Abstract
The cellular polypyrimidine tract-binding protein (PTB) is recruited by the genomic RNAs of picornaviruses to stimulate translation initiation at their internal ribosome entry site (IRES) elements. We investigated the contribution of the individual RNA recognition motif (RRM) domains of PTB to its interaction with the IRES of foot-and-mouth disease virus (FMDV). Using a native gel system, we found that PTB is a monomer, confirming recent reports that challenged the previous view that PTB is a dimer. Mapping the spatial orientation of PTB relative to the bound IRES RNA, we found that the two C-terminal RRM domains III and IV of PTB bind in an oriented way to the IRES. Domain III contacts the IRES stem-loop 2, while domain IV contacts the separate IRES 3′ region. PTB domain I appears not to be involved directly in RNA binding, but domain II stabilizes the RNA binding conferred by domains III and IV. A PTB protein containing only these two C-terminal PTB domains is sufficient to enhance the entry of initiation factor eIF4G to the IRES and stimulate IRES activity, and the long-lived PTB–IRES interaction stabilized by domain II is not a prerequisite for this function. Thus, PTB most likely acts as an RNA chaperone to stabilize IRES structure and, in that way, augment IRES activity.
Keywords: translation, internal ribosome entry site, IRES, picornavirus, PTB, monomer, dimer
INTRODUCTION
The polypyrimidine tract-binding protein (PTB) is a cellular RNA-binding protein that is abundant in many tissues. PTB binds to oligopyrimidine tracts in introns and regulates negative or positive exon definition in differential splicing of various cellular mRNAs, like that for tropomyosin, α-actinin, the proto-oncogene c-src, a GABA receptor subunit, calcitonin/calcitonin gene-related peptide, and fibroblast growth factor-R2 (for reviews, see Valcárcel and Gebauer 1997; Wagner and Garcia-Blanco 2001; Spellman et al. 2005). More recently PTB was described to be also involved in other aspects of RNA metabolism, like the modulation of polyadenylation efficiency (Castelo-Branco et al. 2004) and the expression of nitric oxide synthase mRNA during inflammation (Soderberg et al. 2002). Even a sex-specific role of PTB in the male germline of Drosophila flies was reported (Robida and Singh 2003). Activated cyto-solic PTB appears to be involved in the nutrient-dependent stabilization of mRNAs involved in insulin synthesis and secretion (Knoch et al. 2004; Tillmar and Welsh 2004), and the hormonally regulated cellular protein kinase A promotes nucleocytoplasmic relocalization of PTB (Xie et al. 2003), suggesting a direct role of PTB in the regulation of glucose metabolism.
Moreover, PTB represents the prototype of a heterogeneous group of cellular RNA-binding proteins that are directly involved in the regulation of translation. This function of PTB was discovered when the translation of picornaviruses was investigated (Jang and Wimmer 1990; Luz and Beck 1990). After infection of a cell, the RNA genomes of these small nonenveloped positive-strand RNA viruses are directly used for translation of the viral polyprotein. In contrast to normal cellular mRNAs, recruitment of ribosomes to picornavirus RNAs is facilitated by an internal region of the viral RNA, the internal ribosome entry site (IRES) (Jackson and Kaminski 1995; Niepmann 1999; Ehrenfeld and Teterina 2002). Also the translation of Hepatitis C virus (HCV) and pestiviruses as well as several cellular mRNAs is driven by IRES elements (Hellen and Sarnow 2001).
Picornavirus IRES elements are large cis-acting RNA regions with highly conserved secondary structures that guide the small ribosomal subunit to the starting window at the 3′ border of the IRES, an internal site of the viral RNA that contains an AUG triplet (Pilipenko et al. 1994). According to their sequences and secondary structures, picornaviral IRES elements are classified in three groups, the type I elements of the entero-/rhinovirus group (including poliovirus), the type II elements of the cardio-/aphthovirus group (including foot-and-mouth disease virus, FMDV), and the type III element of hepatitis A virus (HAV) (Jackson and Kaminski 1995).
Ribosome binding to the picornavirus IRES elements is mediated by a number of cellular RNA-binding proteins (Jackson 2002) that fall in two groups. On one hand, all standard eukaryotic translation initiation factors (eIFs) are involved in picornavirus translation, except from the cap-binding protein eIF4E (Pestova et al. 1996). In the 3′ region of the type II IRES elements of FMDV and the related encephalomyocarditis virus (EMCV), these initiation factors bind to a large Y-shaped RNA structure (see Fig. 1A) upstream of the starting window. This IRES stem-loop 4, in particular its subdomain 4-1 (or J in EMCV) and the base and bulge of the stem-loop 4 bind eIF4B (Meyer et al. 1995; Rust et al. 1999) as well as eIF4G and eIF4A (Pestova et al. 1996; Kolupaeva et al. 1998; Lopez de Quinto and Martinez-Salas 2000; Saleh et al. 2001). Within stem-loop 4, a conserved element is essential for initiation factor binding and translation (Clark et al. 2003; Bassili et al. 2004). Also, the corresponding domain V of the type I IRES of polio-virus binds eIF4B and eIF4G (Ochs et al. 2002, 2003).
FIGURE 1.
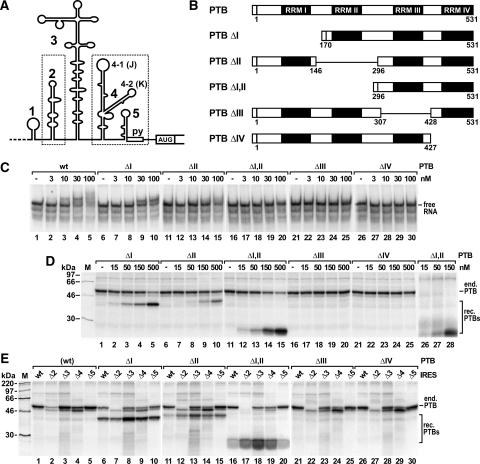
The contribution of PTB domains to strength and specificity of the PTB–IRES interaction. (A) The FMDV IRES with stem-loops 1–5, the oligopyrimidine tract (py), and the first authentic start codon. Dotted boxes mark the two separate PTB-binding regions. (B) The PTB protein with its four RNA-recognition motif (RRM) domains I–IV. Mutants are shown with the amino acid numbers flanking the deletions. The small N-terminal box represents the His6-tag. (C) Electrophoretic mobility shift assays with [α32P]-UTP-labeled FMDV wild-type IRES and the indicated nanomolar concentrations of wild-type (wt) PTB or PTB deletion mutant proteins. (D) The strength of binding of mutant PTB proteins to the FMDV IRES was analyzed in competition UV cross-link assays with rabbit reticulocyte lysate (RRL) containing endogenous PTB (end. PTB) plus increasing concentrations of mutant recombinant PTB proteins (rec. PTBs) as indicated. In lanes 26–28, RRL depleted of endogenous PTB was used. Molecular masses of marker proteins (M) are indicated. (E) The binding specificity of mutant PTB proteins (300 nM) was analyzed in competition UV cross-link assays with RRL and FMDV IRES deletion mutants as indicated. In lanes 1–5, only the endogenous wild-type PTB was analyzed for comparison.
On the other hand, picornavirus IRES RNAs recruit a variety of cellular RNA-binding proteins like PTB that are usually not involved in translation. Poliovirus IRES activity is enhanced by La protein (Meerovitch et al. 1993), poly(rC)-binding protein 2 (PCBP2) (Blyn et al. 1997), and the protein encoded by the gene upstream of N-ras (UNR) (Boussadia et al. 2003), as well as by PTB (Gosert et al. 2000). In the brain, lower levels of PTB may contribute to the attenuation of the poliovirus Sabin strains (Guest et al. 2004) in addition to the impaired binding of initiation factors (Ochs et al. 2003). Rhinovirus IRES activity responds to the synergistic action of UNR and PTB (Hunt et al. 1999). PTB enhances translation driven by the IRES elements of FMDV (Niepmann 1996; Niepmann et al. 1997), EMCV (Kaminski et al. 1995; Kaminski and Jackson 1998) and HAV (Gosert et al. 2000), and a 45-kDa IRES-interacting translation factor (ITAF45) additionally acts on the FMDV IRES (Pilipenko et al. 2000). Also, the activity of certain cellular IRES elements is regulated by PTB, like the IRES in the mRNA for apoptotic protease-activating factor 1, Apaf-1 (Mitchell et al. 2003), and the Bag-1 IRES (Pickering et al. 2004).
In the interaction of PTB with a type II picornavirus IRES element like the FMDV IRES, two binding partners interact with each other, an internal segment of a viral RNA and a cellular RNA-binding protein, each of which provides multiple contact points for the interaction. The FMDV IRES has two separate PTB-binding regions (Fig. 1A). A strong PTB-binding site is stem-loop 2 in the upstream part of the IRES (Luz and Beck 1991); several unpaired pyrimidine residues distributed over almost the entire stem-loop are contacted by PTB. The separate downstream PTB-binding region comprises various dispersed contact points, including the apical loop of stem-loop 4-2 (Kolupaeva et al. 1996; Rust et al. 1999), stem-loop 5, and the oligopyrimidine tract (Luz and Beck 1990, 1991; Luz 1991; Pilipenko et al. 2000).
The other binding partner, PTB, has four RNA recognition motif (RRM) domains (Gil et al. 1991; Kenan et al. 1991; Patton et al. 1991; Ghetti et al. 1992; Fig. 1B), a fact that may be one reason for the versatility of its interactions with RNA. Two of these domains, III and IV, had been reported earlier to be the major RNA-binding domains of PTB (Perez et al. 1997; Oh et al. 1998), whereas more recently also RNA binding by the N-terminal domains of PTB was reported (Simpson et al. 2004; Amir-Ahmady et al. 2005). Although a selection approach identified a pyrimidine-rich consensus sequence for PTB with a slight bias toward C residues interspersed with U and G (Singh et al. 1995), other studies observed binding of PTB to U-rich RNA sequences interspersed with C residues (Wollerton et al. 2001; Yuan et al. 2002; Amir-Ahmady et al. 2005). In picornavirus IRES elements, PTB-binding sites often contain either unstructured oligopyrimidine stretches or stem-loops with a UCUU or related motifs in the apical loop or other exposed single-stranded regions (Luz 1991; Kolupaeva et al. 1996).
Even though the general structure of all four RRM domains is supposed to be similar (Kenan et al. 1991), RRM domains II and III of PTB differ from previously described RRM domains by each having an additional fifth β-sheet (Conte et al. 2000; Simpson et al. 2002, 2004; Yuan et al. 2002). RRM domains I and II presumably also serve functions different from RNA binding since they were found to interact with other proteins involved in RNA metabolism, hnRNP L, hnRNP K, and hnRNP E2 (Hahm et al. 1998; Kim et al. 2000). However, the previous view that RRM domain II of PTB mediates self-interaction and promotes PTB dimerization (Perez et al. 1997; Oh et al. 1998; Kim et al. 2000) was challenged more recently (Simpson et al. 2004; Amir-Ahmady et al. 2005).
In this study, we identified the role of the individual domains of PTB and of the IRES in the PTB–IRES interaction and their spatial orientation relative to each other in the PTB–IRES complex. In addition, we analyzed the contribution of each of the PTB domains to the ability of PTB to enhance FMDV IRES activity as well as the possible self-interaction properties of PTB using a new native gel system.
RESULTS
Analysis of the PTB–IRES interaction
For analyzing the contribution of the individual RRM domains of PTB to the PTB–IRES interaction, we used PTB mutant proteins each lacking only a single RRM domain (Fig. 1B). The remaining parts of the protein most likely assume their correct tertiary structure, since partial PTB proteins containing one or two of the C-terminal domains or the two N-terminal domains correctly assume their native RRM domain structure (Conte et al. 2000; Simpson et al. 2002; Yuan et al. 2002), and also single RRM domains of other proteins fold properly (Nagai et al. 1990; Fleming et al. 2003).
Using these PTB proteins, were analyzed the strength and specificity of the PTB–IRES interaction by two different assays. Both the electrophoretic mobility shift assay (EMSA) (Fig. 1C) and the UV cross-link assay (Fig. 1D, E) detect direct interactions between native protein and native RNA, but each assay has particular advantages and disadvantages. In the shift assay, the protein–RNA interaction is monitored approximately quantitatively and allows a rough estimation of dissociation constants. However, successful detection requires that the interaction of a given protein molecule with a given RNA molecule be stable enough to withstand both the incubation step and the gel electrophoresis. In contrast, in the UV cross-link assay even very short-lived binding events are captured by UV-induced covalent bonds between protein and RNA. However, only 5%–10% of the interactions are covalently fixed by the UV light, while all other interactions are disrupted when loading the samples to a denaturing protein gel.
PTB domain II increases the stability of RNA binding by domains III and IV
In the shift assay (Fig. 1C), interaction of full-length PTB with the FMDV IRES was detected at a PTB concentration as low as 3 nM (Fig. 1C, lane 2). At 10 nM about 20% (Fig. 1C, lane 3), and at 30 nM, nearly 50% of the IRES RNA was present in PTB–IRES complexes (Fig. 1C, lane 4). Accordingly, the dissociation constant of the PTB–IRES interaction can be estimated to be about 30 nM. In previous studies, the Kd of the interaction of PTB with the FMDV IRES was reported to be about 60 nM, and that with the closely related EMCV IRES about 40 nM (Witherell et al. 1993; Conte et al. 2000), while the concentration of PTB required for half-maximal stimulation of FMDV IRES-directed translation was about 20 nM (Niepmann 1996; Niepmann et al. 1997). Consequently, the shift assay can be regarded as a reasonable system for monitoring the strength of the PTB–IRES interaction.
With PTBΔI (Fig. 1C, lanes 6–10), the protein concentration required for 50% RNA binding rose to a value between 30 and 100 nM. Thus, domain I of PTB contributes only very slightly to the PTB–IRES interaction, either directly or indirectly. When domain II was absent from PTB, a more remarkable decrease in affinity was observed (Fig. 1C, lanes 11–15). Only at high PTB concentrations was a diffuse retardation of the RNA detected. This indicates that domain II contributes to the stability of the PTB–IRES interaction either by directly binding to the RNA or by indirectly increasing the stability of the interaction of domains III and/or IV with the RNA. When both domains I and II were absent from PTB, no RNA–protein complexes were detected in the shift assay (Fig. 1C, lanes 16–20). Also, with the domain deletion mutants PTBΔIII and ΔIV, no RNA–protein complexes were detected (lanes 21–30).
In addition to the shift assay, a competition UV cross-link assay was used (Fig. 1D). The mutant PTB proteins were used to compete for the interaction of the wild-type PTB endogenously contained in rabbit reticulocyte lysate (RRL) with the FMDV IRES. This procedure makes the assay completely independent of the aspect if the binding of a mutant PTB protein in question to the IRES is captured by formation of covalent bonds by UV light or not. As long as a variant PTB protein would bind to the RNA it would definitely sequestrate the RNA from binding to the endogenous PTB, regardless of whether the efficiency of label transfer from RNA to protein in the UV cross-link assay may vary among different PTB deletion mutants or even may not at all result in the formation of covalent bonds. In this competition assay, high concentrations of PTBΔI compete well with the interaction of the endogenous PTB (Fig. 1D, lanes 1–5). In contrast, PTBΔII competes more weakly, indicating that the absence of domain II results in a significant loss of affinity to the FMDV IRES. However, to our surprise, the deletion of both domains I and II resulted in strong competition of the interaction of endogenous PTB with the IRES (as well as in strong labeling of the mutant protein) (Fig. 1D, lanes 11–15). Taken together with the above results, this indicates that PTBΔI,II strongly interacts with the FMDV IRES, but the interaction is short-lived and thus can not be detected in the shift assay. In contrast, the PTBΔIII and ΔIV proteins neither bind to the FMDV IRES in the UV cross-link assay nor compete binding of the endogenous full-length PTB (Fig. 1D, lanes 16–25). This rules out that the loss of detection of binding is just due to the loss of radioactive label transfer from RNA to protein and confirms that these two mutant PTB proteins indeed do not bind to the IRES at all.
Mainly domain II, but also domain I, contributes to the specificity of binding to the FMDV IRES
For analyzing the contribution of the individual PTB domains to the specificity of the PTB–IRES interaction, we monitored the ability of the PTB deletion mutants to compete with the wild-type PTB contained endogenously in the reticulocyte lysate for the binding to different FMDV IRES mutants (Fig. 1E). As a control, the binding of endogenous wild-type PTB to these IRES mutants is shown in the absence of competition (Fig. 1E, lanes 1–5). In some cases, an unknown protein migrating slightly faster than PTB binds unspecifically to some of the IRES deletion variants to which PTB binding is reduced, probably due to the exposure of RNA regions that are otherwise covered by PTB in the wild-type IRES.
Confirming our previous data (Luz and Beck 1991; Niepmann 1996), PTB binds fairly to IRES Δ2 since stem-loop 2 is the strongest PTB-binding site in the FMDV IRES (Fig. 1E, lane 2). Also, stem-loop 4 contributes slightly to the interaction (Fig. 1E, lane 4), whereas stem-loop 3 is not required for PTB binding (Fig. 1E, lane 3). The absence of PTB domain I results in significant reduction of binding specificity (Fig. 1E, lanes 6–10). In particular, PTBΔI binds to IRES Δ2, whereas the endogenous full-length PTB does not bind to this RNA under these competitive conditions (Fig. 1E, lane 7). Also, the other IRES deletion mutants are bound better by PTBΔI compared to the endogenous full-length PTB (Fig. 1E, lanes 8–10). An even greater reduction in binding specificity is observed with PTBΔII (Fig. 1E, lanes 11–15), which is nearly unable to compete the binding of wild-type PTB to the wild-type IRES but binds to IRES Δ2. Similar results were obtained with PTBΔI,II (Fig. 1E, lanes 16–20). Binding of the unknown protein migrating faster than PTB is competed well by PTBΔI,II. In contrast, PTBΔIII and ΔIV do not bind to the FMDV IRES. Moreover, they do not compete with binding of the endogenous wild-type PTB to the IRES (Fig. 1E, cf. lanes 21,26 and lane 6), confirming that the lack of detection is actually due to the loss of binding rather than only due to the loss of radioactive label transfer from RNA to protein in the UV cross-link assay. Most likely for the same reason, binding of the unspecifically binding protein is not competed.
In conclusion, mainly PTB domains III and IV serve to actually bind the IRES RNA, while domain II and to a minor extent also domain I contribute to the ability of PTB to specifically recognize the IRES RNA either indirectly or by direct RNA binding.
PTB is a monomer
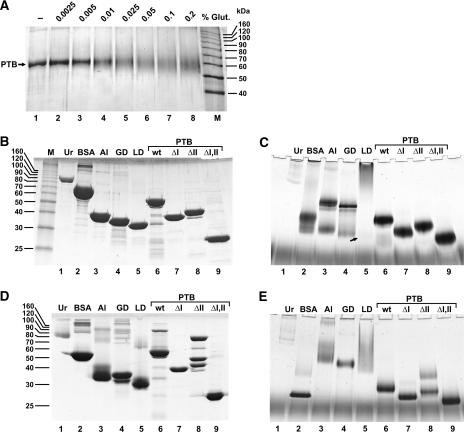
In the following, we investigated the details of the spatial arrangement of the domains of the PTB molecule bound to the IRES. Since PTB had been reported previously to dimerize (Perez et al. 1997; Oh et al. 1998), we first analyzed a possible self-interaction of PTB. When wild-type PTB was incubated in the presence of glutardialdehyde to chemically cross-link the monomers of possible PTB dimers, we were unable to detect a band at the expected dimer position of about 120 kDa even at high glutardialdehyde concentrations. In contrast, severe intramolecular cross-linking of PTB resulted in a diffuse band at the PTB monomer position (Fig. 2A).
FIGURE 2.
Analysis of the oligomerization status of PTB. (A) Chemical cross-linking of PTB. One microgram (1 μg) of wild-type PTB was incubated for 10 min either without or with different concentrations of glutardialdehyde (Glut.) as indicated and analyzed under reducing conditions on an SDS–12% polyacrylamide gel. M, Marker proteins. (B) Analysis of PTB and marker proteins on a standard Laemmli SDS–5%–40% polyacrylamide gel under reducing conditions. One microgram (1 μg) of urease and 6 μg of each of the other proteins were loaded. Marker proteins: Ur, urease; BSA, bovine serum albumin; Al, aldolase; GD, GAPDH; LD, LDH. Positions of size marker proteins are given in kilodaltons on the left. (C) Analysis of wild-type and mutant PTB proteins and marker proteins (as in B) by native gel electrophoresis. Proteins were preincubated for 1 h at room temperature under reducing conditions (10 mM DTT). (D) Analysis of proteins on Laemmli gels as in B but under nonreducing conditions. (E) Analysis of proteins on native gels as in C but under nonreducing conditions.
Then we investigated the migration of wild-type PTB and its deletion mutant variants in question, i.e., the PTB proteins lacking RRM domains I and II either separately (PTBΔI and PTBΔII) or in combination (PTBΔI,II), together with different marker proteins by gel electrophoresis. First, these proteins were analyzed on a standard Laemmli gel (Laemmli 1970) under denaturing and reducing conditions, i.e., boiling of the samples in the presence of 0.1% SDS and 2-mercaptoethanol before gel loading (Fig. 2B). Under these conditions, all proteins migrate approximately at their expected monomer positions, except a minor subfraction of BSA, which was purchased as a mixture of monomers and chemically cross-linked dimers (or oligomers, respectively) (Fig. 2B, lane 2). The urease monomer (Ur) migrates at 90 kDa (Fig. 2B, lane 1), the BSA monomer at 66 kDa (Fig. 2B, lane 2), fructose-1,6-bisphosphate aldolase (Al) at 39 kDa (Fig. 2B, lane 3), glyceralde-hyde-3-phosphate dehydrogenase (GAPDH, GD) at 36 kDa (Fig. 2B, lane 4), and lactate dehydrogenase (LDH, LD) at 32 kDa (Fig. 2B, lane 5). Also, the PTB proteins migrate approximately at their expected sizes, the his-tagged wild-type PTB at 59.1 kDa (Fig. 2B, lane 6), PTBΔI at 40.7 kDa (Fig. 2B, lane 7), and PTBΔII at 43 kDa (Fig. 2B, lane 8). In particular, the 59-kDa wild-type PTB migrates faster than the 66-kDa BSA under these reducing conditions, and the 40.7-kDa PTBΔI migrates marginally slower than the 39-kDa aldolase. Only the 23.7-kDa PTBΔI,II migrates a little slower than expected (Fig. 2B, lane 9), perhaps due to the high relative content of basic amino acids also contained in the His6-tag.
To investigate the size of wild-type PTB and its deletion mutant variants in comparison to the marker proteins under native conditions (i.e., in the absence of denaturing reagents like SDS), we used a newly developed focusing alkaline native gel-electrophoresis system that is also suitable for the analysis of basic proteins. On these gels, the proteins were analyzed after 1 h preincubation under reducing conditions (Fig. 2C). In all cases, the PTB proteins appear approximately at their expected monomer sizes relative to each other and in relation to the marker proteins (Fig. 2, cf. B and C), while no dimer or oligomer bands of wild-type PTB or its deletion mutant variants were detected. In contrast, the marker proteins aldolase (Fig. 2C, lane 3) and GAPDH (Fig. 2C, lane 4) form oligomers that are supposed to be tetramers (Fox and Dandliker 1956; Beernink and Tolan 1994) with molecular masses of 160 kDa and 145 kDa, respectively. The appearance of these tetramers demonstrates that this gel system is suitable to detect the noncovalent interactions between the monomer subunits of well characterized oligomeric proteins. Moreover, small amounts of these proteins appear at their monomer positions of 39 kDa (aldolase; Fig. 2C, lane 3) and 36 kDa (GAPDH; Fig. 2C, lane 4), and the relative positions of the tetramers and monomers of these marker proteins relative to the PTB proteins demonstrate that this gel system is suitable to separate proteins according to their sizes. In contrast to aldolase and GAPDH, most BSA molecules migrate as monomers (Fig. 2C, lane 2), while small amounts of cross-linked BSA are visible as a diffuse band at higher molecular weights (Fig. 2, cf. B, lane 2 and C, lane 2). The fastest urease band (Fig. 2C, lane 1) migrates considerably slower than the aldolase tetramers of 160 kDa and most likely represents the 272-kDa trimer, and also higher molecular weight complexes (Fishbein et al. 1970) appear. LDH appears to form higher molecular weight complexes (Millar 1962) with only minute amounts of monomers appearing at the position of 32 kDa (small arrow in Fig. 2C, lane 5). Upon low pH treatment, all these proteins completely dissociate into their subunits, which then migrate exactly according to their monomer sizes relative to each other and to the PTB proteins in the native gel (data not shown).
Taken together, no oligomers of PTB are detectable under reducing conditions (as present in the cytosol of living cells) in a native gel system that allows formation of tetramers of well-characterized oligomeric proteins.
When these proteins were analyzed on a Laemmli gel under nonreducing conditions (Fig. 2D), some noticeable changes were detected. BSA migrates considerably faster (Fig. 2D, lane 2), since complete unfolding of BSA by SDS under nonreducing conditions is most likely prevented by disulfide bonds similar to the 17 disulfide bonds in the compact structure of human serum albumin (He and Carter 1992). Similarly, almost all molecules of aldolase, which contains eight cysteine residues (Fig. 2D, lane 3), and LDH, which contains four cysteine residues (Fig. 2D, lane 5), as well as a subfraction of GAPDH molecules that contains five cysteine residues (Fig. 2D, lane 4) migrate faster. Interestingly, while most molecules of wild-type PTB migrate at the expected monomer position as before, small amounts of PTB are detected at a position expected for a dimer (Fig. 2D, lane 6). Thus, a minor fraction of PTB molecules appears to be linked by intermolecular disulfide bonds. Cysteine residues possibly involved are present only in RRM domain I (amino acid position 23) and in domain II (positions 250, 251). With PTBΔI no dimers were detected (Fig. 2D, lane 7), whereas a strong dimer band was detected in addition to the monomer with PTBΔII (Fig. 2D, lane 8), indicating that cysteine residue 23 in RRM domain I appears to be involved in formation of disulfide bonds that can link two PTB monomers to form a dimer under nonreducing conditions. Moreover, a third band appeared with PTBΔII at about 50 kDa. We can only speculate whether this band represents a conformation variant of dimeric (or monomeric) PTBΔII. In contrast, with PTBΔI,II no band appeared in addition to the monomer (Fig. 2D, lane 9).
Analysis of the proteins on a native gel under nonreducing conditions shows that BSA migrates faster also in the native gel (Fig. 2E) compared to its migration after reducing treatment (Fig. 2C) according to its compact structure stabilized by disulfide bonds (He and Carter 1992). Aldolase (Fig. 2E, lane 2) and GAPDH (Fig. 2E, lane 3) completely form tetramers, and also LDH forms oligomers starting from the expected tetramer position (Fig. 2E, lane 5). In contrast, even under these nonreducing conditions virtually no dimers or oligomers of PTB are visible (Fig. 2E, lane 6). Only in some experiments were minute amounts of wild-type PTB dimers detected in the native gels under nonreducing conditions (not shown). With PTBΔII, an oligomer band appeared under nonreducing conditions (lane 8), and minute amounts of PTBΔI form dimers (Fig. 2E, lane 7). In conclusion, mainly cysteine residue 23 in RRM domain I may lead to a certain extent of artificial cross-linking of PTB monomers by disulfide bonds in the absence of domain II, but virtually not in the full-length PTB protein.
Taken together, PTB is a monomer under redox conditions similar to those present in the cytosol of living cells as well as under nonreducing conditions.
The PTB molecule contacts both separate binding regions in the FMDV IRES
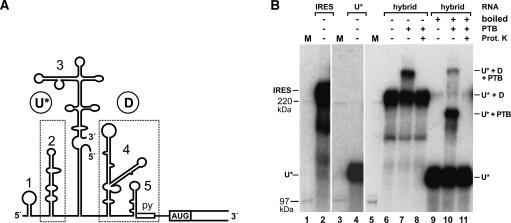
To elucidate the spatial arrangement of the PTB molecule bound to the IRES, we next considered whether separate domains of the PTB molecule would simultaneously bind to separate regions in the IRES and thus connect both binding sites in the IRES. To answer this question, we used an artificial IRES in which the two PTB-binding sites were provided by two different molecules of an IRES hybrid (Fig. 3A). In this composite IRES, the radiolabeled upstream RNA (RNA U*; 435 bases) provides the upstream PTB-binding site, stem-loop 2, while the downstream PTB-binding region is provided by the unlabeled RNA D (317 bases). To allow efficient RNA heteroduplex formation in the hybrid, the sequences in RNA D corresponding to the right base of stem-loop 3, which is not involved in PTB binding, were slightly modified.
FIGURE 3.
The PTB molecule contacts both separate binding regions (boxed in A) in a bipartite FMDV IRES hybrid. (A) Structure of the hybrid IRES. Two RNAs were synthesized. One [α-32P]-UTP-labeled RNA (named RNA U* for upstream) comprises the upstream half of the IRES including three-quarters of the stem-loop 3 sequence. The second, unlabeled RNA (named RNA D for downstream) starts in the remaining quarter of the stem-loop 3 sequence and thus includes the downstream half of the IRES. In the template for transcription of RNA D, the sequence representing the lower stem of stem-loop 3 was slightly modified to allow the formation of a continuous 39 bp heteroduplex of RNA D with RNA U*. Both RNAs were gel-purified after synthesis, hybridized, and the hybrid again gel-purified. (B) UV cross-link with the bipartite IRES hybrid and PTB protein added as indicated. In lanes 6–8, the reactions were loaded to the SDS–5% polyacrylamide gel without boiling, in lanes 9–11 after boiling. Samples in lanes 8 and 11 were treated with proteinase K after UV cross-linking. The asterisk indicates that only RNA U* is labeled. For comparison, a continuous IRES RNA of the same length as the hybrid IRES was loaded in lane 2, and RNA U* alone was loaded in lane 4 to compare RNA migration relative to the 14C-labeled marker proteins (lanes 1,3,5). The interpretation of bands (as discussed in the text) is indicated on the right.
With this hybrid IRES, UV cross-link reactions were performed with or without recombinant wild-type PTB. In a new approach to analyze RNA–protein complexes, the hybrid IRES and IRES–PTB complexes were then resolved on a standard protein gel with low acrylamide concentration (Fig. 3B). When the RNA was not boiled before gel loading, the hybrid IRES (U* + D) migrated as a strong band of about 230 kDa apparent molecular mass (Fig. 3B, lane 6), similar to a continuous IRES RNA (Fig. 3B, lane 2). When PTB was added to the reaction, an additional band appeared (Fig. 3B, lane 7). This band corresponds to an IRES–PTB complex (U* + D + PTB), since it completely disappeared after treatment with proteinase K (Fig. 3B, lane 8). When the hybrid IRES was boiled before gel loading (Fig. 3B, lane 9), the hybrid IRES band almost completely disappeared, while a strong lower molecular weight band appeared. This corresponds to the radio-labeled upstream RNA (U*) (Fig. 3B, cf. lanes 4 and 9), which was detached from its unlabeled counterpart RNA D after boiling, while only minute amounts of the hybrid IRES were detected, which probably re-formed during gel loading. When the sample with PTB bound to the IRES hybrid was separated on the gel after boiling (Fig. 3B, lane 10), a protein–RNA complex appeared, corresponding to a complex of RNA U with PTB (U* + PTB). The amount of this band was ~10% of that of free RNA U*, reflecting the efficiency of covalent bond formation in the UV cross-link assay (see above).
However, most interesting, a second band (U* + D + PTB) migrated after boiling (Fig. 3B, lane 10) at the same position as the complete hybrid plus PTB when not boiled (Fig. 3B, lane 7). This band completely disappeared after proteinase K treatment (Fig. 3B, lane 11). Thus, this band most likely represents a complex of RNA U* and RNA D that are bridged by one PTB molecule covalently linked to each of the RNAs. On one hand, the intensity of this band was lower than that of the RNA U*–PTB complex, reflecting the 10% efficiency of UV-induced covalent bond formation between PTB and RNA U*, multiplied with the 10% efficiency of formation of a second UV-induced covalent bond between the same PTB molecule and RNA D. On the other hand, this band did not originate from hybrids re-formed during gel loading, since it appeared in addition to re-formed hybrids (Fig. 3B, cf. lanes 10 and 9,11) and is much stronger than those. Taken together with the above results, we conclude that a PTB molecule can contact both the upstream and the downstream PTB-binding sites of the IRES RNA.
PTB–IRES binding orientation mapping: PTB domain III contacts IRES stem-loop 2
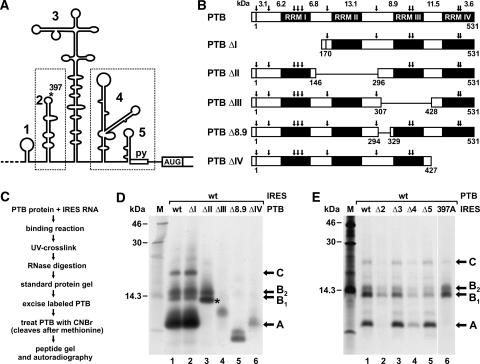
The above result indicates that PTB may bind to the IRES in a stretched conformation and, in that way, may contact both parts of the IRES. To find out which PTB domain actually binds to which binding region in the FMDV IRES, we used an RNA–protein fingerprint assay (Ochs et al. 1999; Fig. 4C). In this assay, the full-length PTB protein was bound to the complete FMDV IRES RNA under native conditions. After binding, UV irradiation was used to covalently attach RNA to protein. Then the samples were treated with RNase, leaving short, trimmed radiolabeled RNA oligonucleotides covalently attached to the protein at the contact sites. This oligonucleotide-labeled PTB was gel purified and subjected to chemical fragmentation by cyanogen bromide (CNBr), which cleaves after methionine (see Fig. 4B), and the cleavage products were resolved on peptide gels and analyzed by autoradiography. The resulting radio-labeled peptides represent the contact sites between native PTB protein and IRES RNA.
FIGURE 4.
Delimitation of PTB–IRES contacts. (A) The FMDV IRES with the two separate PTB-binding regions boxed. (B) PTB proteins used in this assay. Potential CNBr cleavage sites (arrows), the calculated sizes of resulting peptide fragments, and amino acid numbers flanking deletions are indicated. The N-terminal His6-tag is represented by the small box. Note that in PTBΔ8.9 only a small sequence in the N-terminal region of PTB domain III was deleted within the limits of two neighboring methionine residues. (C) Overview of the experimental procedure. (D) Autoradiograph of a peptide gel with CNBr digests of wild-type and mutant PTB proteins UV cross-linked to FMDV wild-type IRES RNA as outlined in C. 14C-labeled marker proteins are shown on the left, and the major labeled peptides are marked on the right. The asterisk marks fragment A*. (E) Autoradiograph of a peptide gel with CNBr digests of wild-type PTB protein UV cross-linked to FMDV wild-type and mutant RNAs as indicated.
When the contact of PTB to the IRES was analyzed in this assay, a peptide of <10 kDa obtained most radioactive label (Fig. 4D, lane 1). Thus, this fragment (designated fragment A) represents the major cross-link site between PTB and FMDV IRES. In addition, two less intensively labeled fragments in the range of 14 kDa (B1 and B2) and a larger product (C) were detected. The peptides could not be identified solely according to their molecular masses. Complete CNBr cleavage of PTB should result in peptides not larger than 13.1 kDa (see Fig. 4B), but obviously not every methionine residue in the PTB protein is accessible to complete chemical cleavage even after repeated CNBr treatments. Moreover, a possible change in the migration of the labeled peptides in the gel could not be precisely predicted, since the short, attached RNA oligonucleotides add an unknown amount of mass but also additional negative charges.
To identify from which domain within the PTB protein fragment A originates, we used the PTB domain deletion mutants. With PTBΔI, the pattern of radiolabeled peptides remained unchanged (Fig. 4D, lane 2), indicating that RRM domain I does not contact the IRES in a way that allows formation of covalent bonds upon UV irradiation. With PTBΔII (Fig. 4D, lane 3), the pattern of labeled peptides changed significantly. Fragment A disappeared from its previous position, and an additional fragment (A*, marked by an asterisk) appeared migrating slightly faster than fragment B1. This new fragment A* is supposed to represent a fusion of the C-terminal portion of domain I with the N-terminal portion of domain III, since the deletion in PTBΔII just reaches beyond the methionine residue located between domains II and III (see Fig. 4B). This interpretation is supported by the result obtained with PTBΔIII (Fig. 4D, lane 4), in which the CNBr cleavage sites flanking domain II were recovered. Now fragments A, A*, and B disappeared completely, while another fragment of intermediate size appeared. Since fragment A does not appear with PTBΔIII even though the methionine residues flanking domain II are retained, fragment A cannot be assigned to domain II but must be assigned to domain III. To confirm this interpretation, we generated PTBΔ8.9, in which only a small portion of RRM domain III was deleted without affecting any flanking methionine residues (Fig. 4B). Also with this deletion mutant, fragment A disappeared from its original position but migrated faster (Fig. 4D, lane 5), confirming that fragment A originates from domain III. With PTBΔIV, fragment A was weakly detected, while fragments B1 and B2 disappeared (Fig. 4D, lane 6). In conclusion, fragments B1 and B2 most likely represent the 11.5-kDa cleavage fragment (and/or longer fusion products thereof due to incomplete CNBr cleavage) near the C terminus of PTB, which covers the N-terminal portion of domain IV.
To analyze which part of the FMDV IRES is contacted by the PTB amino acid sequences represented by fragments A and B, we used wild-type PTB in assays with IRES deletion mutants (Fig. 4E). When the major PTB binding site in the IRES, stem-loop 2, was absent from the IRES (Fig. 4E, lane 2), labeling of fragment A was completely abolished, while fragments B were labeled weaker. To confirm that fragment A (representing PTB domain III) contacts IRES stem-loop 2, we used an IRES mutant in which the integrity of stem-loop 2 was maintained but only one U residue in its apical loop was mutated to A (FMDV position 397, asterisk in Fig. 4A) (Luz 1991). With this IRES 397A, labeling of peptide fragment A was completely lost (Fig. 4E, lane 6), while labeling of peptides B was retained, indicating that mainly the label transfer from IRES stem-loop 2 to the amino acids contained in fragment A was impaired. Deletion of IRES stem-loop 3 had no effect (Fig. 4E, lane 3). In contrast, deletion of the downstream PTB-binding site in IRES Δ4 resulted in weaker labeling of all peptides (Fig. 4E, lane 4), and deletion of stem-loop 5 sequences (Fig. 4E, lane 5) did not affect the labeling of fragment A but decreased labeling of fragments B. Thus, the downstream PTB-binding site of the IRES is contacted by PTB domain IV.
Taken together, these results show that the PTB domain III contacts stem-loop 2 of the FMDV IRES, while PTB domain IV contacts the downstream PTB-binding region of the IRES. Most likely these contacts actually represent the most important interactions between the PTB protein and the IRES RNA, even if possibly additional contacts of domain II (or I) with the IRES may be present (but are not detectable in the UV cross-link).
PTB domains III and IV are sufficient to stimulate initiation factor entry and FMDV translation
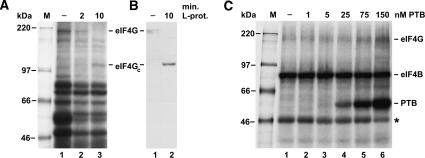
To identify the molecular link between the mere binding of PTB and the stimulation of IRES function, we investigated whether PTB influences the interaction of translation initiation factors with the viral RNA. eIF4G and eIF4B bind to the 3′ region of the FMDV IRES (Meyer et al. 1995; Ochs et al. 1999; Rust et al. 1999; Lopez de Quinto and Martinez-Salas 2000; Saleh et al. 2001), and the most plausible explanation for the action of PTB is that it stimulates the interaction of these factors with the IRES, which in turn results in an increased rate of ribosome recruitment to the viral RNA.
Binding of eIF4G and eIF4B was analyzed by the UV cross-link assay using radiolabeled FMDV wild-type IRES RNA as a probe. The identity of the eIF4G band was confirmed using its cleavage by the FMDV leader protease and subsequent detection of eIF4G in the UV cross-link and immunoblot (Fig. 5A,B) as performed previously (Saleh et al. 2001). Using the PTB-depleted reticulocyte lysate described previously (Niepmann 1996; Niepmann et al. 1997), eIF4B and an unknown protein migrating at about 47 kDa (asterisk) were strongly labeled in the absence of any supplemented PTB (Fig. 5C, lane 1). Most importantly, the weak double band of eIF4G was detected at about 200 kDa. When increasing concentrations of recombinant native wild-type PTB were added (Fig. 5C, lanes 2–6), the band of the recombinant PTB appeared clearly at a concentration of 25 nM and was very strong at 150 nM. In parallel, the eIF4G band appeared with increased intensity (Fig. 5C, lanes 5,6), whereas the intensity of the 47-kDa band was reduced. In conclusion, these results indicate that PTB stimulates binding of eIF4G to the FMDV IRES.
FIGURE 5.
PTB stimulates binding of eIF4G to the FMDV IRES. (A,B) Identification of eIF4G. (A) UV cross-link assay with normal RRL (−) or RRL treated with FMDV leader protease (L-prot.). L-protease RNA transcribed from plasmid pFMDV14 was preincubated with RRL at 30°C for the times (min.) as indicated. Samples were then used for UV cross-link reactions with [α-32P]-UTP-labeled wild-type FMDV IRES. Reactions were incubated at 30°C for 10 min and UV irradiated for 20 min, and excess RNA was digested with RNase A. Fifty percent of each sample was applied to an SDS–8% polyacrylamide gel and analyzed by autoradiography. Molecular masses of marker proteins (M) are given in kilodaltons. (B) The other 50% of the samples prepared in A were separated on a gel, proteins transferred to nitrocellulose, and eIF4G was immunostained using anti-eIF4G anti-serum. 4G, eIF4G; eIF4GC, C-terminal fragment of eIF4G. (C) UV cross-link assay with [α-32P]-CTP-labeled wild-type FMDV IRES and PTB-depleted rabbit reticulocyte lysate (Niepmann et al. 1997). In lanes 2–6, recombinant wild-type PTB was added in the increasing concentrations indicated on the top. Molecular masses of marker proteins (M) are indicated. The asterisk marks an unknown protein mentioned in the text. (D) UV cross-link assay as in C, but with the PTBΔI,II mutant protein supplemented instead of wild-type PTB (lanes 2–5).
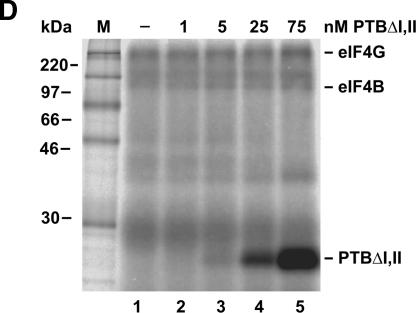
To find out whether RRM domains III and IV of PTB that are preferentially engaged in RNA binding are sufficient to mediate this stimulation effect on eIF4G binding to the IRES, we used PTBΔI,II in the same assay (Fig. 5D). Also, this PTB protein is able to stimulate binding of eIF4G to the IRES (Fig. 5D, lanes 4,5). In conclusion, a PTB protein providing domains III and IV is sufficient to promote initiation factor entry at the FMDV IRES. eIF4B is labeled weaker in this case, most likely due to the use of a different batch of PTB-depleted lysate.
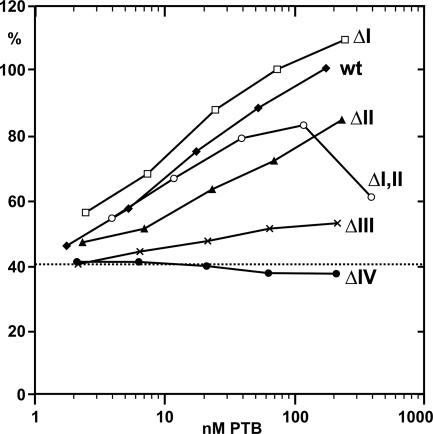
The functional contribution of the individual PTB domains on the stimulation of IRES activity was analyzed in translation reactions (Fig. 6) using PTB-depleted lysate. PTBΔI was even more active than wild-type PTB in the stimulation of IRES activity. This effect was observed with up to 240 nM of PTBΔI, which is in the range of the concentration of endogenous PTB in untreated RRL (Niepmann et al. 1997). Thus, PTB domain I is not required for FMDV translation stimulation. With PTBΔII, a slightly lower efficiency of FMDV translation stimulation was observed. At the highest concentration of 240 nM, still 85% stimulation was achieved.
FIGURE 6.
Stimulation of FMDV IRES-dependent translation by wild-type PTB and PTB deletion mutants. The dotted line indicates the basal translation efficiency of 40.6% obtained with PTB-depleted RRL in the absence of supplemented PTB. To the PTB-depleted RRL, either wild-type PTB (wt) or the corresponding mutants PTBΔI, ΔII, ΔI,II, ΔIII, or ΔIV (see Fig. 1), respectively, was added before translation. The X-axis indicates the final concentrations of added PTB protein in the reaction. Readings show the efficiency of translation of the IRES-dependent luciferase gene (pM12 RNA), standardized by setting the reaction with 100 ng wild-type PTB as 100%. All curves originate from the same value at about 40% stimulation (lines not shown due to the logarithmic scale on the X-axis).
In accordance with the results shown above, a stimulation of IRES driven translation was also obtained with PTBΔI,II. At low concentrations, stimulation was similar to that obtained with wild-type PTB. We can only speculate whether this is due to the loss of both the negative effect of domain I and the positive effect of domain II in the ΔI,II protein. Only at the very high concentration of 390 nM did PTBΔI,II inhibit FMDV translation, most likely due to unspecific effects at those high protein concentrations. In contrast, PTBΔIII caused only a very slight increase in translation efficiency, whereas no stimulation was detected with PTBΔIV.
In conclusion, stimulation of FMDV IRES-driven translation can be achieved with a PTB protein that only contains the two RNA-binding domains III and IV.
DISCUSSION
Using different assays for RNA–protein interactions and functional assays, we investigated the contribution of the four RNA recognition motif domains of PTB to three aspects of the PTB–IRES interaction: (1) the strength, specificity, and orientation of binding; (2) the possible oligomeric status of PTB; and (3) the role in stimulating FMDV translation.
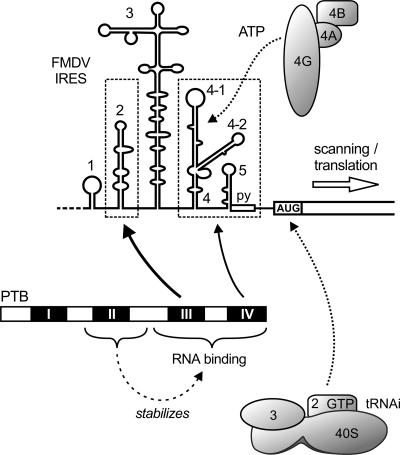
The FMDV IRES contains two binding regions for PTB that are separated by a large RNA stem-loop secondary structure, IRES stem-loop 3. Mapping the spatial orientation of PTB relative to the bound FMDV IRES, we found that the PTB molecule contacts the IRES in a way such that each of the two C-terminal RRM domains of the protein attaches to one of the two separate binding regions in the IRES RNA. Most likely, the spatial orientation of PTB relative to the IRES is organized by these two sets of contacts (Fig. 7). An important interaction is made between PTB domain III and IRES stem-loop 2. PTB contacts several nucleotides in this compact RNA structure, providing the strongest PTB–IRES interaction (Luz and Beck 1990, 1991; Luz 1991). Another interaction is made by PTB domain IV to the separate IRES 3′ region including stem-loop 4-1, stem-loop 5, and the oligopyrimidine tract (Luz and Beck 1991; Kolupaeva et al. 1996; Niepmann 1996; Rust et al. 1999). Accordingly, the two PTB-binding sites in a hybrid IRES are bridged by a PTB molecule. Only to a minor extent may the two PTB-binding regions of the IRES be bound independently by two molecules of PTB (data not shown). The idea that a rather linear arrangement of the RNA-binding domains of PTB may serve to organize the IRES tertiary structure is supported by the results of small-angle X-ray scattering studies that show that full-length PTB is a rod-shaped rather than a globular molecule in solution (Simpson et al. 2004).
FIGURE 7.
Schematic representation of the interaction of PTB with the FMDV IRES (details are discussed in the text).
The RRM domain II of PTB stabilizes the interaction of domains III and IV with the IRES. In particular, deletion of domain II results in a remarkable decrease in stability of the PTB–IRES interaction detected in the shift assay, whereas deletion of domain I causes only a slight decrease in RNA binding stability. However, the interaction of a PTB protein lacking both domains I and II with the FMDV IRES was still well detected in the UV cross-link in which also short-lived interactions are immediately captured by UV-induced covalent bonds. This indicates that even though domain II stabilizes a long-lived interaction of PTB domains III and IV with the IRES (Fig. 7), the short-lived but strong interaction of PTB domains III plus IV with the IRES in the absence of domain II is sufficient for stimulating IRES activity (Fig. 6).
Since no RNA–protein contacts could be assigned to these domains in the orientation mapping assay, we do not know if domain II makes direct contacts to the RNA or not. On one hand, domain II could indirectly stabilize the interaction of domains III and/or IV with the IRES. On the other hand, the observed stabilization of the RNA binding by domains III and IV to the IRES could be accomplished by direct contacts of domain II with the IRES. In this case, the geometry of its interaction with the IRES may just not allow the formation of covalent bonds upon UV irradiation. Recent studies imply that the N-terminal domains of PTB are indeed able to bind RNA. However, the actual involvement of a given PTB domain in RNA binding as well as the affinity of each PTB domain to a given RNA depends on the nature of that RNA and may vary between different substrate RNAs. Various studies show that the N-terminal PTB domains may bind RNA, but they bind more weakly than the C-terminal domains. While the C-terminal portion of PTB comprising domains III and IV obtains most label after UV cross-linking to splice repression substrate RNAs, also a peptide assigned to the N-terminal domain of PTB obtained some label (Amir-Ahmady et al. 2005). Also, in another study, the two N-terminal domains were shown to bind weakly to RNA (Oh et al. 1998). Similarly, a PTB protein lacking domains I and II binds to a splice repression site RNA substrate with a KM increased only less than threefold compared to the wild-type PTB protein, whereas a PTB protein lacking domains III and IV bound to the RNA with a KM increased more than 10-fold (Liu et al. 2002), suggesting that the C-terminal domains bind much more strongly to RNA than the N-terminal domains. In contrast, Simpson and coworkers (Simpson et al. 2004) showed that both N-terminal RRM domains as well as the two C-terminal domains of PTB directly bind the IRES RNA of EMCV with a slight bias even toward stronger RNA binding by the N-terminal domains, indicating that all domains of PTB are directly involved in binding of PTB to the EMCV IRES.
We found here using the FMDV IRES–RNA that deletion of either a single domain III or IV, respectively, suffices to completely abolish binding of the remainder of the PTB molecule to the IRES. In conclusion, domains I or II do not bind strongly enough to the FMDV IRES to contribute to the stability of the PTB–FMDV–IRES interaction in a way that could compensate for the loss of even only one of domains III or IV. Accordingly, deletion of domain II results only in a modest decrease in RNA-binding efficiency. These results were obtained using a binding competition assay in translation-competent reticulocyte lysate (Fig. 1D), thereby ruling out any assay-specific limitations in the detection of an interaction that come along with, e.g., the actual formation of covalent bonds in the UV cross-link reaction or with the need for long-lasting binding stability in the shift assay. Finally, both domains I and II can be deleted without losing the stimulatory effect of the C-terminal remainder of the PTB protein on initiation factor entry and IRES activity, indicating that domains I and II most likely do not provide any essential protein–RNA or protein–protein contacts involved in initiation factor recruitment.
Despite the obvious similarities in the proposed secondary structures of the two different viral IRES elements of FMDV and EMCV (Jackson and Kaminski 1995), the extent of the contribution of the N-terminal domains of PTB to the stability of the PTB–IRES interaction most likely is different between the two different viral RNAs. This idea is supported by two different studies. Kaminski and coworkers (Kaminski et al. 1995) found in UV cross-link experiments that a PTB protein containing domains II–IV bound efficiently to the EMCV IRES, whereas a PTB protein containing only domains III plus IV bound more weakly. Similarly, Conte and coworkers (Conte et al. 2000) showed that a deletion of the two N-terminal domains of PTB resulted in an eightfold decrease in binding affinity to the EMCV IRES but only in a twofold decrease in binding affinity to the FMDV IRES in a filter-binding assay. Since a filter-binding assay can be assumed to detect both long-lived and short-lived interactions, the latter result can be regarded to be similar to the result obtained from our competition UV cross-link in Figure 1D, which also shows that deletion of domains I and II does not cause a marked decrease in binding affinity to the FMDV IRES. This difference in the contribution of the N-terminal domains of PTB to IRES binding affinity most likely originates from differences between the RNAs used. Both Kaminski and coworkers (Kaminski et al. 1995) and Conte and coworkers (Conte et al. 2000) used an EMCV IRES that also includes the very 5′ region of the EMCV IRES, the so-called EMCV IRES domain I. That EMCV IRES domain I includes stem-loop H, which is the strongest PTB binding site in FMDV (there named stem-loop 2). However, EMCV IRES domain I additionally contains several upstream RNA stem-loops with U/C-rich apical loops (Kaminski et al. 1995), which are excellent binding sites for PTB. An RNA containing only this EMCV domain I binds to PTB as strongly as the complete EMCV IRES (Witherell et al. 1993) and, like the complete EMCV IRES, it also displays an eightfold decrease in binding affinity to a PTB molecule lacking the two N-terminal domains compared with the full-length PTB (Conte et al. 2000). In the FMDV IRES, however, these additional stem-loops are not included. In conclusion, the contribution of the N-terminal domains of PTB to its binding to the EMCV IRES (Conte et al. 2000) may be mainly driven by a region of the RNA substrate that is not contained in the FMDV IRES. This discrepancy between the structures and properties of the two different IRES RNAs may also account for the fact that Kaminski and coworkers (Kaminski et al. 1995) reported that a construct containing domains II–IV stimulated EMCV translation, but a construct containing only domains III plus IV did not. Besides the differences in the RNA substrates, a second reason for this discrepancy may be that the construct of Kaminski and coworkers (1995) containing domains III plus IV included only amino acids 330–531. In contrast, our PTBΔI,II includes amino acids 296–531, thereby including additional amino acids in the N-terminal region of domain III that possibly are required for the stimulatory activity.
The rather rodlike structure of PTB (Simpson et al. 2004) and the oriented binding of PTB to the FMDV IRES (see above) also lead us to the idea that PTB domains I and II probably do not point toward the 3′ region of the IRES where initiation factors bind. Accordingly, we would not necessarily expect an interference of the binding of PTB with the simultaneous binding of initiation factors. In turn, this arrangement of PTB-binding regions in the IRES supports the idea that PTB probably exerts its stimulatory effect on IRES activity in an RNA chaperone function, i.e., by stabilizing the IRES secondary and tertiary structures in a way that serves to better expose those RNA determinants involved in initiation factor binding. However, we cannot exclude that protein–protein contacts may also contribute to the stimulatory effect of PTB on initiation factor entry and ribosome recruitment. Interestingly, PTB appears to be able to guide ribosomes to an RNA in a yet unknown way (directly or indirectly) if PTB is artificially recruited to that RNA (Mitchell et al. 2005). In contrast, in certain cellular IRES systems PTB appears to be involved in the induction of structural changes in the RNA, often together with other RNA-binding proteins, like in the Apaf-1 IRES (Mitchell et al. 2003) and in the Bag-1 IRES (Pickering et al. 2004), suggesting that these structural changes in the RNA may lead to the exposure of RNA determinants that then are involved in direct or indirect ribosome recruitment.
In the FMDV IRES, the PTB binding determinants in the 3′ PTB-binding region are interspersed between the contact points for the canonical initiation factors. PTB binds to the apical loop of IRES stem-loop 4-2 (termed K in EMCV), while eIF4G and eIF4B bind to IRES stem-loop 4-1 (J in EMCV) and to the base of stem-loop 4 (Kolupaeva et al. 1996, 1998; Rust et al. 1999; Lopez de Quinto and Martinez-Salas 2000; Pilipenko et al. 2000; Saleh et al. 2001; Stassinopoulos and Belsham 2001; Bassili et al. 2004). This arrangement of binding sites would be consistent with the idea that PTB bridges the two PTB-binding regions in the IRES and in this way favors a certain tertiary structure conformation of the IRES to expose those RNA determinants that facilitate initiation factor entry. In the EMCV IRES, only one additional A residue in the bulge immediately downstream of the K domain (corresponding to stem-loop 4-2 in the FMDV IRES) renders the activity of the IRES PTB dependent (as do heterologous reporter sequences), indicating that PTB can help to achieve that tertiary structure conformation of the IRES that is optimal for initiation factor entry in cases in which the IRES structure is distorted (Kaminski and Jackson 1998).
The previous conclusion that PTB is a dimer originated mainly from two previous studies, in which dimerization was supposed to be driven mainly by RRM domain II based on coprecipitation and cross-linking experiments (Perez et al. 1997; Oh et al. 1998). In one of these studies (Perez et al. 1997), PTB was also found to migrate approximately at the size of a dimer in gel filtration. Supporting the dimer hypothesis, the two N-terminal domains of PTB were found to facilitate PTB self-interaction in two-hybrid studies (Hahm et al. 1998; Oh et al. 1998; Kim et al. 2000). However, the self-interaction of PTB was quite weak in coprecipitation assays (Oh et al. 1998; Kim et al. 2000), whereas the interaction of PTB with other hnRNP proteins like hnRNP E2 as well as the self-interaction of other hnRNP proteins like C1 and L were considerably stronger (Kim et al. 2000), suggesting that a possible self-interaction of PTB is not very strong. Furthermore, a more recent study suggests that the previously observed migration characteristic of full-length PTB in gel filtration experiments (Perez et al. 1997) may be aberrant due to its rodlike rather than globular structure (Simpson et al. 2004). It was noted in that latter study that N-terminal residues of PTB somehow cause the aberrant migration of PTB in the gel filtration, since full-length PTB migrates at about 100–115 kDa (Perez et al. 1997; Simpson et al. 2004), whereas a PTB protein that was N-terminally truncated by 55 amino acids migrates at about 70 kDa in the gel filtration, much closer to the size of a monomer (Simpson et al. 2004). Moreover, Simpson et al. (2004) and Amir-Ahmady et al. (2005) also used analytical ultracentrifugation to calculate the size of PTB in solution, and both groups concluded that PTB is supposed to be a monomer.
By the use of a different analytical method, a newly developed native gel system, here we visualize directly and in this way confirm that PTB is a monomer under reducing conditions and even under nonreducing conditions. This gel system allows a good estimation of the size of PTB when compared to marker proteins like BSA (Fig. 2E, cf. lanes 2 and 6) under conditions that allow the formation of the known oligomers of glycolysis enzymes under nonreducing and under reducing conditions as described (Fox and Dandliker 1956; Millar 1962; Beernink and Tolan 1994).
In summary, the two C-terminal RRM domains of a monomeric PTB molecule bind in an oriented way to the IRES and most likely act as an RNA chaperone to stabilize IRES structure and in that way augment IRES activity. Short-lived PTB–IRES interactions suffice for this stimulation of IRES function, while a long-lived PTB–IRES interaction stabilized by domain II is not required for this action.
MATERIALS AND METHODS
Plasmids
pSP449 (Luz and Beck 1991) contains downstream of an SP6 promoter the FMDV O 1 K IRES and coding sequences from positions 363 to 831. In plasmids pSP449Δ2, pSP449Δ3, and pSP449Δ4, the sequences of the corresponding IRES stem-loops had been precisely removed (Meyer et al. 1995). pM12 (Ochs et al. 1999) contains an SP6 promoter followed by FMDV IRES sequences from positions 185 to 804 with the 11th AUG of FMDV (position 805) fused to the firefly luciferase gene. pMSal-5 was derived from pM12 by PCR mutagenesis. It contains an SP6 promoter followed by a linker sequence and FMDV IRES nucleotides 185–599 (ending in GT-599), followed by CGAC to create an artificial SalI cleavage sequence that is located in the region corresponding to the right half of stem-loop 3 of the FMDV IRES. pMSal-3 was derived from pM13 (Ochs et al. 1999). It contains an SP6 promoter followed by the linker sequence GAA TAGATCTCGA and FMDV nucleotides 600–888 fused to the luciferase coding sequence. Moreover, those nucleotides in the FMDV sequence 600–641 that are predicted not to base pair in the lower stem of IRES stem-loop 3 (Pilipenko et al. 1989; Jackson and Kaminski 1995) were mutated in this plasmid in a way that a sequence of 39 bases could form a perfectly matching heteroduplex RNA with its left counterpart sequence at the base of IRES stem-loop 3 (cf. Figs. 5A and 1A).
pQE-PTB (Ochs et al. 2002) contains the PTB sequence with 16 additional N-terminal codons, including the His 6 sequence. All PTB deletion mutants were derived from pQE-PTB; the amino acid numbers flanking the deletions are shown in Figures 1 and 6. For deletion of RRM domain I sequences (pQE-PTBΔI), an internal BamHI–EagI fragment was excised from the PTB sequence in pQE-PTB, and the sticky ends were filled and religated. For construction of pQE-PTBΔII, RRM domain II sequences were excised with ApaI and SacII, and the 3′ overhangs removed by T4 DNA polymerase and religated. For construction of pQE-PTBΔI,II, pQE-PTB was cleaved with BamHI in the linker and with SmaI in the PTB sequence, and the sticky ends were filled and religated. For pQE-PTBΔIII, RRM domain III sequences were excised using NarI and EcoNI, and sticky ends were filled and religated. For pQE-PTBΔIV, pQE-PTB was cleaved in the PTB sequence with EcoNI and in the downstream linker with HindIII, and sticky ends were filled and religated. For pQE-PTBΔ8.9, pQE-PTB was cleaved in the PTB sequence with SacII and the ends recessed with T4 DNA polymerase. Then the DNA was cleaved with XmaI and the ends filled and religated.
Preparation of RNAs
pSP449 was linearized with SmaI downstream of the FMDV IRES sequence. The RNA obtained after in vitro transcription contains 28 nt of linker sequence, FMDV IRES sequences 363–804, and the first 27 nt of the FMDV coding sequence. RNAs from pSP449Δ2, pSP449Δ3, and pSP449Δ4 were obtained correspondingly. RNA IRES Δ5 was obtained from pSP449 linearized with NgoMIV in the stem-loop 5 sequence. pM12 was linearized with BbsI immediately downstream of the luciferase start codon. The RNA obtained after in vitro transcription contains the linker sequence GAATAGATCTCGATG, FMDV IRES sequences 185–804, and the first 15 nt of the luciferase sequence. pMSal-5 was linearized at the artificial SalI site directly downstream of FMDV IRES position 599. The RNA obtained after in vitro transcription (named RNA U for upstream; 435 nt) contains a 17-nt linker sequence, IRES sequences starting at position 185 and ending at position 599 in the right half of the IRES stem-loop 3, plus the 3′-terminal sequence CGA due to the artificial SalI sequence (see Fig. 5A). pMSal-3 was linearized with BbsI downstream of the luciferase start codon. The in vitro-transcribed RNA (named RNA D, for downstream) contains the linker sequence GAATAGATCTCGA, followed by FMDV nucleotides 600–888, plus the first 15 nt of the firefly luciferase coding sequence. For in vitro transcription of RNAs for translation, pM12 was linearized with SmaI downstream of the luciferase sequence. pFMDV14 (Saleh et al. 2001) was linearized with BamHI in the FMDV 1D (VP1) coding region.
Labeled RNAs were synthesized using SP6 RNA polymerase in the presence of 2.5 μM [α-32P]-UTP or -CTP (400 Ci/mmol; Amersham) plus 10 μM nonradioactive labeling nucleotide. Unincorporated nucleotides were removed on Sephadex G-25 columns. Unlabeled RNAs were synthesized in the presence of 500 μM nucleotides.
Proteins
Full-length His6-PTB or deletion mutants were expressed in Escherichia coli XL1blue and purified with Ni2+-loaded metal affinity beads (Qiagen) according to the protocol provided by the supplier. Native marker proteins were purchased from Sigma-Aldrich: urease (from jack bean, EC 3.5.1.5; isoelectric point [pI] = 5.0), bovine serum albumin (BSA, including monomers and chemically cross-linked dimers; pI = 4.7), fructose-1,6-bisphosphate aldolase (aldolase, from rabbit muscle, EC 4.1.2.13; pI = 8.2), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, from rabbit muscle, EC 1.2.1.12; pI = 8.5), and lactate dehydrogenase (LDH, from rabbit muscle, EC 1.1.1.27; pI = 8.5). Unlabeled marker proteins (Benchmark Protein Ladder) are from Invitrogen; 14C-labeled marker proteins are from Amersham Biosciences.
Electrophoretic mobility shift assays
Shift assays (Konarska and Sharp 1986) were performed in EMSA-150 buffer (5 mM HEPES-OH at pH 7.4, 2 mM MgCl2, 150 mM KCl, 0.1 mM EDTA, 3.8% glycerol, 2 mM DTT) and 50 μg/mL tRNA. Reactions were supplemented with PTB proteins at 0°C as indicated. After adding 0.01 pmol of [α-32P]-UTP-labeled FMDV IRES RNA, reactions were incubated at 30°C for 10 min, stopped on ice, supplemented with 5% glycerol, and immediately loaded onto 4% polyacrylamide gels containing 5% glycerol, 45 mM Tris-borate at pH 8.3 and 1.25 mM EDTA. Gels were run at 4°C, dried, and analyzed by autoradiography.
UV cross-link assays
UV cross-links were performed with 4.4 μL RRL in a volume of 10 μL in the presence of 10 mM HEPES at pH 7.9, 2 mM MgCl2, 1 μg/μL tRNA, 1 mM dithiothreitol (DTT), 10% glycerol, and 0.05% Nonidet-P40 (Saleh et al. 2001), a final potassium concentration adjusted to 120 mM with KCl, and 0.2 pmol of [α-32P]-UTP or -CTP-labeled IRES RNA. Reactions were incubated at 30°C and irradiated with UV light. Excess RNA was digested with 0.1 mg/mL RNase A at 37°C for 60 min. Proteins were separated on SDS-10% polyacrylamide gels and analyzed by autoradiography.
Focusing alkaline native gel electrophoresis
For analyzing the oligomerization status of PTB, we developed a new focusing alkaline native gel electrophoresis system. This gel system (M. Niepmann, in prep.) combines features of the Blue native electrophoresis (Schägger and von Jagow 1991) and a focusing system (Laemmli 1970) using chloride as the fast migrating ion in the gel but employing histidine instead of glycine as the slowly following ion in the cathode buffer. Proteins were mixed with native loading buffer (100 mM Tris-Cl at pH 8.0, 40% glycerol, 0.5% Serva Blue G) and loaded to a 5%–40% polyacrylamide gradient gel containing 200 mM Tris-Cl at pH 8.8. The cathode buffer contained 100 mM histidine adjusted to pH 8.0 using Tris-base (without chloride) and 0.002% Serva Blue G. The anode buffer contained 100 mM Tris-Cl at pH 8.0. Gels were run for 1 h at 4 °C and 12 V/cm and for 6–8 h at 30 V/cm. After the run, the gels were fixed and destained with four changes of 7.5% acetic acid, 5% ethanol.
RNA–protein fingerprint assay
UV cross-link reactions with native, complete PTB protein and FMDV IRES RNA were performed as above. After RNase A treatment and SDS-polyacrylamide gel electrophoresis, the PTB proteins labeled by the trimmed, covalently attached RNA oligonucleotides were identified by autoradiography. Gel slices containing the labeled proteins were excised and the minced gel pieces incubated for 24 h with 10 mg CNBr (Sigma) in 700 μL 70% formic acid at room temperature in the dark (Nikodem and Fresco 1979). After the samples were lyophilized, the chemical treatment was repeated to achieve complete cleavage as far as possible. The samples were lyophilized several times, separated on peptide gels (Schägger and von Jagow 1987), and radiolabeled peptides analyzed by autoradiography (Ochs et al. 1999).
In vitro translation
RRL was purchased from Promega. PTB-depleted RRL was prepared using poly(U)-Sepharose (Niepmann 1996) or with an RNA-affinity column using the EMCV IRES domain I (Kaminski et al. 1995). Ten-microliter in vitro translations usually contained 44% untreated RRL or PTB-depleted RRL, a final potassium concentration of 120 mM, and 0.15 μg of pM12 reporter RNA. Samples were supplemented with full-length PTB or PTB deletion mutants, incubated for 60 min at 30°C, stopped on ice, and luciferase activity was measured (Niepmann et al. 1997).
Acknowledgments
We thank C. Juenemann for reading the manuscript and E. Beck for stimulating discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft (GK 370 and SFBs 535, 547).
Abbreviations
PTB, polypyrimidine tract-binding protein
RRM, RNA recognition motif
IRES, internal ribosome entry site
FMDV, foot-and-mouth disease virus
EMCV, encephalomyocarditis virus
RRL, rabbit reticulocyte lysate
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7430405.
REFERENCES
- Amir-Ahmady, B., Boutz, P.L., Markovtsov, V., Phillips, M.L., and Black, D.L. 2005. Exon repression by polypyrimidine tract binding protein. RNA 11: 699–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassili, G., Tzima, E., Song, Y., Saleh, L., Ochs, K., and Niepmann, M. 2004. Sequence and secondary structure requirements in a highly conserved element for foot-and-mouth disease virus internal ribosome entry site activity and eIF4G binding. J. Gen. Virol. 85: 2555– 2565. [DOI] [PubMed] [Google Scholar]
- Beernink, P.T. and Tolan, D.R. 1994. Subunit interface mutants of rabbit muscle aldolase form active dimers. Protein Sci. 3: 1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn, L.B., Towner, J.S., Semler, B.L., and Ehrenfeld, E. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71: 6243–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussadia, O., Niepmann, M., Creancier, L., Prats, A.C., Dautry, F., and Jacquemin-Sablon, H. 2003. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 77: 3353–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco, P., Furger, A., Wollerton, M., Smith, C., Moreira, A., and Proudfoot, N. 2004. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 24: 4174– 4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A.T., Robertson, M.E., Conn, G.L., and Belsham, G.J. 2003. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J. Virol. 77: 12441–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte, M.R., Grune, T., Ghuman, J., Kelly, G., Ladas, A., Matthews, S., and Curry, S. 2000. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 19: 3132–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenfeld, E. and Teterina, N.L. 2002. Initiation of translation of picornavirus RNAs: Structure and function of the internal ribosome entry site. In Molecular biology of picornaviruses (eds. B.L. Semler and E. Wimmer), pp. 159–169. ASM Press, Washington, DC.
- Fishbein, W.N., Nagarajan, K., and Scurzi, W. 1970. Urease catalysis and structure. VI. Correlation of sedimentation coefficients and electrophoretic mobilities for the polymeric urease isozymes. J. Biol. Chem. 245: 5985–5992. [PubMed] [Google Scholar]
- Fleming, K., Ghuman, J., Yuan, X., Simpson, P., Szendroi, A., Matthews, S., and Curry, S. 2003. Solution structure and RNA interactions of the RNA recognition motif from eukaryotic translation initiation factor 4B. Biochemistry 42: 8966–8975. [DOI] [PubMed] [Google Scholar]
- Fox Jr., J.B. and Dandliker, W.B. 1956. A study of some of the physical properties of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 218: 53–57. [PubMed] [Google Scholar]
- Ghetti, A., Pinol Roma, S., Michael, W.M., Morandi, C., and Dreyfuss, G. 1992. hnRNP I, the polypyrimidine tract-binding protein: Distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 20: 3671–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, A., Sharp, P.A., Jamison, S.F., and Garçia-Blanco, M.A. 1991. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes & Dev. 5: 1224–1236. [DOI] [PubMed] [Google Scholar]
- Gosert, R., Chang, K.H., Rijnbrand, R., Yi, M., Sangar, D.V., and Lemon, S.M. 2000. Transient expression of cellular polypyrimidine- tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 20: 1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest, S., Pilipenko, E., Sharma, K., Chumakov, K., and Roos, R.P. 2004. Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3. J. Virol. 78: 11097–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm, B., Cho, O.H., Kim, J.E., Kim, Y.K., Kim, J.H., Oh, Y.L., and Jang, S.K. 1998. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 425: 401–406. [DOI] [PubMed] [Google Scholar]
- He, X.M. and Carter, D.C. 1992. Atomic structure and chemistry of human serum albumin. Nature 358: 209–215. [DOI] [PubMed] [Google Scholar]
- Hellen, C.U. and Sarnow, P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Hunt, S.L., Hsuan, J.J., Totty, N., and Jackson, R.J. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes & Dev. 13: 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J. 2002. Proteins involved in the function of picornavirus internal ribosomal entry sites. In Molecular biology of picornaviruses (eds. B.L. Semler and E.Wimmer), pp. 171–183. ASM Press, Washington, DC.
- Jackson, R.J. and Kaminski, A. 1995. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA 1: 985–1000. [PMC free article] [PubMed] [Google Scholar]
- Jang, S.K. and Wimmer, E. 1990. Cap-independent translation of encephalomyocarditis virus RNA: Structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes & Dev. 4: 1560–1572. [DOI] [PubMed] [Google Scholar]
- Kaminski, A. and Jackson, R.J. 1998. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski, A., Hunt, S.L., Patton, J.G., and Jackson, R.J. 1995. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1: 924–938. [PMC free article] [PubMed] [Google Scholar]
- Kenan, D.J., Query, C.C., and Keene, J.D. 1991. RNA recognition: Towards identifying determinants of specificity. Trends Biochem. Sci. 16: 214–220. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Hahm, B., Kim, Y.K., Choi, M., and Jang, S.K. 2000. Protein–protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298: 395–405. [DOI] [PubMed] [Google Scholar]
- Knoch, K.P., Bergert, H., Borgonovo, B., Saeger, H.D., Altkruger, A., Verkade, P., and Solimena, M. 2004. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell. Biol. 6: 207–214. [DOI] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Hellen, C.U., and Shatsky, I.N. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2: 1199–1212. [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Pestova, T.V., Hellen, C.U., and Shatsky, I.N. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273: 18599–18604. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M. and Sharp, P.A. 1986. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46: 845–855. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Liu, H., Zhang,W., Reed, R.B., Liu,W., and Grabowski, P.J. 2002. Mutations in RRM4 uncouple the splicing repression and RNA-binding activities of polypyrimidine tract binding protein. RNA 8: 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S. and Martinez-Salas, E. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6: 1380–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz, N. 1991. “Untersuchungen zur bindung eines 57 kD proteins an die interne translationsstartstelle des maul- und klauenseuche virus.” Ph.D. thesis. University of Heidelberg, Germany.
- Luz, N. and Beck, E. 1990. A cellular 57 kDa protein binds to two regions of the internal translation initiation site of foot-and-mouth disease virus. FEBS Lett. 269: 311–314. [DOI] [PubMed] [Google Scholar]
- ———. 1991. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 65: 6486–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch, K., Svitkin, Y.V., Lee, H.S., Lejbkowicz, F., Kenan, D.J., Chan, E.K., Agol, V.I., Keene, J.D., and Sonenberg, N. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67: 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K., Petersen, A., Niepmann, M., and Beck, E. 1995. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J. Virol. 69: 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, D.B. 1962. Physico-chemical properties of lactic dehydrogenase. J. Biol. Chem. 237: 2135–2139. [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Coldwell, M.J., Jackson, R.J., and Willis, A.E. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11: 757–771. [DOI] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Bushell, M., Evans, J.R., Stoneley, M., Le Quesne, J.P., Spriggs, R.V., and Willis, A.E. 2005. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes & Dev. 19: 1556–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, K., Oubridge, C., Jessen, T.H., Li, J., and Evans, P.R. 1990. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature 348: 515–520. [DOI] [PubMed] [Google Scholar]
- Niepmann, M. 1996. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett. 388: 39–42. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Internal initiation of translation of picornaviruses, hepatitis C virus and pestiviruses. Recent Res. Devel. Virol. 1: 229–250. [Google Scholar]
- Niepmann, M., Petersen, A., Meyer, K., and Beck, E. 1997. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and- mouth disease virus. J. Virol. 71: 8330–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikodem, V. and Fresco, J.R. 1979. Protein fingerprinting by SDS-gel electrophoresis after partial fragmentation with CNBr. Anal. Biochem. 97: 382–386. [DOI] [PubMed] [Google Scholar]
- Ochs, K., Rust, R.C., and Niepmann, M. 1999. Translation initiation factor eIF4B interacts with a picornavirus internal ribosome entry site in both 48S and 80S initiation complexes independently of initiator AUG location. J. Virol. 73: 7505–7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs, K., Saleh, L., Bassili, G., Sonntag, V.H., Zeller, A., and Niepmann, M. 2002. Interaction of translation initiation factor eIF4B with the poliovirus internal ribosome entry site. J. Virol. 76: 2113–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs, K., Zeller, A., Saleh, L., Bassili, G., Song, Y., Sonntag, A., and Niepmann, M. 2003. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol. 77: 115– 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, Y.L., Hahm, B., Kim, Y.K., Lee, H.K., Lee, J.W., Song, O., Tsukiyama- Kohara, K., Kohara, M., Nomoto, A., and Jang, S.K. 1998. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem. J. 331: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.G., Mayer, S.A., Tempst, P., and Nadal Ginard, B. 1991. Characterization and molecular cloning of polypyrimidine tract-binding protein: A component of a complex necessary for pre-mRNA splicing. Genes & Dev. 5: 1237–1251. [DOI] [PubMed] [Google Scholar]
- Perez, I., McAfee, J.G., and Patton, J.G. 1997. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry 36: 11881–11890. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U., and Shatsky, I.N. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16: 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering, B.M., Mitchell, S.A., Spriggs, K.A., Stoneley, M., and Willis, A.E. 2004. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell. Biol. 24: 5595–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Blinov, V.M., Chernov, B.K., Dmitrieva, T.M., and Agol, V.I. 1989. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 17: 5701–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Gmyl, A.P., Maslova, S.V., Belov, G.A., Sinyakov, A.N., Huang, M., Brown, T.D., and Agol, V.I. 1994. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J. Mol. Biol. 241: 398–414. [DOI] [PubMed] [Google Scholar]
- Pilipenko, E.V., Pestova, T.V., Kolupaeva, V.G., Khitrina, E.V., Poperechnaya, A.N., Agol, V.I., and Hellen, C.U. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes & Dev. 14: 2028–2045. [PMC free article] [PubMed] [Google Scholar]
- Robida, M.D. and Singh, R. 2003. Drosophila polypyrimidine-tract binding protein (PTB) functions specifically in the male germline. EMBO J. 22: 2924–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust, R.C., Ochs, K., Meyer, K., Beck, E., and Niepmann, M. 1999. Interaction of eukaryotic initiation factor eIF4B with the internal ribosome entry site of foot-and-mouth disease virus is independent of the polypyrimidine tract-binding protein. J. Virol. 73: 6111–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, L., Rust, R.C., Füllkrug, R., Beck, E., Bassili, G., Ochs, K., and Niepmann, M. 2001. Functional interaction of translation initiation factor eIF4G with the foot-and-mouth-disease virus internal ribosome entry site. J. Gen. Virol. 82: 757–763. [DOI] [PubMed] [Google Scholar]
- Schägger, H. and von Jagow, G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379. [DOI] [PubMed] [Google Scholar]
- ———. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199: 223–231. [DOI] [PubMed] [Google Scholar]
- Simpson, P.J., Davydova, N., Curry, S., and Matthews, S. 2002. Resonance assignment and topology of the 2H, 13C, 15N labelled 29 kDa N-terminal fragment of the polypyrimidine tract binding protein (PTB). J. Biomol. NMR 24: 79–80. [DOI] [PubMed] [Google Scholar]
- Simpson, P.J., Monie, T.P., Szendroi, A., Davydova, N., Tyzack, J.K., Conte, M.R., Read, C.M., Cary, P.D., Svergun, D.I., Konarev, P.V., et al. 2004. Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure (Camb.) 12: 1631–1643. [DOI] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J., and Green, M.R. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176. [DOI] [PubMed] [Google Scholar]
- Soderberg, M., Raffalli-Mathieu, F., and Lang, M.A. 2002. Inflammation modulates the interaction of heterogeneous nuclear ribonucleoprotein (hnRNP) I/polypyrimidine tract binding protein and hnRNP L with the 3′untranslated region of the murine inducible nitric-oxide synthase mRNA. Mol. Pharmacol. 62: 423–431. [DOI] [PubMed] [Google Scholar]
- Spellman, R., Rideau, A., Matlin, A., Gooding, C., Robinson, F., McGlincy, N., Grellscheid, S.N., Southby, J., Wollerton, M., and Smith, C.W. 2005. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 33: 457–460. [DOI] [PubMed] [Google Scholar]
- Stassinopoulos, I.A. and Belsham, G.J. 2001. A novel protein–RNA binding assay: Functional interactions of the foot-and-mouth disease virus internal ribosome entry site with cellular proteins. RNA 7: 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmar, L. and Welsh, N. 2004. Glucose-induced binding of the polypyrimidine tract-binding protein (PTB) to the 3′-untranslated region of the insulin mRNA (ins-PRS) is inhibited by rapamycin. Mol. Cell. Biochem. 260: 85–90. [DOI] [PubMed] [Google Scholar]
- Valcárcel, J. and Gebauer, F. 1997. Post-transcriptional regulation: The dawn of PTB. Curr. Biol. 7: 705–708. [DOI] [PubMed] [Google Scholar]
- Wagner, E.J. and Garcia-Blanco, M.A. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21: 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell, G.W., Gil, A., and Wimmer, E. 1993. Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry 32: 8268–8275. [DOI] [PubMed] [Google Scholar]
- Wollerton, M.C., Gooding, C., Robinson, F., Brown, E.C., Jackson, R.J., and Smith, C.W. 2001. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA 7: 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., Lee, J.A., Kress, T.L., Mowry, K.L., and Black, D.L. 2003. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. 100: 8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, X., Davydova, N., Conte, M.R., Curry, S., and Matthews, S. 2002. Chemical shift mapping of RNA interactions with the polypyrimidine tract binding protein. Nucleic Acids Res. 30: 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]