5′-decapping step in mRNA degradation.
Keywords: mRNA processing body, mRNA decay, autoantigen
INTRODUCTION
Gene expression is initiated in the cell nucleus, where RNA transcripts are produced and processed to mRNA. Mature mRNAs traverse nuclear pores and are translated in the cytoplasm. A frequently overlooked step in the regulation of gene expression is the degradation of mRNA. Two important pathways of mRNA degradation have been described (for review, see Coller and Parker 2004; Parker and Song 2004). In both pathways, mRNA degradation is initiated by shortening of the poly(A) tail followed by removal of poly(A) binding protein (PABP). In the 3′ → 5′ pathway of mRNA destruction, the cytoplasmic exosome, a complex containing multiple exonucleases, degrades mRNA in the 3 ′ → 5′ direction, resulting in an oligonucleotide cap structure that is hydrolyzed by the scavenger decapping enzyme, DcpS. In the 5′ → 3′ pathway of mRNA degradation, shortening of the 3′-poly(A) tail and removal of PABP is followed by cleavage of the 5′-mRNA cap by a complex containing decapping enzymes 1a and 2 (DCP1a/DCP2). The mRNA molecule is then subjected to 5′ → 3′ degradation mediated by exoribonuclease enzyme 1 (Xrn1). In both yeast and mammalian cells, the proteins involved in 5′ → 3′ mRNA decay are concentrated in cytoplasmic structures that have been designated mRNA “processing bodies” (P-bodies, also known as “cytoplasmic foci” and “GW182 bodies”) (Eystathioy et al. 2003b; Sheth and Parker 2003; Cougot et al. 2004). In addition to DCP1a/DCP2 and Xrn1, other proteins localize to P-bodies. These proteins include Sm-like proteins 1–7 (Lsm1–7), the DEAD box family helicase Rck/ p54, and the autoantigen GW182 (Bouveret et al. 2000; Coller et al. 2001; Eystathioy et al. 2003b; Cougot et al. 2004). The Lsm proteins enhance assembly of the decapping complex, and Rck/p54 increases the efficiency of mRNA decapping. GW182 is a putative RNA-binding protein of unknown function.
Studies in yeast and mammalian cells showed that P-bodies are sites of active mRNA degradation (Sheth and Parker 2003; Cougot et al. 2004). Treatment of cells with cyclohexamide, which inhibits translation elongation and traps mRNAs on polysomes, decreases the flow of mRNA to P-bodies and causes rapid loss of these structures. In contrast, inhibition of Xrn1 in yeast or mammalian cells blocks the 5′ → 3′ mRNA degradation step, increases the size and number of P-bodies, and results in accumulation of mRNAs within these structures. The observation that P-bodies are modified by changes in mRNA metabolism suggests that these structures are actively involved in mRNA decay.
In mammalian cells, exposure to environmental stress results in the formation of cytoplasmic structures known as “stress granules” (SGs) (for review, see Kedersha and Anderson 2002). SGs contain mRNAs, translation initiation factors, the mRNA-binding proteins TIA and TIAR, and 40S ribosome subunits. The accumulation and retention of the pre-stress or “housekeeping” pool of mRNAs in these structures permits mRNAs encoding stress and repair proteins to gain access to the cellular translation machinery. If the cell survives the environmental stress, SGs disappear and housekeeping mRNAs may return to active translation. The precise relationship between P-bodies and TIA-containing SGs has not been determined. Cougot et al. (2004) reported that P-bodies did not co-localize with SGs and that P-bodies only rarely appeared to be adjacent to SGs. More recently, Wilczynska et al. (2005) and Kedersha et al. (2005) observed a dynamic link between these two cytoplasmic structures.
Primary biliary cirrhosis (PBC) is an autoimmune disease of unknown etiology characterized by the progressive destruction of intrahepatic biliary ductules, leading to hepatic fibrosis and liver failure (for review, see Kaplan 1996). Patients with PBC may develop autoantibodies directed against a spectrum of cellular structures including mitochondria, PML nuclear bodies, and nuclear pore complexes. In a recent study of 492 PBC patients, we observed that ~5% of PBC patients have antibodies directed against P-bodies (Bloch et al. 2005). In this study, we used serum from a patient with PBC to show that autoantigen Ge-1 is a component of P-bodies. We delineated the portion of Ge-1 that localizes the protein to P-bodies and examined the cellular location of Ge-1 relative to SGs during the course of oxidative stress and in the recovery period. We show that Ge-1 has a critical role in maintaining the integrity of mRNA P-bodies.
RESULTS AND DISCUSSION
Antibodies in serum from a patient with primary biliary cirrhosis react with a novel component of cytoplasmic mRNA processing bodies
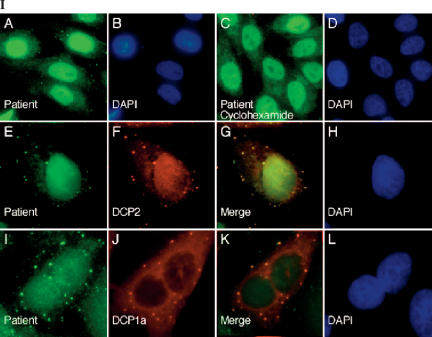
Using indirect immunofluorescence and the human Hep-2 cell line as substrate, we observed that antibodies in serum from PBC patient 0020 reacted with 5–20 cytoplasmic foci per cell (Fig. 1, panel I-A,E). To test the possibility that these foci were P-bodies, Hep-2 cells were treated with cyclohexamide and then stained with patient 0020 serum. Treatment of cells with cyclohexamide, which disrupts P-bodies, resulted in loss of cytoplasmic dot staining by 0020 serum (Fig. 1, panel I-C,D). These results suggest that at least one constituent of P-bodies is recognized by autoantibodies in 0020 serum.
FIGURE 1.
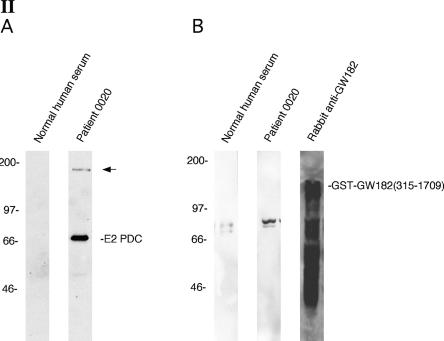
Antibodies in the serum of a primary biliary cirrhosis patient react with a novel P-body component. (Panel I) Patient 0020 serum contained antibodies that reacted with 5–20 dots in the cytoplasm of Hep-2 cells as determined by indirect immunofluorescence (I-A,E). The patient’s serum also contained antibodies directed against E2-pyruvate dehydrogenase complex (E2-PDC), which produced a cytoplasmic, linear staining pattern, and antibodies directed against an unknown antigen that was present diffusely throughout the nucleus. Treatment of Hep-2 cells with cyclohexamide resulted in disappearance of cytoplasmic dots with little effect on the staining of mitochondria or nuclei (I-C). (I-E) Patient antibodies (green) co-localized with (I-F) anti-DCP2 antibodies (red). To further confirm that patient antibodies reacted with P-bodies, Hep-2 cells were transfected with a plasmid encoding the Flag-epitope fused to P-body component DCP1a. Cells were fixed and stained with (I-I) patient serum (green) and (I-J) anti-Flag antibodies (red). (I-G,K) Overlap of red and green staining is shown in yellow. DAPI staining in I-B,D,H,L indicates the location of cell nuclei. (Panel II) Immunoblotting was used to characterize the putative P-body autoantigen. Antibodies in patient serum reacted with 70-kDa (E2-PDC) and ~160-kDa proteins in an extract prepared from Hep-2 cells. (II-A) Normal human serum did not react with either of these two proteins. To specifically test the patient serum for anti-GW182 antibodies, recombinant GST-GW182 was prepared, fractionated by PAGE, and transferred to nitrocellulose membranes. (II-B) Rabbit anti-GW182 antiserum reacted strongly with the recombinant protein and with associated breakdown products. Neither patient serum nor normal human serum contained antibodies directed against GW182. Both human sera reacted with an ~80-kDa protein in the extract that is likely to be residual, contaminating E. coli protein.
Direct confirmation of anti-P-body antibodies in 0020 serum was obtained using two-color indirect immunofluorescence. Rabbit anti-DCP2 antibodies and human antibodies co-localized in cytoplasmic foci (Fig. 1, panel I-E–H). In addition, using a commercial Hep-2000 substrate, which contains detectable levels of P-body component GW182, it was found that rabbit anti-GW182 antibodies also co-localized with antibodies in 0020 serum (data not shown). To further confirm that the autoantibodies reacted with P-bodies, Hep-2 cells were transfected with a eukaryotic expression vector encoding Flag-DCP1a. Human antibodies co-localized in cytoplasmic dots with anti-Flag antibodies (Fig. 1, panel I-I–L). These results confirmed that 0020 serum reacts with P-bodies.
To begin the characterization of the P-body antigen recognized by patient serum, an immunoblot was prepared using Hep-2 cell lysates. Antibodies in 0020 serum reacted with ~70-kDa and ~160-kDa proteins in Hep-2 cells (Fig. 1, panel II-A). The smaller protein was presumed to be the E2 component of pyruvate dehydrogenase complex (E2-PDC), the predominant autoantigen recognized by antibodies in PBC patients. The larger band was similar in size to the previously identified P-body autoantigen GW182. To test whether patient 0020 serum contained anti-GW182 antibodies, recombinant GST-GW182(315–1709) fusion protein was prepared. This portion of GW182 was previously shown to contain the epitopes recognized by serum from patients with autoimmune disease (Eystathioy et al. 2003a). Neither 0020 serum nor normal human serum reacted with GST-GW182. In contrast, rabbit anti-GW182 antiserum reacted strongly with the recombinant protein (Fig. 1, panel II-B). These results show that 0020 serum reacts with an ~160-kDa component of P-bodies that is not GW182.
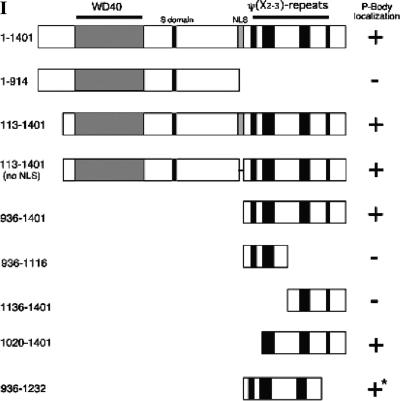
To identify the novel P-body autoantigen, serum from patient 0020 was used to screen a λGT11 cDNA expression library. A single clone producing immunoreactive recombinant protein was identified and the plaque was purified. This clone contained nucleotides 1949–4659 of a cDNA encoding autoantigen Ge-1 (also known as RCD-8; Gen-Bank # NM_014329). Ge-1 was previously identified as an ~160-kDa autoantigen recognized by a patient (Ge) with autoimmune disease (Bloch et al. 1994).5 The full-length cDNA encoding Ge-1 has a 4.2-kb coding region and predicts a 1401-amino-acid polypeptide.6 The N-terminus of Ge-1 is notable for the presence of seven WD40 repeats (amino acids 172–409). This motif forms a seven-bladed β-propeller that, when present in other proteins, functions as a protein–protein interaction domain (for review, see Smith et al. 1999). The middle of Ge-1 contains a serine-rich region, in which 16 of 17 consecutive residues are Ser (amino acids 613–629), and a bipartite nuclear localization sequence (NLS; amino acids 910–927). The Ge-1 C-terminus contains four regions that have novel repeating hydrophobic residue periodicity characterized by two residues with long aliphatic side chains (Leu, Val, Ile, or Met) separated by two or three amino acids. These regions, here-after referred to as “ψ(X2–3)-repeat domains,” span residues 971–994, 1023–1074, 1188–1231, and 1313–1328. The structural features of Ge-1 are shown schematically in Figure 2, panel I.
FIGURE 2.
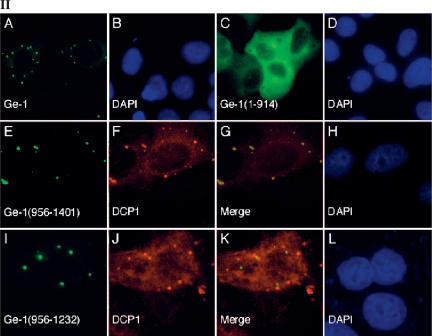
Structure of Ge-1 and identification of the portion of Ge-1 that mediates localization to P-bodies. (Panel I) The predicted amino acid sequence of Ge-1 contained seven WD40 domains (amino acids 172–409), a serine-rich region (S domain; amino acids 611–629), and a nuclear localization sequence (NLS; amino acids 910–927). The C-terminal portion of Ge-1 contained four regions with a repeating ψ(X2–3) motif (amino acids 971–994, 1023–1074, 1192–1236, and 1313–1328.) The portions of Ge-1 that were capable of localizing green fluorescent protein (GFP) to DCP1a-containing P-bodies are indicated by +. Representative indirect immunofluorescence results are shown in panel II. Note that GFP-Ge-1(936–1232) localized to cytoplasmic dots that were indistinguishable from P-bodies, but these structures did not contain DCP1a (indicated by +*; see panel II-I–L). (Panel II) Two-color immunofluorescence was used to identify the portion of Ge-1 that was necessary and sufficient for localization to DCP1a-containing P-bodies. (II-A) Full-length Ge-1 localized to cytoplasmic dots (green) and co-localized with DCP1a (data not shown). (II-C) An N-terminal portion of Ge-1 (amino acids 1–914) was distributed diffusely throughout the cytoplasm (green). (II-F) The C-terminal portion of Ge-1 (amino acids 936–1401) localized to cytoplasmic dots and co-localized with DCP1a (red). Merge of red and green staining appears yellow in II-G. (II-I) A fragment of Ge-1 (amino acids 936–1232), which lacks the fourth ψ(X2–3)-rich domain, localized to cytoplasmic dots (green) but did not co-localize with (II-J) DCP1a (red). The merge of II-I and II-J is shown in II-K. Successful production of the GFP-Ge-1 fragment fusion proteins was confirmed by immunoblot using anti-GFP antiserum (data not shown). DAPI staining identified the location of nuclei in II-B,D,H,L.
The C-terminus of Ge-1 directs the protein to P-bodies
In a previous study, Ge-1 was reported to be a nuclear protein, in part because of the presence of a functional NLS (amino acids 910–927) (Bloch et al. 1994). Garcia-Lozano et al. (1997) subsequently suggested that Ge-1 was present in the cytoplasm. To further define the cellular location of Ge-1, a fusion protein containing GFP attached to the N-terminus of full-length Ge-1 was coexpressed with Flag-DCP1a in Hep-2 cells. Antibodies directed against GFP localized to cytoplasmic dots (Fig. 2, panel II-A) and co-localized with anti-Flag antibodies (data not shown), confirming that Ge-1 is a component of P-bodies. To identify the portion of Ge-1 that directs the protein to P-bodies, expression vectors encoding GFP fused to various portions of Ge-1 were transfected into Hep-2 cells. The results are summarized in Figure 2, panel I, and representative photomicrographs are shown in Figure 2, panel II. GFP fused to Ge-1 amino acids 113–1401 or amino acids 113–1401 lacking residues 914–935 localized to cytoplasmic foci, demonstrating that the N-terminal 112 amino acids and the putative NLS were not required for P-body localization. A fusion protein containing the Ge-1 N-terminus (amino acids 1–914) did not localize to cytoplasmic dots, but was distributed diffusely throughout the cytoplasm (Fig. 2, panel II-C,D). These results showed that the WD40 repeats as well as the Ser-rich region were not sufficient to direct Ge-1 to P-bodies. In contrast, the Ge-1 C-terminal domain, which contains the four ψ(X2–3)-repeat domains (936–1401), was sufficient to target GFP to cytoplasmic dots, and these dots co-localized with cotransfected Flag-DCP1a (Fig. 2, panel II-E–H). Fusion proteins that contained only the first two (936–1116) or last two (1136– 1401) ψ(X2–3)-repeat domains were distributed diffusely throughout the cytoplasm and did not localize to P-bodies (data not shown). GFP-Ge-1(1020–1401), which contains the last three ψ(X2–3)-repeat domains, localized to DCP1a-containing P-bodies (data not shown). GFP-Ge-1(936–1232), which contains the first three ψ(X2–3)-repeat domains, localized to cytoplasmic dots that appeared indistinguishable from P-bodies. However, these dots were distinct from DCP1a-containing P-bodies (Fig. 2, panel II-I–L).
The results of these studies show that Ge-1 residues 1020–1401, encompassing the last three ψ(X2–3)-repeat domains, are necessary and sufficient to direct this protein to DCP1a-containing P-bodies. The ψ(X2–3)-repeat domains in Ge-1 are distinct from previously described leucine-containing motifs, including the leucine zipper (LX6)3–6 and “leucine-rich repeats” (LX2LXLX2N/CXL) (for review, see Bornberg-Bauer et al. 1998; Kobe and Kajava 2001). Of note, none of the ψ(X2–3)-repeat domains in Ge-1 contain the leucine zipper or leucine-rich repeat consensus sequences. The ψ(X2–3)-repeat domains in Ge-1 are similar to the consensus sequence for a nuclear export sequence (NES; ψX2–3ψX2–3ψXψ). The third ψ(X2–3)-repeat domain in Ge-1 contains two potential NES, which may explain, in part, the predominant cytoplasmic location of Ge-1.
A smaller C-terminal region in Ge-1, which contains only the first three ψ(X2–3)-repeat domains, localized to cytoplasmic dots that were similar in appearance to, but clearly distinct from, DCP1a-containing P-bodies. The presence of these alternative cytoplasmic structures indicates that cytoplasmic dots may be heterogeneous. We speculate that the C-terminal portion of Ge-1 may serve to link two distinct cytoplasmic domains, one containing the decapping complex proteins, and the other containing as yet uncharacterized components.
The Ge-1 N-terminus, which contains seven WD40 domains, was not required for localization of the protein to P-bodies. It is of interest that the exosome, which is the protein complex that mediates 3′ → 5′ mRNA decay, also has a protein (Ski8p) that, like Ge-1, contains seven WD40 domains (Madrona and Wilson 2004). The crystal structure of Ski8p from Saccharomyces cerevisiae reveals a seven-bladed propeller, and this portion of Ski8p is likely to mediate interaction with exosome component Ski3p (Cheng et al. 2004). The WD40 domains in Ge-1, like those in Ski8p, are likely to recruit other protein components to P-bodies.
The relationship between Ge-1-containing P-bodies and stress granules
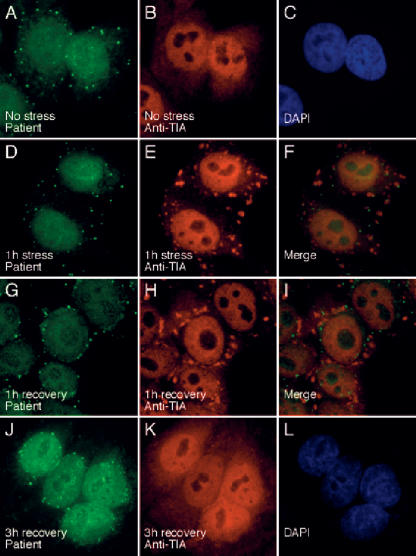
Stress granules (SGs) are cytoplasmic domains that form in response to environmental stress, and TIA is a marker for these cellular structures. To investigate the relationship between Ge-1-containing P-bodies and SGs, SGs were induced in Hep-2 cells using arsenite, and cells were subsequently fixed and stained with patient 0020 serum and anti-TIA antiserum. Prior to treatment with arsenite, TIA localized to nuclei and Ge-1 was detected in P-bodies that were distributed diffusely throughout the cytoplasm (Fig. 3A–C). After the cells were exposed to arsenite for 60 min, TIA was present in SGs, and Ge-1-containing P-bodies were detected adjacent to these structures (Fig. 3D–F). When cells were exposed to arsenite for 1 h and then permitted to recover for 1 h, TIA-containing SGs persisted in the cell cytoplasm, but many Ge-1-containing foci were detected in a perinuclear distribution. A few Ge-1-containing P-bodies remained adjacent to SGs (Fig. 3G–I). Three hours after treatment with arsenite, SGs were no longer present in the cytoplasm and TIA was again detected in nuclei. Ge-1-containing P-bodies, however, remained in a perinuclear distribution (Fig. 3J–L). Using rabbit anti-DCP1a antiserum, we observed that DCP1a also localized to the perinuclear region 1 and 3 h after arsenite treatment and co-localized with Ge-1 (data not shown). Twelve hours after arsenite-induced stress, Ge-1-and DCP1a-containing P-bodies were again distributed diffusely throughout the cytoplasm (data not shown).
FIGURE 3.
The effect of arsenite-induced oxidative stress on the cellular location of Ge-1. In resting Hep-2 cells, Ge-1-containing P-bodies were distributed throughout the cytoplasm (A), and TIA was detected diffusely throughout the nucleus, but excluded from nucleoli (B). After 1 h of exposure to sodium arsenite, TIA was detected in (E) cytoplasmic granules (red), and (D) Ge-1-containing P-bodies (green) were frequently detected adjacent to TIA-containing SGs (merge of D and E shown in F). After 1 h of exposure to arsenite and 1 h of recovery in culture medium, (H) TIA-containing SGs persisted in the cell cytoplasm (red). Ge-1-containing P-bodies, however, were detected in a (G) perinuclear location (green). After 1 h of stress and 3 h of recovery, TIA was detected in cell nuclei (K), and Ge-1-containing P-bodies persisted in a perinuclear distribution (J). DAPI staining was used to indicate the location of cell nuclei in C and L.
The significance of the changing cellular location of P-bodies during arsenite-induced stress is uncertain. It is possible that P-bodies localize to SGs, because these structures have similar functions: P-bodies, like SGs, may store housekeeping mRNAs in an inactive state during cellular stress, thereby permitting mRNAs encoding repair proteins to have access to the cell’s mRNA translation machinery. Alternatively, P-bodies may associate with SGs to facilitate degradation of mRNAs that have been irreversibly damaged by oxidative stress.
During the period of recovery from oxidative stress, TIA returns to the nucleus and Ge-1-containing P-bodies localize to the perinuclear domain. Possibly, perinuclear P-bodies are part of a counter-regulatory process: Late in the course of cellular recovery, stress-related proteins may no longer be needed by the cell. The active destruction of stress-related mRNAs in recovering cells may permit housekeeping mRNAs, which are released from SGs during the recovery period, to return to polysomes and resume active translation. In this model, perinuclear P-bodies would function to intercept and degrade stress-related mRNAs that continue to leave the nucleus after the recovery process is nearly complete.
Decreasing the cellular level of Ge-1 alters the structure of P-bodies
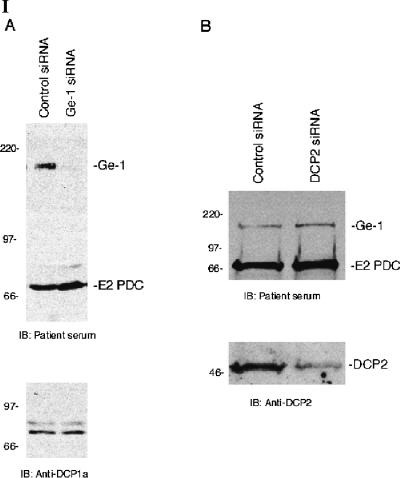
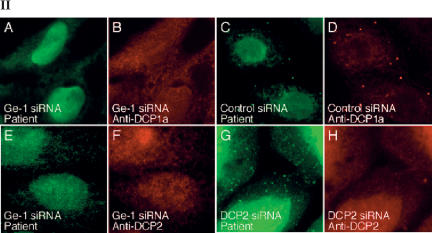
To investigate the effect of lowering the cellular level of Ge-1 on the composition of P-bodies, siRNAs specific for Ge-1 were introduced into Hep-2 cells. Ge-1 siRNA decreased Ge-1 mRNA to ~20% of control levels as determined by quantitative RT-PCR (data not shown) and decreased the level of Ge-1 protein in Hep-2 cellsasdeter-mined by immunoblot (Fig. 4, panel I-A). Note that the level of P-body component DCP1a was not altered by Ge-1 siRNA. On examination by indirect immunofluorescence, >80% of Ge-1 siRNA-transfected cells lacked Ge-1-containing P-bodies (Fig. 4, panel II-A,E). A second Ge-1 siRNA produced similar results (data not shown). Control siRNA did not decrease the cellular level of Ge-1 protein (Fig. 4, panel I-A) and did not alter Ge-1-containing P-bodies (Fig. 4, panel II-C). To determine the effect of decreasing Ge-1 on the cellular location of another P-body component, Ge-1 siRNA-transfected cells were stained for Ge-1 and DCP1a. In cells that lacked Ge-1, DCP1a was no longer detected in P-bodies. Similar results were observed when cells were stained for Ge-1 and DCP2 (Fig. 4, panel II-E–H).
FIGURE 4.
The effect of Ge-1 knock-down on P-bodies. (Panel I) Immunoblotting was used to determine the effect of siRNAs on the level of Ge-1, DCP1a, and DCP2 proteins in Hep-2 cells. Ge-1 siRNA decreased the level of Ge-1, but did not alter the level of E2 pyruvate dehydrogenase complex (E2 PDC) or DCP1a in Hep-2 cells (I-A). Note that in Hep-2 cells, DCP1a migrated as a doublet. DCP2 siRNA decreased DCP2 but did not alter the level of Ge-1 or E2 PDC (I-B). The levels of Ge-1, DCP1a, DCP2, and E2 PDC were unaffected by transfection with control siRNA. Ge-1 and E2 PDC were detected using patient serum. DCP1a and DCP2 were detected using rabbit anti-DCP1a and anti-DCP2 antiserum, respectively. (Panel II) After transfection of Ge-1 siRNA into Hep-2 cells, neither (II-A) Ge-1 (green) nor (II-B) DCP1a (red) were detected in P-bodies. Control siRNA did not alter the P-body location of either protein (II-C,D). Transfection of Ge-1 siRNA into Hep-2 cells also displaced DCP2 from P-bodies (II-E,F). After transfection of Hep-2 cells with DCP2 siRNA, (II-H) DCP2 (red) was no longer detected in P-bodies, but (II-G) Ge-1-containing P-bodies (green) persisted.
To examine the effect of lowering the cellular level of DCP2 on staining for Ge-1, DCP2 siRNA was transfected into cells. DCP2 siRNA decreased DCP2 mRNA to ~25% of control levels, as determined by quantitative RT-PCR (data not shown) and decreased the level of DCP2 protein as determined by immunoblot. The level of Ge-1 protein was not altered by exposure of cells to DCP2 siRNA (Fig. 4, panel I-B). Although DCP2-containing P-bodies were no longer detected in >80% of the cells, nearly all of the cells had Ge-1-containing P-bodies (Fig. 4, panel II-G,H). These results indicate that the P-body localization of decapping complex components DCP1a and DCP2 is dependent on Ge-1, but that the P-body localization of Ge-1 is not dependent on DCP2.
Previous investigators showed that the loss of an early component of the 5′ → 3′ mRNA decay pathway may lead to disruption of the entire structure. In contrast, interruption of later stages in the process resulted in accumulation of early components in P-body-like structures (Sheth and Parker 2003; Andrei et al. 2005). Thus, siRNA-mediated knock-down of components involved in the early steps of 5′ → 3′ mRNA degradation, including Lsm1 and Rck/p54, prevented the accumulation of other degradation factors in P-bodies (Yang et al. 2004b; Andrei et al. 2005). Inhibition of P-body components involved in later stages of mRNA degradation, such as DCP2 or XrnI, resulted in P-body accumulation of components involved in earlier stages, such as Lsm1 or Rck/p54 (Sheth and Parker 2003; Andrei et al. 2005). Because Ge-1 knock-down prevents DCP1a and DCP2 from accumulating in P-bodies, but DCP2 knockdown does not prevent Ge-1 accumulation in P-bodies, it seems likely that Ge-1 acts prior to decapping of mRNA and exonuclease activity.
The mechanism by which knock-down of Ge-1 results in loss of P-bodies is uncertain. One possibility is that P-bodies are physically unable to assemble in the absence of Ge-1. Alternatively, the lack of Ge-1 may alter the rate of mRNA degradation, thereby potentially favoring disassembly of these structures.
CONCLUSIONS
In this study, we identified Ge-1 as a component of mammalian P-bodies. The C-terminal ψ(X2–3)-repeat domains mediated targeting of Ge-1 to P-bodies. The N-terminal WD40 domains were not required for P-body localization and may recruit other proteins to these structures.
Ge-1 containing P-bodies localized adjacent to SGs in the setting of oxidative stress, but localized near the nuclear envelope during the recovery period. In this perinuclear location, P-bodies may participate in a counter-regulatory process that limits the cellular response to stress. Localization of DCP1a and DCP2 to P-bodies was dependent on the presence of Ge-1, but Ge-1-containing P-bodies were present in the absence of DCP2. These results suggest that Ge-1 may act before decapping and exonucleolytic activity in the 5′ → 3′ mRNA degradation pathway.
MATERIALS AND METHODS
Plasmids and antisera
A plasmid encoding Ge-1 was previously described (Bloch et al. 1994). A full-length cDNA encoding Ge-1 was ligated in pEGFP (Clontech) to produce a green fluorescent protein (GFP) fusion protein (GFP-Ge-1). The full-length cDNA was treated with XmaI, ligated into pEGFP, and used to produce GFP-Ge-1(113–1401). A plasmid encoding N-terminal Ge-1 amino acids 1–914 was prepared by treating GFP-Ge-1 with BamHI and religating the DNA, resulting in removal of DNA encoding Ge-1 amino acids 915–1401. To prepare a GFP-Ge-1 fusion protein that lacked the putative nuclear localization sequence (NLS), the BamHI fragment of Ge-1 encoding amino acids 936–1401 was ligated into the BamHI site of GFP-Ge-1(113–914). The resulting plasmid lacked DNA encoding amino acids 914–935 (GFP-Ge-1 [113–1401 ΔNLS]) and fortuitously recreated the open reading frame. A plasmid encoding Ge-1 amino acids 936–1401 fused to GFP was prepared by ligating the 3′-BamHI fragment of Ge-1 into pEGFP to produce GFP-Ge-1(936–1401). DNA fragments encoding Ge-1 amino acids 936–1116, 1136–1401, 1020–1401, and 936–1232 were prepared using oligonucleotides and PCR and were ligated into pEGFP to encode GFP-Ge-1 fragment fusion proteins.
EST clone KIAA1460, encoding GW182 amino acids 315–1709, was obtained from Kazusa DNA Research Institute and was ligated into pGEX (Pharmacia Biotech, Inc.) so as to encode GST-GW182(315–1709) fusion protein. Recombinant GST-GW182 was prepared in Escherichia coli as described (Smith and Johnson 1988). A plasmid encoding DCP1a fused to a Flag epitope was provided by J. Lykke-Andersen (University of Colorado, Boulder, CO).
Serum from a patient with PBC (patient 0020) was identified in a study to determine the clinical significance of autoantibodies in PBC (Yang et al. 2004a). Mouse anti-Flag antibodies were purchased from Sigma-Aldrich. Goat anti-TIA antiserum was obtained from Santa Cruz Biotechnology. Rabbit anti-GW182, anti-DCP2, and anti-DCP1a antisera were provided by M. Fritzler (University of Calgary, Calgary, Alberta, Canada), M. Kiledjian (Rutgers University, Piscataway, NJ), and J. Lykke-Andersen, respectively. FITC- or rhodamine-conjugated, species-specific antisera were obtained from Jackson ImmunoResearch Laboratories.
Cell culture and immunohistochemistry
Hep-2 cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in DMEM supplemented with 10% fetal calf serum, L-glutamine (2 mM), penicillin (200 U/mL), and streptomycin (200 μg/mL). To induce stress granule formation, Hep-2 cells were exposed to sodium arsenite (0.5 mM) for 1 h at 37 °C. For immunofluorescent staining, Hep-2 cells were grown in tissue culture chambers (Nunc Inc.), fixed with 4% paraformaldehyde in PBS, and permeabilized with methanol. Cells were stained with primary and secondary antisera as previously described (Bloch et al. 2005).
Expression library screening
Serum from patient 0020 was diluted 1:1000 in blotting solution (PBS containing 5% nonfat dry milk) and used to screen a λGT11 cDNA expression library prepared from HepG2 (human hepatocellular carcinoma) cells (Clontech) according to the method of Young and Davis (1983). Bound antibodies were detected using chemiluminescence. A single clone encoding immunoreactive protein was identified from among 1 million bacteriophages and was isolated by the method of plaque purification.
SDS-PAGE and immunoblotting
Hep-2 cell or E. coli protein extracts were fractionated in SDS-8% polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were incubated in blocking solution and then with human serum, rabbit anti-GW182, anti-DCP1a, or anti-DCP2 antiserum diluted in blotting solution. Bound human or rabbit antibodies were detected using HRP-conjugated goat anti-human or anti-rabbit antiserum (Jackson ImmunoResearch) and chemiluminescence.
Mammalian cell transfection
To identify the portion of Ge-1 that mediates localization to P-bodies, plasmids encoding GFP fused to portions of Ge-1 were transfected into Hep-2 cells using the Effectene transfection system (QIAGEN) as directed by the manufacturer. Cells were fixed and stained 24 h after transfection.
siRNAs were used to decrease the level of Ge-1 and DCP2 in Hep-2 cells. siRNAs for Ge-1, DCP2, and controls were obtained from Ambion Inc. (RCD-8/Ge-1, cat# 21122; DCP2, cat# 16708; and control, cat# 4605). A second Ge-1 siRNA was obtained from Dharmacon Research Inc. (RCD-8/Ge-1, cat# D-016635-01). Double-stranded siRNA was transfected into Hep-2 cells using oligofectamine (Invitrogen) as directed by the manufacturer, and cells were fixed and stained 48 h after transfection.
Acknowledgments
The authors thank M. Kiledjian and M. Fritzler for providing anti-DCP2 and anti-GW182 antiserum. J. Lykke-Andersen provided rabbit anti-DCP1a antiserum and a plasmid encoding Flag-tagged DCP1a. The authors also thank P. Anderson, N. Kadersha, and K.J. Bloch for advice. K.D.B. was supported by grants from the National Institutes of Health (HL070896 and HL074352). D.B.B. was supported by grants from the Arthritis Foundation, the William F. Milton Fund, and the National Institutes of Health (DK051179). D.B.B. is a recipient of an Established Investigator Grant from the American Heart Association.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2142405.
Footnotes
Patient Ge was initially believed to have Sjögren’s syndrome. She was subsequently found to have abnormal liver function tests and anti-mitochondrial antibodies, and the diagnosis of primary biliary cirrhosis was established.
In the original report, a sequencing error at nt 3671 resulted in a frameshift and failure to appreciate the C-terminal 213 amino acids of Ge-1.
REFERENCES
- Andrei, M.A., Ingelfinger, D., Heintzmann, R., Achsel, T., Rivera-Pomar, R., and Luhrmann, R. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, D.B., Rabkina, D., Quertermous, T., and Bloch, K.D. 1994. The immunoreactive region in a novel autoantigen contains a nuclear localization sequence. Clin. Immunol. Immunopathol. 72: 380–389. [DOI] [PubMed] [Google Scholar]
- Bloch, D.B., Yu, J.H., Yang, W.H., Graeme-Cook, F., Lindor, K.D., Viswanathan, A., Bloch, K.D., and Nakajima, A. 2005. The cytoplasmic dot staining pattern is detected in a subgroup of patients with primary biliary cirrhosis. J. Rheumatol. 32: 477–483. [PubMed] [Google Scholar]
- Bornberg-Bauer, E., Rivals, E., and Vingron, M. 1998. Computational approaches to identify leucine zippers. Nucleic Acids Res. 26: 2740–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret, E., Rigaut, G., Shevchenko, A., Wilm, M., and Seraphin, B. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19: 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Liu, Y., Wang, C., Parker, R., and Song, H. 2004. Crystal structure of Ski8p, a WD-repeat protein with dual roles in mRNA metabolism and meiotic recombination. Protein Sci. 13: 2673–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J. and Parker, R. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73: 861–890. [DOI] [PubMed] [Google Scholar]
- Coller, J.M., Tucker, M., Sheth, U., Valencia-Sanchez, M.A., and Parker, R. 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7: 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., and Seraphin, B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy, T., Chan, E.K., Takeuchi, K., Mahler, M., Luft, L.M., Zochodne, D.W., and Fritzler, M.J. 2003a. Clinical and serological associations of autoantibodies to GW bodies and a novel cytoplasmic autoantigen GW182. J. Mol. Med. 81: 811–818. [DOI] [PubMed] [Google Scholar]
- Eystathioy, T., Jakymiw, A., Chan, E.K., Seraphin, B., Cougot, N., and Fritzler, M.J. 2003b. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lozano, J.R., Gonzalez-Escribano, M.F., Wichmann, I., and Nunez-Roldan, A. 1997. Cytoplasmic detection of a novel protein containing a nuclear localization sequence by human autoantibodies. Clin. Exp. Immunol. 107: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, M.M. 1996. Primary biliary cirrhosis. N. Engl. J. Med. 335: 1570–1580. [DOI] [PubMed] [Google Scholar]
- Kedersha, N. and Anderson, P. 2002. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30: 963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fitzler, M.J., Scheuner, D., Kaufman, R.J., Golan, D.E., and Anderson, P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B. and Kajava, A.V. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11: 725–732. [DOI] [PubMed] [Google Scholar]
- Madrona, A.Y. and Wilson, D.K. 2004. The structure of Ski8p, a protein regulating mRNA degradation: Implications for WD protein structure. Protein Sci. 13: 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R. and Song, H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11: 121–127. [DOI] [PubMed] [Google Scholar]
- Sheth, U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.B. and Johnson, K.S. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40. [DOI] [PubMed] [Google Scholar]
- Smith, T.F., Gaitatzes, C., Saxena, K., and Neer, E.J. 1999. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185. [DOI] [PubMed] [Google Scholar]
- Wilczynska, A., Aigueperse, C., Kress, M., Dautry, F., and Weil, D. 2005. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 118: 981–992. [DOI] [PubMed] [Google Scholar]
- Yang, W.H., Yu, J.H., Nakajima, A., Neuberg, D., Lindor, K., and Bloch, D.B. 2004a. Do antinuclear antibodies in primary biliary cirrhosis patients identify increased risk for liver failure? Clin. Gastroenterol. Hepatol. 2: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Jakymiw, A., Wood, M.R., Eystathioy, T., Rubin, R.L., Fritzler, M.J., and Chan, E.K. 2004b. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 117: 5567–5578. [DOI] [PubMed] [Google Scholar]
- Young, R.A. and Davis, R.W. 1983. Efficient isolation of genes by using antibody probes. Proc. Natl. Acad. Sci. 80: 1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]