Abstract
We used nuclear extracts from Drosophila Kc cells to characterize 3′ end processing of Drosophila histone pre-mRNAs. Drosophila SLBP plays a critical role in recruiting the U7 snRNP to the pre-mRNA and is essential for processing all five Drosophila histone pre-mRNAs. The Drosophila processing machinery strongly prefers cleavage after a fourth nucleotide following the stem–loop and favors an adenosine over pyrimidines in this position. Increasing the distance between the stem–loop and the HDE does not result in a corresponding shift of the cleavage site, suggesting that in Drosophila processing the U7 snRNP does not function as a molecular ruler. Instead, SLBP directs the cleavage site close to the stem–loop. The upstream cleavage product generated in Drosophila nuclear extracts contains a 3′ OH, and the downstream cleavage product is degraded by a nuclease dependent on the U7 snRNP, suggesting that the cleavage factor has been conserved between Drosophila and mammalian processing. A 2′O-methyl oligonucleotide complementary to the first 17 nt of the Drosophila U7 snRNA was not able to deplete the U7 snRNP from Drosophila nuclear extracts, suggesting that the 5′ end of the Drosophila U7 snRNA is inaccessible. This oligonucleotide selectively inhibited processing of only two Drosophila pre-mRNAs and had no effect on processing of the other three pre-mRNAs. Together, these studies demonstrate that although Drosophila and mammalian histone pre-mRNA processing share common features, there are also significant differences, likely reflecting divergence in the mechanism of 3′ end processing between vertebrates and invertebrates.
Keywords: histone pre-mRNAs, 3′ end processing, Drosophila, SLBP, U7 snRNP
INTRODUCTION
Metazoan replication-dependent histone pre-mRNAs do not contain introns, and the only processing reaction necessary to generate mature histone mRNAs is a single endonucleolytic cleavage of the mRNA precursors (pre-mRNAs) to form the 3′ end. Studies on 3′ end processing were initially carried out in Xenopus oocytes using synthetic pre-mRNAs (Krieg and Melton 1984) and sea urchin histone genes (Birnstiel and Schaufele 1988) and later were facilitated by the development of an in vitro system based on nuclear extracts from mammalian cells (Gick et al. 1986; Mowry and Steitz 1987a). Replication-dependent histone pre-mRNAs contain two cis elements required for 3′ end processing: a highly conserved stem–loop structure consisting of a 6-bp stem and a 4-nt loop and a less conserved histone downstream element (HDE) located ~15 nt 3′ of the stem–loop (Birnstiel and Schaufele 1988; Mowry et al. 1989). Mammalian histone pre-mRNAs are cleaved between the two elements, 5 nt downstream of the stem–loop. The stem–loop is recognized by the stem–loop binding protein (SLBP) (Wang et al. 1996), also referred to as the hairpin binding protein (HBP) (Martin et al. 1997). The HDE interacts with the U7 snRNP, which contains an ~60-nt U7 snRNA (Galli et al. 1983; Mowry and Steitz 1987b), and this interaction is primarily mediated by base-pairing between the HDE and the 5′ end of U7 snRNA (Schaufele et al. 1986; Bond et al. 1991). In vitro studies in mammalian nuclear extracts suggest that SLBP stabilizes binding of the U7 snRNP to the pre-mRNA (Dominski et al. 1999) and is essential in processing of only those pre-mRNAs that do not form sufficiently stable duplexes with the U7 snRNA (Streit et al. 1993; Spycher et al. 1994). This role of SLBP in mammalian processing is most likely mediated by ZFP100, a 100-kDa zinc finger protein associated with the U7 snRNP and interacting with the SLBP/stem–loop complex (Dominski et al. 2002a). In addition to bridging the two factors bound to their respective sequence elements, ZFP100 may also play other roles in 3′ end processing, possibly including the recruitment of the cleavage factor.
Purification of the U7 snRNP from mammalian cells resulted in identification of two novel Sm-like proteins: Lsm10 (Pillai et al. 2001) and Lsm11 (Pillai et al. 2003), which replace the D1 and D2 Sm proteins present in the spliceosomal snRNPs. Lsm11 interacts in vitro with ZFP100 (Azzouz et al. 2005) and plays a key role in recognizing the unique sequence of the Sm binding site in U7 snRNA (Grimm et al. 1993; Pillai et al. 2003). Orthologs of Lsm10 and Lsm11 are also found in the Drosophila U7 snRNP, demonstrating that the unique structure of the U7 snRNP in vertebrates and invertebrates is conserved (Azzouz and Schumperli 2003). A counterpart of ZFP100 has not been yet identified in the Drosophila genome, suggesting that ZFP100 is either weakly conserved between vertebrates and invertebrates or processing of histone pre-mRNAs in Drosophila does not require this protein.
We recently reported that nuclear extracts from Drosophila S-2 and Kc cultured cells and embryos are capable of 3′ end processing of presynthesized Drosophila histone pre-mRNAs (Dominski et al. 2002b) and identified the Drosophila U7 snRNA (Dominski et al. 2003b). Nuclear extracts from Kc cells are also capable of cotranscriptional processing of histone pre-mRNAs (Adamson and Price 2003). Unlike the auxiliary role played by SLBP in mammalian in vitro processing, Drosophila SLBP is indispensable for processing of all Drosophila histone pre-mRNAs (Dominski et al. 2002b). This observation suggests that Drosophila SLBP plays a much more important role in recruiting the U7 snRNP to the pre-mRNA than it does in the mammalian processing. Here we used the in vitro system based on Drosophila nuclear extracts to characterize 3′ end processing of Drosophila histone pre-mRNAs and to define differences and similarities in processing between this model invertebrate processing system and processing in mammalian nuclear extracts.
RESULTS
Mapping the cleavage site in Drosophila 3′ end processing
To characterize 3′ end processing of histone pre-mRNAs in Drosophila we used an in vitro system based on nuclear extracts from Drosophila Kc cells. A simplification of the Drosophila system is that there are only five different histone genes (each repeated multiple times) compared with the more than 60 nonallelic histone genes present in mammals (Marzluff et al. 2002). As substrates in most of these studies we used hybrid pre-mRNAs consisting of the stem–loop and cleavage site from the mouse H2a-614 pre-mRNA and the HDE from each of the five different Drosophila histone pre-mRNAs (Dominski et al. 2002b). The two elements were joined by introducing an EcoRI restriction site 8–13 nt downstream of the stem–loop of the H2a-614 DNA clone, in a region that does not play an important role in 3′ end processing (Fig. 1A). The H2a/RI pre-mRNA, a derivative of the H2a-614 pre-mRNA containing the EcoRI sequence, is processed in a mouse nuclear extract with the same efficiency as the parental pre-mRNA (Dominski et al. 1999).
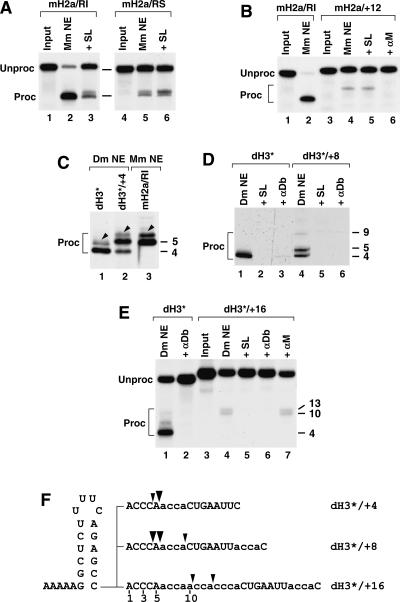
FIGURE 1.
Mapping of the cleavage site in Drosophila 3′ end processing. (A) Sequences of pre-mRNA substrates used in this study. The histone downstream element (HDE) specific to each pre-mRNA was fused to the stem–loop (SL) and the flanking sequences (including the cleavage site) from the mouse H2a-614 pre-mRNA using the EcoRI restriction site (underlined) generated between the two sequence elements. The nucleotides are numbered starting from the first residue after the stem–loop. (B) The upstream cleavage products of the dH3*, dH2a*, and dH2b* pre-mRNAs generated in a Drosophila nuclear extract (lanes 1,3,4) were analyzed in a high-resolution polyacrylamide gel next to the upstream cleavage product of the mouse H2a/RI pre-mRNA generated in a mouse nuclear extract (lane 2). The unprocessed input pre-mRNAs are not shown. The minor bands (arrow heads) that migrate over the major cleavage product visible in this and subsequent panels result from processing of the substrates longer at the 5′ end by 1 or 2 nt due to heterogeneity of the transcription start site selection. (C) Upstream cleavage products of the dH3* and mH2a/RI pre-mRNAs (lanes 1,2) were incubated with the 3′hExo (lanes 3,4) or a Drosophila nuclear extract (lanes 5,6). (D) Cleavage products of the mutant dH3* pre-mRNAs (indicated in E) generated in a Drosophila nuclear extract and separated in a high-resolution polyacrylamide gel. Mutated nucleotides are indicated by low-ercase letters. The cleavage product of the wild-type dH3* pre-mRNA is shown in lane 1. The unprocessed input pre-mRNAs are not shown. (E) The sequence of the pre-mRNA substrates analyzed in D. The major and minor cleavage sites are marked with large and small arrowheads, respectively.
All the five hybrid pre-mRNAs, designated dH1*, dH2a*, dH2b*, dH3*, and dH4*, are processed equally well in Drosophila nuclear extracts (Dominski et al. 2002b). The mouse H2a-614 pre-mRNA, containing the original mouse-specific HDE, was also processed in the Drosophila nuclear extract, although with much lower efficiency (Dominski et al. 2002b). We previously showed using high-resolution electrophoresis capable of detecting single nucleotide differences in RNA length that the upstream cleavage product generated from the dH3* pre-mRNA in a Drosophila nuclear extract migrated faster than the product of the mouse H2a pre-mRNA generated in a mouse nuclear extract. To establish whether generation of the faster migrating processing product is a general property of Drosophila nuclear extracts, we tested two other Drosophila-specific pre-mRNAs, dH2a* and dH2b*. These pre-mRNAs were incubated in a Drosophila nuclear extract and their cleavage products analyzed in a high-resolution gel next to products of the dH3* pre-mRNA generated in the Drosophila nuclear extract and of the mouse H2a/RI pre-mRNA, generated in a mouse myeloma nuclear extract. Processing of dH2a* and dH2b* generated the upstream cleavage product that had the same increased mobility as the dH3* product (Fig. 1B, lanes 1,3,4). Note that the minor band migrating more slowly than the major processing product in this and other lanes is the cleavage product of the longer substrate containing an additional nucleotide at the 5′ end as a result of an alternative transcription start site. The other two Drosophila-specific pre-mRNAs, dH1* and dH4* pre-mRNAs, were also cleaved to an identical product migrating slightly faster than the upstream processing product generated in a mouse nuclear extract from the mouse H2a/RI pre-mRNA (not shown). The processing product of the mouse H2a/RI pre-mRNA generated in Drosophila nuclear extracts also had a higher mobility, demonstrating that this difference is due to a unique property of Drosophila extracts rather than processing signals in Drosophila histone pre-mRNAs (Dominski et al. 2002b).
The difference in migration could indicate that Drosophila nuclear extracts cleave pre-mRNAs 1 nt closer to the stem, thus producing slightly shorter mRNAs, or cleave at the same site as mammalian extracts but generate a 3′ end containing a phosphate group, which would increase mobility of the RNA (Sollner-Webb et al. 2001). Alternatively, Drosophila nuclear extracts might contain a 3′ exonucleolytic activity resistant to EDTA that shortens the upstream cleavage product. The upstream cleavage product generated in mammalian nuclear extracts ends with the ACCCA following the stem–loop and contains a 3′ hydroxyl group (Scharl and Steitz 1994; Furger et al. 1998; Dominski et al. 2002b). To characterize the differences in the 3′ end, we isolated the cleavage products generated in mouse or Drosophila nuclear extracts and treated these products with the baculovirus-expressed 3′hExo (Dominski et al. 2003a), a 3′–5′ exonuclease that removes the single-stranded tail from histone mRNAs containing a hydroxyl group at the 3′ end but not a phosphate (our unpubl. results). Incubation of each processing product with 50 pmol of 3′hExo resulted in formation of identical 44-nt RNAs ending at the base of the stem, demonstrating that Drosophila processing product is sensitive to the exonuclease and hence must terminate with a hydroxyl group (Fig. 1C, lanes 3,4). To test whether Drosophila nuclear extract contains a 3′ exonucleolytic activity capable of removing 1 nt from the cleavage product ending five nucleotides after the stem–loop, we incubated the purified mouse cleavage product in the Drosophila nuclear extract. After 90-min incubation in the presence of EDTA, this product was unchanged (Fig. 1C, lanes 5,6). Based on these results we conclude that the upstream cleavage product generated in Drosophila nuclear extracts is shorter by 1 nt than the cleavage product of the mouse processing and contains a 3′ hydroxyl group (Fig. 1E).
Mammalian nuclear extracts preferentially cleave histone pre-mRNAs after an adenosine and less efficiently after a cytosine (Scharl and Steitz 1994; Furger et al. 1998). Processing of the Drosophila-specific pre-mRNAs and the mouse H2a pre-mRNA in Drosophila nuclear extracts after the cytosine 4 nt 3′ of the stem–loop was surprising since there is an adenosine 1 nt further downstream (Fig. 1A). To test whether Drosophila processing has a different nucleotide preference than mammalian processing, we created mutants of the dH3* pre-mRNA by changing the ACCCA to ACaaA (the sequence in the genuine Drosophila histone H3 pre-mRNA), ACaCA, ACCaA, or ACCuA. The ACaaA pre-mRNA was predominantly cleaved after the adenosine 4 nt downstream of the stem–loop (position +4). Processing of this substrate also generated a minor product with the cleavage site after the adenosine at +3 (Fig. 1D, lane 2). In contrast, the ACaCA pre-mRNA was predominantly cleaved after the adenosine at +3, with a significant amount of the substrate being cleaved after the next cytidine (Fig. 1D, lane 3). Both the ACCaA and the ACCuA pre-mRNAs were processed at the normal site, after the fourth nucleotide, and processing of the ACCaA pre-mRNA was more efficient than processing of the ACCuA or the wild-type dH3* pre-mRNAs (Fig. 1D, lanes 4–6). Thus, Drosophila processing has a strong preference to cleave pre-mRNA after an adenosine located 4 nt downstream of the stem–loop. Efficient cleavage also occurs after an adenosine at position +3 but not after an adenosine located at position +5 (Fig. 1E).
Effects of increasing the distance between the stem–loop and the HDE on Drosophila processing
Removal of SLBP from a mammalian nuclear extract or sequestering SLBP by addition of an excess of the stem–loop RNA has a variable effect on 3′ end processing of mammalian histone pre-mRNAs and this effect depends on the sequence of the HDE. Processing of pre-mRNAs containing an HDE that allows only weak base-pairing with the U7 snRNA is completely inhibited in the absence of SLBP, whereas processing of pre-mRNAs with strong complementarity to U7 snRNA proceeds under these conditions with only partially reduced efficiency (Streit et al. 1993; Spycher et al. 1994; Dominski et al. 1999). In vitro processing of the mouse H2a/RI pre-mRNA is reduced but not abolished in the presence of the excess stem–loop RNA (Fig. 2A, lanes 2,3). The presence of the stem–loop RNA, in addition to reducing the processing efficiency at the major site after the ACCCA, allowed cleavage at an additional site, which is located 2 nt further downstream, as determined in a high-resolution gel (not shown). The same reduction in processing efficiency and activation of the cryptic site was achieved by reversing the sequence of the stem in the pre-mRNA, abolishing binding of SLBP to the substrate (Fig. 2A, lane 5). The efficiency of processing of this substrate was not further reduced by addition of the stem–loop RNA, indicating that the reverse stem mutation and addition of the stem–loop competitor have the same effect on processing, preventing interaction between SLBP and the pre-mRNA (Fig. 2A, lane 6).
FIGURE 2.
Effects of increasing the distance between the stem–loop and the HDE on Drosophila processing. (A) The mouse histone mH2a/RI pre-mRNA (lanes 1–3) or its derivative mH2a/RS with the stem sequence reversed (lane 4–6) was incubated in a nuclear extract from mouse myeloma cells in the absence (lanes 2,5) or presence (lanes 3,6) of an excess of stem–loop RNA. (B) The mH2a/RI pre-mRNA (lanes 1,2) or the mH2a/+12 pre-mRNA that contains a 12-nt insertion (F) between the stem–loop and the HDE (lanes 3–6) was incubated in a mouse nuclear extract under control conditions (lanes 2,4), in the presence of excess stem–loop RNA (lane 5), or in the presence of the αM 2′O-methyl oligonucleotide blocking the mouse U7 snRNA (lane 6). (C) The dH3* pre-mRNA (lane 1), the dH3*/+4 pre-mRNA (lane 2), or the mH2a/RI pre-mRNA (lane 3) was incubated in the indicated nuclear extracts and the cleavage products analyzed in a high-resolution polyacrylamide gel. The minor band migrating over the major cleavage product (arrow heads) is a result of processing of a 1-nt-longer pre-mRNA. The unprocessed input pre-mRNAs are not shown. (D) The dH3* pre-mRNA (lanes 1–3) and the dH3*/+8 pre-mRNA (lanes 4–6) were incubated in a Drosophila nuclear extract under control conditions (lanes 1,4) or in the presence of excess stem–loop RNA (lanes 2,5) or the αDb oligonucleotide (lanes 3,6). The cleavage products were analyzed in a high-resolution polyacrylamide gel. The unprocessed input pre-mRNAs are not shown. (E) The dH3* pre-mRNA (lanes 1,2) and the dH3*/+16 pre-mRNA (lanes 3–7) were incubated in a Drosophila nuclear extract under control conditions (lanes 1,4) or in the presence of excess stem–loop RNA (lane 5), the αDb oligonucleotide (lane 6), or the αM oligonucleotide complementary to the mouse U7 snRNA (lane 7). (F) Sequences of the dH3* mutant pre-mRNAs analyzed in C–E and cleavage sites detected in the processing reactions.
The cleavage site in mammalian pre-mRNA is determined by the position of the HDE; moving the HDE away from the stem–loop results in a corresponding shift of the cleavage site and in a reduction of processing efficiency (Fig. 2B, lanes 2,4; Scharl and Steitz 1994 Scharl and Steitz 1996). This observation suggests that U7 snRNP functions as a molecular ruler in processing (Scharl and Steitz 1994), and binding of U7 snRNP to the HDE, in contrast to binding SLBP to the stem–loop, is the critical event for determining the cleavage site in mammalian histone pre-mRNA 3′ end processing. A possible explanation for the reduced processing efficiency of the mutant pre-mRNAs was that increasing the spacing between the stem–loop and the HDE abolishes an interaction between SLBP and U7 snRNP required for stabilizing the U7 snRNP on the pre-mRNA. Indeed, the residual processing of H2a/+12, which contains a 12-nt insertion within the EcoRI sequence (Dominski et al. 1999), is SLBP independent, since it is not affected by addition of a molar excess of the stem–loop RNA but is fully inhibited by the αM 2′O-methyl oligonucleotide, which is complementary to the first 17 nt of the mouse U7 snRNA (Fig. 2B, lanes 5,6).
We determined whether the same rules apply to histone pre-mRNA processing in Drosophila nuclear extracts. We increased the distance between the stem–loop and the HDE by inserting 4, 8, or 16 nt after the ACCCA of the dH3* pre-mRNA and tested cleavage of the resulting pre-mRNAs in Drosophila nuclear extract. The inserted sequence consisted of adenosines and cytidines and provided favorable cleavage sites at a constant distance from the HDE (Fig. 2F). Overall processing of the dH3 */+4 pre-mRNA was as efficient as processing of the parental dH3 * pre-mRNA and generated two products: the minor product that was identical to the product of processing of the dH3* pre-mRNA, hence resulting from cleavage after the cytidine at +4, and the major product that was 1 nt longer, resulting from cleaving the dH3 */+4 pre-mRNA after the ACCCA (Fig. 2C, lanes 1,2). Increasing the distance between the two sequence elements by 8 nt (Fig. 2F) yielded a similar result (Fig. 2D, lane 4). Processing of the resultant dH3*/+8 occurred with equal efficiency at the normal +4 cleavage site and after the ACCCA (Fig. 2D, lane 4). There was also a small amount of a product resulting from cleavage after an adenosine at the position +9. Processing at all three sites was dependent on both SLBP and U7 snRNP, since it was abolished by excess of the stem–loop RNA or a 2′O-methyl oligonucleotide, αDb (Fig. 5A, below), complementary to 20 nt of the Drosophila U7 snRNA (Fig. 2D, lanes 4–6). The shift of the processing site by only 1 nt together with retaining efficient processing at the original site upon inserting 4 and 8 nt was in clear contrast to mammalian processing, which is strongly determined by the position of the HDE, and insertions as small as 4 nt in the H2a/RI pre-mRNA result in the complete shift of the cleavage site by the corresponding number of nucleotides (Dominski et al. 1999).
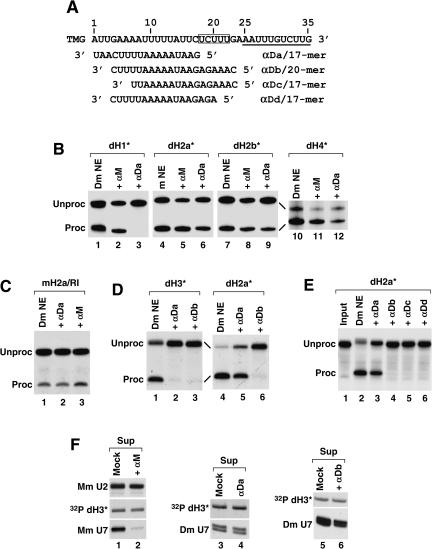
FIGURE 5.
Variable effects of 2′O-methyl oligonucleotides complementary to the Drosophila U7 snRNA on Drosophila histone pre-mRNA processing. (A) The sequence of the first 35 nt of the Drosophila U7 snRNA (written in 5′–3′ orientation) extending from the 5′ terminal tri methyl guanosine cap (TMG) to the end of the Sm binding site (underlined) and the sequences of the four 2′O-methyl oligonucleotides complementary to the 5′ end of the U7 snRNA (written in 3′–5′ orientation). The highly conserved UCUUU sequence in the U7 snRNA is boxed. (B) Processing of the dH1*, dH2a*, dH2b*, and dH4* histone pre-mRNAs in a Drosophila nuclear extract under control processing conditions (lanes 1,4,7,10) or after preincubation of the nuclear extract with either the αM (lanes 2,5,8,11) or the αDa oligonucleotide (lanes 3,6,9,12). (C) Processing of the mouse histone H2a/RI pre-mRNA in a Drosophila nuclear extract under control processing conditions (lane 1) or after preincubation of the nuclear extract with either the αDa (lane 2) or the αM oligonucleotide (lane 3). (D) Processing of the dH3* and dH2a* pre-mRNAs in a Drosophila nuclear extract under control processing conditions (lanes 1,4) or after preincubation of the nuclear extract with either the αDa (lanes 2,5) or the αDb oligonucleotide (lanes 3,6). (E) Processing of the dH2a* pre-mRNA in a Drosophila nuclear extract under control processing conditions (lane 2) or after preincubation of the nuclear extract with the indicated oligonucleotides complementary to the Drosophila U7 snRNA (lanes 3–6). Lane 1 contains the input pre-mRNA. (F) The indicated biotinylated 2′-O-methyl oligonucleotides were incubated with either a mouse (lane 1) or a Drosophila (lanes 4,6) nuclear extract and collected on streptavidin beads. The level of U7 snRNA left in the supernatant was determined by Northern blotting. In the mock experiments (lanes 1,3,5), the nuclear extracts were preincubated with streptavidin beads in the absence of any oligonucleotide. A small amount of the labeled pre-mRNA was added to the extract to control for the efficiency of precipitation (32P dH3*). The mouse U2 snRNA (Mm U2) was detected by Northern blotting to determine the specificity of depletion of the mouse U7 snRNA (Mm U7) in the mouse nuclear extract (lanes 1,2).
In contrast to the 4- and 8-nt insertions, insertion of 16 nt into the dH3 *pre-mRNA (Fig. 2F) abolished processing at the normal site and activated two weak cleavage sites further downstream (Fig. 2E, lane 4). Although we have not precisely mapped the cleavage site in dH3 */+16, comparison with the processing product of the mouse mH2a/+12 pre-mRNA generated in the mouse nuclear extract indicates that in both pre-mRNAs the cleavage sites have moved up to 9–12 nt from the regular cleavage site (not shown). Interestingly, while processing of the mH2a/+12 pre-mRNA in a mouse nuclear extract was independent of SLBP (Fig. 2B, lanes 4,5), processing of the dH3 */+16 pre-mRNAs remained sensitive to excess of stem–loop RNA (Fig. 2E, lane 5), indicating that binding of SLBP to the stem–loop is indispensable in Drosophila processing even if the cleavage site is located 13 nt distal to the stem–loop.
The role of SLBP and the HDE in Drosophila 3′ end processing
SLBP functions in mammalian histone pre-mRNA processing by facilitating recruitment of U7 snRNP to the pre-mRNA (Streit et al. 1993; Spycher et al. 1994; Dominski et al. 1999). We previously used an anti-SLBP antibody to precipitate processing complexes containing the U7 snRNP formed in a mouse nuclear extract on the mouse H2a-614 pre-mRNA (Dominski et al. 1999). However, we were unable to directly demonstrate that SLBP stimulates binding of the U7 snRNP to the pre-mRNA, since depleting or sequestering SLBP from the nuclear extract precludes subsequent precipitation of processing complexes by anti-SLBP. Recently we developed a new approach for isolating proteins (Dominski et al. 2003a) or processing complexes associated with histone pre-mRNAs (Dominski et al. 2003b) that allows direct testing of the role of SLBP in recruitment of the U7 snRNP. In this method, a pre-mRNA is annealed to an adapter 2′O-methyl oligonucleotide containing biotin on the 3′ end (Fig. 3A). The oligonucleotide is complementary to the first 17 nt of the pre-mRNA and formation of the duplex does not interfere with processing reaction but allows subsequent recovery of the processing complexes on streptavidin beads. We previously successfully used this method to isolate and identify the Drosophila U7 snRNA (Dominski et al. 2003b) and to determine binding affinities of 3′hExo to various mutants of the mature H2a-614 mRNAs (Dominski et al. 2003a).
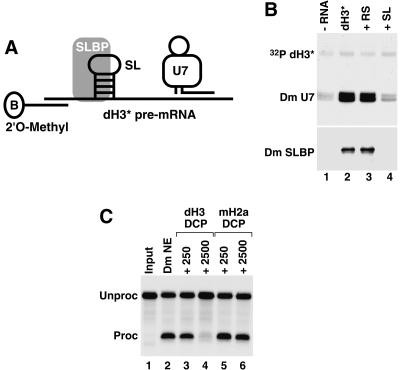
FIGURE 3.
The role of SLBP and the HDE in Drosophila processing. (A) The scheme for purification of processing complexes containing SLBP and U7 snRNP assembled on the dH3 * pre-mRNA. The adapter 2 O-methyl oligonucleotide complementary to the first 17 nt of the pre- ′ mRNA contains biotin (B) on the 3 ′end. (B) Detection of the Drosophila U7 snRNA (Dm U7) by Northern blotting (top) and SLBP (Dm SLBP) by Western blotting (bottom ) in Drosophila processing complexes formed on the dH3 * pre-mRNA in the absence of any competitor RNA (lane 2 ), in the presence of the reverse stem RNA (lane 3), or the stem–loop RNA (lane 4). The background amount of Dm SLBP and Dm U7 snRNA bound to streptavidin beads in the absence of the dH3 *pre-mRNA is shown in lane 1. A small amount of a radioactive dH3 *pre-mRNA ( 32P dH3 *) was added to each processing reaction to monitor the efficiency of isolation of substrate RNA and to control for ethanol precipitation. (C) Processing of the dH3 * pre-mRNA in a Drosophila nuclear extract under control conditions (lane 2 ) or in the presence of a 250 or 2500 M excess of the downstream cleavage product (DCP) from the dH3 pre-mRNA (lanes 3 ,4) or the mouse H2a-614 pre-mRNA (lanes 5,6).
To form processing complexes we incubated 50 ng of the dH3* pre-mRNA annealed to the 2′O-methyl oligonucleo-tide with either 250 μL of a normal Drosophila nuclear extract or the same amount of extract preincubated with a large excess of the stem–loop RNA to sequester all SLBP. As a negative control we used the same preparation of the nuclear extract preincubated with the reverse stem RNA (RS) unable to bind SLBP. Each sample contained a small amount of radioactively labeled dH3* pre-mRNA to monitor recovery of the substrate. The amount of SLBP and U7 snRNA in processing complexes was determined by using Western and Northern blotting, respectively. Upon brief incubation at room temperature with the Drosophila nuclear extract, the dH3* pre-mRNA formed processing complexes containing SLBP and the U7 snRNA, whereas only trace amounts of both factors were detected on streptavidin beads in the absence of the bulk of unlabeled dH3* pre-mRNA (Fig. 3B, lanes 1,2). Addition of a large molar excess of the RS RNA did not affect the amount of both factors detected in the processing complexes assembled on the dH3* pre-mRNA (Fig. 3B, lane 3). However, in the presence of the SL RNA, only background amounts of both SLBP and the U7 snRNA were collected on streptavidin beads, whereas the amount of the labeled dH3* pre-mRNA was constant, indicating that the pre-mRNA was efficiently recovered from the nuclear extract (Fig. 3B, lane 4). Based on these results we conclude that Drosophila SLBP is essential for binding of U7 snRNP to histone pre-mRNA.
The HDE from the mouse H2a-614 pre-mRNA at 50-fold molar excess fully inhibited processing of the H2a-614 pre-mRNA, whereas the HDE from the mouse H1t pre-mRNA at the same concentration had no effect on processing of this pre-mRNA (Dominski et al. 1999). This result demonstrated that the HDE from the H2a-614 pre-mRNA, but not from the H1t pre-mRNA, can efficiently interact with the U7 snRNP in the absence of the stem–loop. We tested an ability of the dH3-specific HDE to compete processing of the dH3* pre-mRNA. As a competitor in this experiment we used a 48-nt RNA corresponding to the downstream cleavage product (DCP) generated during processing of the Drosophila H3 pre-mRNA (Fig. 4A). The dH3 DCP RNA begins with the nucleotide that follows the cleavage site in the genuine Drosophila H3 pre-mRNA and contains the entire U7 binding site including the purine core GAGA. As a control we used the DCP generated during processing of the mouse H2a-614 pre-mRNA (Materials and Methods). This substrate is processed in Drosophila nuclear extracts with very low efficiency (Dominski et al. 2002b) and forms a weaker duplex with the Drosophila U7 snRNP than does the Drosophila H3 pre-mRNA (Dominski et al. 2003b). The dH3 DCP only slightly reduced processing of the labeled dH3* pre-mRNA at 250-fold molar excess but nearly completely inhibited processing of this pre-mRNA at 2500 molar excess (Fig. 3C, lanes 3,4). Even at this higher concentration, the mouse H2a-614 DCP had only a slight effect on processing of the dH3* substrate (Fig. 3C, lanes 5,6). Altogether, these results demonstrate that in Drosophila processing, the HDE separated from the stem–loop cannot efficiently interact with the U7 snRNA, and SLBP plays the key role in recruiting the U7 snRNP to the pre-mRNA.
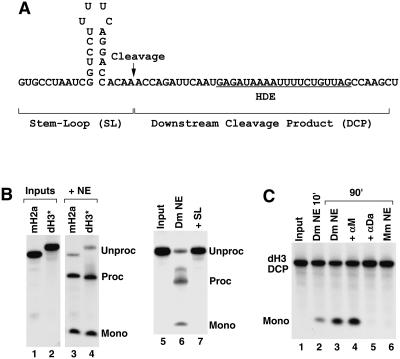
FIGURE 4.
Degradation of the Drosophila DCP by a 5′–3′ exonuclease. (A) The sequence of the genuine Drosophila H3 pre-mRNA encompassing the stem–loop and the HDE. The arrow indicates the cleavage site. The upstream and the downstream cleavage products are indicated. (B) Processing of the uniformly labeled mH2a/RI (lane 3) or dH3* pre-mRNA (lanes 4,6,7) in a mouse or a Drosophila nuclear extract, respectively, showing generation of the upstream cleavage product and the release of mononucleotides from degradation of the DCP. (C) Degradation of the dH3-specific DCP incubated during the indicated time in a Drosophila nuclear extract (lanes 2–5) under control conditions (lanes 2,3) or in the presence of the αM or αDa oligonucleotides (lanes 4,5). The dH3 DCP incubated for 90 min in a mouse nuclear extract is shown in lane 6.
Degradation of the Drosophila DCP by a 5′–3′ exonuclease
Following cleavage of mammalian histone pre-mRNA, the 3′ downstream cleavage product (DCP) is subjected to exonucleolytic degradation by an activity that is resistant to EDTA and dependent on U7 snRNP (Walther et al. 1998). Synthetic RNAs encompassing the DCP from the mouse H4-12 pre-mRNA (Walther et al. 1998) or the mouse H2a-614 pre-mRNA (our unpubl. results) are also rapidly degraded in mammalian nuclear extracts by the same activity, demonstrating that the exonucleolytic degradation of the DCP can be uncoupled from the endonucleolytic cleavage.
We analyzed the fate of the DCP generated during processing of the uniformly labeled dH3* pre-mRNA in Drosophila nuclear extracts. After 90-min incubation, in many preparations of the Drosophila nuclear extract the dH3* substrate was cleaved to form the upstream product containing the stem–loop, whereas the downstream cleavage product was not detectable and the remaining radioactivity was present as mononucleotides (Fig. 4B, lane 4). The radioactive mononucleotides were also genearted during processing of the uniformly labeled mouse H2a/RI pre-mRNA in a mouse nuclear extract (Fig. 4B, lane 3). Accumulation of the mononucleotides during processing of the dH3* pre-mRNA was inhibited by the presence of the SL RNA (Fig. 4B, lane 7) or an oligonucleotide blocking Drosophila U7 snRNA (not shown) and thus was strictly related to the processing activity and not a result of nonspecific degradation of the dH3* pre-mRNA. We tested whether 5′–3′ exonucleolytic activity can degrade a synthetic DCP from the Drosophila H3 pre-mRNA (Fig. 4A). The 39-nt dH3 DCP RNA was labeled at the 5′ end and incubated in a Drosophila nuclear extract under the same conditions as used for histone pre-mRNA processing and the release of the radioactive mononucleotide and disappearance of the input RNA monitored in denaturing gels. A small amount of the mononucleotide was generated after 10-min incubation at room temperature, and after 90 min ~50% of the input was degraded (Fig. 4C, lanes 2,3). The degradation was inhibited by the αDa oligonucleotide complementary to the first 17 nt of the Drosophila U7 snRNA but was unaffected by the same concentration of the αM complementary to the first 17 nt of the mouse U7 snRNA (Fig. 4C, lanes 4,5). Thus, release of the mononucleotide was dependent on the ability of the Drosophila U7 snRNP to bind the dH3 HDE. The Drosophila H3 DCP was stable during a 90-min incubation in a mouse nuclear extract (Fig. 3D, lane 6), and the mouse H2a-614 specific DCP was not degraded in the Drosophila nuclear extract (not shown), further demonstrating that degradation of the RNA substrate is U7 dependent and is not catalyzed by nonspecific nucleases resistant to EDTA. These results demonstrate that degradation of the DCP by the U7-dependent activity is a universal feature of 3′ end processing of histone pre-mRNAs that has been conserved between vertebrates and invertebrates.
Effects of 2′O-methyl oligonucleotides complementary to the Drosophila U7 snRNA on 3′ end processing
Drosophila U7 snRNA is unusual in having a long 5′ region that can potentially base pair with the HDE over a 24-nt region. At the 3′ end of this region (nt 18–22) there is a UCUUU sequence, which is complementary to the purine core of the HDE and is conserved in vertebrate and sea urchin U7 snRNAs. A 2′O-methyl oligonucleotide complementary to the first 17 nt of the U7 snRNA (αDa), which does not overlap with the UCUUU sequence (Fig. 5A), inhibits processing of the dH3* pre-mRNA (Fig. 5E), providing evidence that the base-pairing between the U7 snRNA and the HDE is essential in Drosophila processing (Dominski et al. 2003b). We tested the effect of the same oligonucleotide on processing of the substrates containing the HDE from the other four Drosophila histone pre-mRNAs: H1, H2a, H2b, and H4 (Fig. 1A). As a negative control we used a 2′O-methyl oligonucleotide complementary to the first 17 nt of the mouse H2a-614 pre-mRNA (αM). Surprisingly, processing of only one of these pre-mRNAs, dH1*, was inhibited by the αDa oligonucleotide, whereas processing of the three other substrates, dH2a*, dH2b*, and dH4* pre-mRNAs, was only partially reduced (Fig. 5B). Processing of the mouse H2a/RI pre-mRNA in the Drosophila nuclear extract was not affected by this oligonucleotide (Fig. 5C).
The sensitivity of dH3* and dH1* pre-mRNAs and the resistance of the four other pre-mRNAs were independent of the cell line (Kc or S2) or batch of Drosophila nuclear extract, suggesting that it reflects a fundamental difference in how U7 snRNP recognizes these two groups of substrates. We tested three additional 2′O-methyl oligonucleo-tides: αDb complementary to a 20-nt region of the Drosophila U7 snRNA that extends into the UCUUU conserved sequence; αDc, a 17-mer with the same 5′ end as αDb; and αDd, a 17-mer with the same 3′ end as αDb (Fig. 5A). Each of these oligonucleotides inhibited processing of the dH2a* pre-mRNA (Fig. 5D, lane 6; Fig. 5E, lanes 4–6) and the mouse H2a/RI pre-mRNA (not shown).
One possibility explaining the failure of a large molar excess of the αDa oligonucleotide to inhibit processing of dH2a*, dH2b*, dH4*, and the mouse H2a/RI pre-mRNAs was that the region located closer to the 5′ end of the Drosophila U7 snRNA is bound by proteins or structured and thus inaccessible to the oligonucleotide until the processing reaction has been initiated. We determined whether a large molar excess of the αDa and αDb oligonucleotides could efficiently deplete the U7 snRNA from a Drosophila nuclear extract. As a negative control we carried out a parallel experiment in the absence of any oligonucleotide. We also tested the ability of the αM oligonucleotide to deplete the mouse U7 snRNA from a mouse nuclear extract. Mouse or Drosophila nuclear extracts were incubated with the appropriate oligonucleotide, each containing biotin on the 5′ end, followed by absorption of the oligonucleotide and the bound U7 snRNP on streptavidin beads. The efficiency of depletion was determined by analyzing the amount of the U7 snRNAs remaining in each supernatant using Northern blotting with either mouse- or Drosophila-specific probes. Incubation of the mouse nuclear extract with the αM oligonucleotide reduced the amount of the U7 snRNA to ~20% of the amount detected in the control supernatant (Fig. 5F, lanes 1,2). The amount of the U2 snRNA (Mm U2) in each supernatant was similar, indicating that the αM oligonucleotide specifically removed the mouse U7 snRNA. In contrast, the αDa oligonucleotide did not reduce the amount of the Drosophila U7 snRNP in the Drosophila nuclear extract (Fig. 5F, lanes 3,4), and the αDb removed only ~50% of the U7 snRNP (Fig. 5F, lanes 5,6). Thus, the region near the 5′ end of the U7 snRNA in Drosophila U7 snRNP is not readily accessible.
Effects of mutations within the HDEs of the dH3* and H2a/RI pre-mRNAs on 3′ end processing
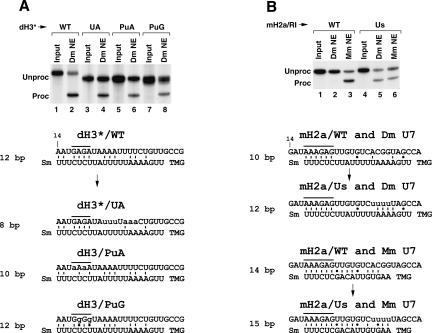
A feature of the extended 5′ end of the Drosophila U7 snRNA is a stretch of four adenosines followed by five uridines (Fig. 5A). All the Drosophila HDEs contain a stretch of uridines, and all, except H2b pre-mRNA, contain an adjacent stretch of adenosines, which could potentially base pair to this region of the U7 snRNA. We previously showed that two 3-nt mutations within the HDE of the dH3* replacing the AAA or the UUU located 23–25 and 27–29 nt downstream of the stem–loop with the complementary sequences had no effect on processing (Dominski et al. 2002b). When these two mutations were combined in the dH3*/UA pre-mRNA, processing was less efficient but not abolished, suggesting that base-pairing in this region is not essential for processing (Fig. 6, lane 4). In contrast, a 3-nt mutation within the AGA purine core (nt 18–20 downstream of the stem–loop) nearly completely inhibited processing (Dominski et al. 2002b). To investigate what features of the purine core are important for processing, we changed the GAGA in the dH3* pre-mRNA (overlined in Figs. 6, 7) to either four adenosines or four guanosines and tested the resulting mutant pre-mRNAs, dH3/PuA and dH3/PuG, for processing in a Drosophila nuclear extract. Both mutations in the purine core of the HDE had a moderate effect on processing, reducing efficiency from 60% for the wild type to ~25% for each mutant pre-mRNA (Fig. 6A). Thus, the presence of both adenosines and guanosines in the purine core of the HDE is important for maximum efficiency of processing.
FIGURE 6.
Effects of mutations within the HDEs of the dH3* and H2a/RI pre-mRNAs on 3′ end processing. (A) Processing of the uniformly labeled wild-type dH3* histone pre-mRNA (lanes 1,2) and its mutants, UA, PuA, and PuG (lanes 3–8), in a Drosophila nuclear extract. The sequence of the pre-mRNAs and their likely base-pairing with the Drosophila U7 snRNA are shown at the bottom. GU base pairs are indicated with dots. In each duplex, the pre-mRNA sequence beginning with the 14th nucleotide following the stem–loop is shown at the top and the U7 snRNA sequence is shown at the bottom. The purine core is overlined and the mutated nucleotides are written in lowercase letters. (B) Processing of the wild-type mouse H2a/RI pre-mRNA (lanes 1–3) and the mutant mH2a/Us pre-mRNA containing four Us in the HDE (lanes 4–6) in a Drosophila (lanes 2,5) or a mouse (lanes 3,6) nuclear extract. Likely base-pairing schemes between the pre-mRNAs and either the Drosophila U7 snRNA or mouse U7 snRNA are shown at the bottom.
FIGURE 7.
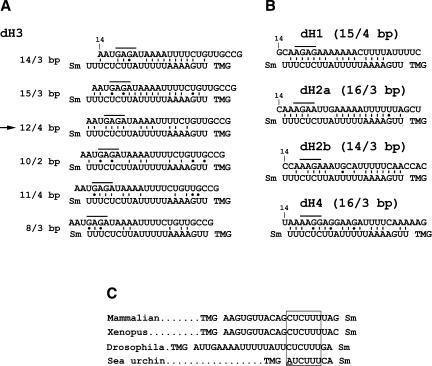
Potential base-pairing schemes between Drosophila pre-mRNAs and the Drosophila U7 snRNA. (A) Several possible base-pairing alignments between the dH3-specific HDE (top sequence) and the first 21 nt of the Drosophila U7 snRNA (bottom sequence). The sequence of the HDE starts 14 nt downstream of the stem–loop (10 nt after the cleavage site). The purine core located between nt 17 and 20 is overlined. The Sm indicates the Sm binding site and the TMG indicates the tri-methyl guanosine cap at the 5′ end of the U7 snRNA. There are two additional nucleotides between the last indicated nucleotide of the U7 snRNA and the Sm binding site. Due to the proximity to the Sm binding site these nucleotides are unlikely to base pair with the pre-mRNA and are not included. The arrow indicates the most favorable alignment that forms a relatively stable duplex and contains the highest number of base pairs between the purine core of the HDE and the UCUUU sequence of the U7 snRNA. (B) The most favorable alignments between the HDE from the four remaining Drosophila histone pre-mRNAs, H1, H2a, H2b, and H4, and the Drosophila U7 snRNA. The numbers in parentheses indicate the total number of base pairs within the duplex and the number of the base pairs formed between the purine core of the HDE and the CUCUUU sequence of the U7 snRNA, respectively. (C) The sequences located between the trimethyl guanosine (TMG) cap and the Sm binding site of the U7 snRNAs from evolutionarily distant organisms. The highly conserved CUCUUU sequence is boxed. The adenosine in the sea urchin U7 snRNA departing from the consensus is underlined.
The mouse H2a/RI pre-mRNA is processed in Drosophila nuclear extracts with very low efficiency. The HDE of this pre-mRNA also contains the purine-rich core, which can efficiently base pair with the Drosophila U7 snRNA. However, the mouse H2a/RI pre-mRNA can only form a total of 10 bp with the Drosophila U7 snRNA, compared to the 12 bp formed by the dH3*. In particular the mouse H2a HDE cannot base pair with the stretch of adenosines and uridines in the Drosophila U7 snRNA. We replaced the ACGG in the mouse H2a HDE with four uridines to allow additional base-pairing to the 5′ end of the Drosophila U7 snRNA (Fig. 6B). This nucleotide substitution should increase the number of base pairs formed between the pre-mRNA and the Drosophila U7 snRNA from 10 to 12 without disrupting the base-pairing with the mouse U7 snRNA. We compared processing of the original mouse H2a/RI pre-mRNA and the mutant mH2a/Us pre-mRNA in both Drosophila and mouse nuclear extracts. The wild-type H2a/RI pre-mRNA was not detectably processed in this preparation of a Drosophila nuclear extract, although the same preparation of the nuclear extract processed ~45% of the mutant pre-mRNA with the improved base-pairing to the 5′ end of the Drosophila U7 snRNA (Fig. 6B, lanes 2,5). In a mouse nuclear extract the wild-type H2a/RI pre mRNA was processed with ~60% efficiency, and substitution of the ACGG with the UUUU reduced the efficiency to 35% (Fig. 6B, lanes 3,6), possibly because the mutation incorporated an additional mismatch to the duplex with the mouse U7 snRNA or the UUUU sequence in the HDE is less favorable in mammalian processing. Thus, by increasing the base-pairing potential of the mouse H2a pre-mRNA to the Drosophila U7 snRNA without significantly changing the base-pairing potential of the mutant pre-mRNA to the mouse U7 snRNA, we created a substrate that, unlike any of the wild-type Drosophila histone pre-mRNAs, undergoes efficient processing in both Drosophila and mouse nuclear extracts.
DISCUSSION
We carried out in vitro studies using nuclear extracts from Drosophila Kc cells and mouse myeloma cells to compare 3′ end processing of histone pre-mRNAs in Drosophila and mammalian systems. Our studies demonstrate that although Drosophila and mammalian histone pre-mRNA processing occur with similar chemistry and both require SLBP and the U7 snRNP, the two mechanisms differ significantly in the relative importance of these trans-acting factors and in the specification of the cleavage site.
The stem–loop and SLBP play a dominant role in Drosophila processing
Drosophila nuclear extracts cleave histone pre-mRNAs after the fourth nucleotide following the stem–loop and prefer an adenosine preceding the cleavage site. Consistent with this, all natural Drosophila histone pre-mRNAs contain an adenosine in this position. If the fourth nucleotide is changed to a pyrimidine, cleavage is also efficient after an adenosine at the third position but not after an adenosine located 5 nt downstream of the stem–loop, i.e., at the site exclusively utilized during mammalian processing. Sea urchin histone mRNAs, the only other invertebrate histone mRNAs with the characterized 3′ ends, terminate with an ACCA consensus sequence (Birnstiel and Schaufele 1988). Thus, cleavage after the fourth nucleotide following the stem–loop may be a general feature of 3′ end processing of invertebrate histone pre-mRNAs. Both Drosophila and mammalian processing machineries are similar in their extreme resistance to EDTA, generation of a 3′ hydroxyl group at the end of the upstream cleavage product, and degradation of the downstream cleavage product by a U7 snRNP dependent activity. These results suggest that both processing machineries utilize the same or a highly related cleavage factor in 3′ end processing of histone pre-mRNAs.
In mammalian processing, the site of cleavage is determined by the position of the HDE, and moving the HDE, and, hence, the U7 snRNP, away from the stem–loop by as few as 4 nt results in a corresponding shift of the cleavage site (Scharl and Steitz 1994, 1996; Dominski et al. 1999). This observation led to the hypothesis that U7 snRNP recruits the cleavage factor to the pre-mRNA and acts as a molecular ruler to specify the cleavage site (Scharl and Steitz 1994, 1996). SLBP bound to the stem–loop facilitates binding of the U7 snRNP to the HDE but does not play a direct role in recruitment of the cleavage factor. Consistent with this model, removal of SLBP, or using a substrate that cannot bind SLBP, reduces processing activity but does not abolish it.
In contrast to mammalian processing, processing of Drosophila histone pre-mRNA is absolutely dependent on SLBP. In addition, increasing the distance between the stem–loop and the HDE by 4 or 8 nt in Drosophila histone pre-mRNA moved the cleavage site only 1 nt upstream from its normal position and did not abolish processing at the normal site. Larger insertions between the stem–loop and the HDE resulted in low efficiency cleavage further away from the stem–loop, but cleavage at these sites was still dependent on SLBP. This is in direct contrast to mammalian histone processing, where cleavage at the distant sites is independent of SLBP. Thus, in Drosophila processing the U7 snRNP does not function as a molecular ruler, but instead SLBP plays the critical role in specifying the cleavage site.
To explain the observed differences between processing in Drosophila and mammalian nuclear extracts, we propose that within the Drosophila processing complex SLBP tightly interacts with the U7 snRNP, and this interaction is essential for bringing the U7 snRNP to the pre-mRNA. The two factors remain associated even if their respective binding sites are separated by a larger distance, likely by looping out the inserted nucleotides. The mutant pre-mRNAs are preferentially cleaved close to the stem–loop, reflecting the critical role of SLBP in forming the processing complex, although the precise position of the cleavage site and efficiency of processing depends on the size of the insert. In mammalian processing, the region between the stem–loop and the HDE is either rigidified, thus precluding looping out the inserted nucleotides, as previously suggested (Scharl and Steitz 1994, 1996), or the interaction between SLBP and the U7 snRNP is relatively weak and disrupted by larger insertions, so binding of the U7 snRNP to the pre-mRNA depends solely on the base-pairing interaction. It is likely that in Drosophila processing the cleavage factor is recruited to histone pre-mRNA by interaction with both the U7 snRNP and SLBP, and neither factor is competent to carry out this function individually.
The 5′ end of Drosophila U7 snRNA is not accessible
In mammalian nuclear extracts processing of histone pre-mRNAs is efficiently inhibited by relatively short 2′O-methyl oligonucleotides complementary to the 5′ end of the mammalian U7 snRNA (Cotten et al. 1991). These oligonucleotides, including a 10-mer, are also very efficient in depleting the U7 snRNP from nuclear extracts and were successfully used to affinity purify U7 snRNP from mammalian cells, demonstrating that the 5′ end of the mammalian U7 snRNA is readily accessible (Smith et al. 1991; Pillai et al. 2001, 2003). In contrast, two relatively long oligonucleotides, αDa, complementary to the first 17 nt of the Drosophila U7 snRNA, and αDb, complementary to nt 4–23, were not effective in depleting the U7 snRNP from Drosophila nuclear extracts. These results suggest that the 5′ end of U7 snRNA is not accessible in the Drosophila U7 snRNP.
Surprisingly, the αDa 2′O-methyl oligonucleotide abolished processing of the dH3* and dH1* pre-mRNAs but did not significantly affect processing of the other three Drosophila histone pre-mRNAs. Three additional oligonucleotides complementary to the regions of the U7 snRNP located closer to the Sm binding site effectively blocked processing of all five histone pre-mRNAs. We do not understand why processing of only two Drosophila pre-mRNAs was affected by the αDa oligonucleotide and which features of the HDEs make processing of the Drosophila pre-mRNAs either sensitive or resistant to this oligonucleotide. Selective inhibition of processing by the αDa oligonucleotide depending on the type of pre-mRNA used in the reaction suggests that blocking of the U7 snRNA must occur during processing. One possibility is that the U7 snRNP is initially recruited to the pre-mRNA solely by SLBP bound to the pre-mRNA, and later this interaction is followed by formation of a duplex between the HDE and the U7 snRNA, as a result of unmasking of the 5′ end of U7 snRNA. The αDa oligonucleotide might block binding of the U7 snRNA to the HDE in the dH1* and dH3* pre-mRNAs, but not in the other pre-mRNAs, during this later step, while the other oligonucleotides block binding to all the HDEs.
Overall, our studies indicate that the structure of the 5′ end of the Drosophila U7 snRNA and the mechanism of its initial interactions with the HDE differ significantly from the recognition of the HDE in processing of mammalian histone pre-mRNAs.
Base-pairing between U7 snRNA and HDE in Drosophila processing
In vitro processing of all five Drosophila histone pre-mRNAs is absolutely dependent on SLBP (Dominski et al. 2002b). Here we demonstrated that SLBP is essential for recruitment of the U7 snRNP to the pre-mRNA. The necessity of SLBP for recruitment of the U7snRNP to the Drosophila pre-mRNAs suggests that either Drosophila HDEs are unable to form a strong duplex with the U7 snRNA or that the interaction of the U7 snRNP with the SLBP/pre-mRNA complex is necessary to promote base-pairing by making the 5′ end of U7 snRNA accessible.
Both the 5′ end of the Drosophila U7 snRNA and Drosophila HDEs are AU rich, allowing a number of possible base-pair schemes for making a duplex between the two RNAs. We hypothesize that the most likely alignment used during processing is the one that allows formation of the largest number of base pairs between the purine core of the HDE and the CUCUUU sequence in the U7 snRNA and not necessarily the alignment, which allows formation of the overall most stable duplex (Fig. 7). The CUCUUU sequence is highly conserved among all known U7 snRNAs and is involved in recognition of the purine core in sea urchin and mammalian histone pre-mRNAs. A 3-nt mutation within the purine core of the dH3* pre-mRNA abolished processing (Dominski et al. 2002b), whereas a 6-nt mutation within the AU-rich region immediately downstream of the purine core only partially inhibited processing. These results support our interpretation that base-pairing between the U7 snRNA and the purine core is critical, whereas formation of additional base in other regions increases the efficiency of Drosophila processing. It is also possible that the base-pairing interaction is limited to the purine core and the CUCUUU sequence in the U7 snRNA, whereas the AU-rich sequences in the U7 snRNA and the HDE are brought together by protein–protein interactions.
We demonstrated that the HDE of the dH3* pre-mRNA can abolish processing of the full-length substrate, presumably by sequestering the U7 snRNP, only when present at very high concentrations. Interestingly, this weak interaction of Drosophila HDEs with the U7 snRNP is sufficient to recruit a 5′–3′ exonuclease that specifically degrades the downstream cleavage product in a U7 dependent manner. Thus, the endonucleolytic cleavage must require much stronger binding of the U7 snRNP to the pre-mRNA, while degradation of the DCP by an exonuclease may require only loose association of the HDE with the U7 snRNP.
Conclusions
The most notable difference between histone pre-mRNA processing in Drosophila and mammalian nuclear extracts is the absolute dependence of Drosophila processing on SLBP and the role of SLBP in specifying the cleavage site close to the stem–loop. The Drosophila U7 snRNP does not function as a molecular ruler in processing and this feature most likely reflects a critical role of SLBP in recruiting the cleavage factor as well as the U7 snRNP, to histone pre-mRNA. Our data suggest that SLBP and the U7 snRNP may form a tight complex on the histone pre-mRNA, and this complex remains stable even in the presence of large insertions between the stem–loop and the HDE.
The similarities in the chemistry of the cleavage reaction, including preference for an adenosine preceding the cleavage site and generation of the 3′OH group in the presence of EDTA, as well as degradation of the downstream cleavage product by a U7-dependent 5′–3′ exonuclease suggest that the cleavage factor has been conserved between Drosophila and mammalian processing. It will be of interest to determine whether there are factors unique to only one of these two types of organisms emphasizing long evolutionary distance and the divergence between vertebrates and invertebrates.
MATERIALS AND METHODS
RNA
RNA oligonucleotides were synthesized by Dharmacon. The sequences of the 2′O-methyl oligonucleotides complementary to the Drosophila U7 snRNA are shown in Figure 5. Other 2′O-methyl oligonucleotides had the following sequences (written in 5′–3′ orientation):
AAAGAGCUGUAACACUU (αM), CGAGCUCGAAUUCGCCC (adapter oligonucleotide with biotin on the 3′ end), ACCAGAUUCAAUGAGAUAAAAUUUUCUGUUAGCCAAGCU (Drosophila H3 DCP), and CUGAAUCAGAUAAAGAGUUGUGUCACGGUAGCCAAGCU (mouse H2a-614 DCP).
Drosophila and mouse-specific pre-mRNA substrates were generated by T7 transcription. In most cases the pre-mRNA substrates were first synthesized in the presence of unlabeled nucleotides, gel purified, dephosphorylated by calf intestinal phosphatase, and labeled at the 5′ end using T4 polynucleotide kinase (NEB) and [32P]-γATP. Internally labeled RNA substrates were synthesized by incorporation of [32P]-αUTP, as described (Dominski et al. 1999).
Nuclear extract preparation and histone pre-mRNA processing
Nuclear extracts were prepared from Drosophila Kc cells and mouse myeloma cells, and processing of histone pre-mRNAs was carried out as previously described (Dominski et al. 1995, 2002b). Each processing reaction contained in a final volume of 10 μL the following: 7.5 μL nuclear extract (10 mg/mL protein), 20 mM EDTA (pH 8), and 0.1 pmol of a radioactively labeled substrate (Dominski et al. 1999, 2002b). Drosophila and mouse processing reactions were incubated for 90 min at 25°C (room temperature) or 32°C, respectively. The reactions were then treated with 5 μg of proteinase K, diluted with 4 volumes of 7 M urea dye, and the processing products analyzed in 8%/7 M polyacrylamide gels.
Formation of Drosophila processing complexes
Drosophila processing complexes were assembled and isolated as described (Dominski et al. 2003b). The mouse and Drosophila U7 snRNAs were analyzed as described (Dominski et al. 1999, 2003b).
Acknowledgments
This work was supported by NIH grant GM58921 to W.F.M.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2179305.
REFERENCES
- Adamson, T.E. and Price, D.H. 2003. Cotranscriptional processing of Drosophila histone mRNAs. Mol. Cell Biol. 23: 4046–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz, T.N. and Schumperli, D. 2003. Evolutionary conservation of the U7 small nuclear ribonucleoprotein in Drosophila melanogaster. RNA 9: 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz, T.N., Gruber, A., and Schumperli, D. 2005. U7 snRNP-specific Lsm11 protein: Dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucleic Acids Res. 33: 2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel, M.L. and Schaufele, F.J. 1988. Structure and function of minor snRNPs. In Structure and function of major and minor small ribonucleoprotein particles (ed. M.L. Birnstiel), pp. 155–182. Springer-Verlag, Berlin.
- Bond, U.M., Yario, T.A., and Steitz, J.A. 1991. Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes & Dev. 5: 1709–1722. [DOI] [PubMed] [Google Scholar]
- Cotten, M., Oberhauser, B., Brunar, H., Holzner, A., Issakides, G., Noe, C.R., Schaffner, G., Wagner, E., and Birnstiel, M.L. 1991. 2′- O-methyl, 2′-O-ethyl oligoribonucleotides and phosphorothioate oligodeoxyribonucleotides as inhibitors of the in vitro U7 snRNP-dependent mRNA processing event. Nucleic Acids Res. 19: 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Sumerel, J., Hanson, R.J., and Marzluff, W.F. 1995. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA 1: 915–923. [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Zheng, L.-X., Sanchez, R., and Marzluff, W.F. 1999. The stem-loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol. Cell. Biol. 19: 3561–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Erkmann, J.A., Yang, X., Sanchez, R., and Marzluff, W.F. 2002a. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes & Dev. 16: 58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X., Raska, C.S., Santiago, C.S., Borchers, C.H., Duronio, R.J., and Marzluff, W.F. 2002b. 3′ end processing of Drosophila histone pre-mRNAs: Requirement for phosphorylated dSLBP and co-evolution of the histone pre-mRNA processing system. Mol. Cell. Biol. 22: 6648–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X., Kaygun, H., and Marzluff, W.F. 2003a. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell 12: 295–305. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X.C., Purdy, M., and Marzluff, W.F. 2003b. Cloning and characterization of the Drosophila U7 small nuclear RNA. Proc. Natl. Acad. Sci 100: 9422–9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger, A., Schaller, A., and Schümperli, D. 1998. Functional importance of conserved nucleotides at the histone RNA 3′ processing site. RNA 4: 246–256. [PMC free article] [PubMed] [Google Scholar]
- Galli, G., Hofstetter, H., Stunnenberg, H.G., and Birnstiel, M.L. 1983. Biochemical complementation with RNA in the Xenopus oocyte: A small RNA is required for the generation of 3′ histone mRNA termini. Cell 34: 823–828. [DOI] [PubMed] [Google Scholar]
- Gick, O., Kramer, A., Keller, W., and Birnstiel, M.L. 1986. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 5: 1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, C., Stefanovic, B., and Schümperli, D. 1993. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 12: 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg, P.A. and Melton, D.A. 1984. Formation of the 3′ end of histone mRNA by post-transcriptional processing. Nature 308: 203–206. [DOI] [PubMed] [Google Scholar]
- Martin, F., Schaller, A., Eglite, S., Schümperli, D., and Müller, B. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16: 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff, W.F., Gongidi, P., Woods, K.R., Jin, J.P., and Maltais, L. 2002. The human and mouse replication-dependent histone genes. Genomics 80: 487–498. [PubMed] [Google Scholar]
- Mowry, K.L. and Steitz, J.A. 1987a. Both conserved signals on mammalian histone pre-mRNAs associate with small nuclear ribonucleoproteins during 3′ end formation in vitro. Mol. Cell. Biol. 7: 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1987b. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone pre-messenger RNA’s. Science 238: 1682–1687. [DOI] [PubMed] [Google Scholar]
- Mowry, K.L., Oh, R., and Steitz, J.A. 1989. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3′-end processing reaction. Mol. Cell. Biol. 9: 3105–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Will, C.L., Lührmann, R., Schümperli, D., andMüller, B. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20: 5470–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Grimmler, M., Meister, G., Will, C.L., Luhrmann, R., Fischer, U., and Schümperli, D. 2003. Unique Sm core structure of U7 snRNPs: Assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes & Dev. 17: 2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl, E.C. and Steitz, J.A. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 13: 2432–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1996. Length suppression in histone messenger RNA 3′-end maturation: Processing defects of insertion mutant pre-messenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc. Natl. Acad. Sci. 93: 14659–14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele, F., Gilmartin, G.M., Bannwarth, W., and Birnstiel, M.L. 1986. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature 323: 777–781. [DOI] [PubMed] [Google Scholar]
- Smith, H.O., Tabiti, K., Schaffner, G., Soldati, D., Albrecht, U., and Birnstiel, M.L. 1991. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotiny-lated 2′-O-methyl oligoribonucleotides. Proc. Natl. Acad. Sci. 88: 9784–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb, B., Cruz-Reyes, J., and Rusche, L.N. 2001. Direct sizing of RNA fragments using RNase-generated standards. Methods Enzymol. 342: 378–383. [DOI] [PubMed] [Google Scholar]
- Spycher, C., Streit, A., Stefanovic, B., Albrecht, D., Koning, T.H.W., and Schümperli, D. 1994. 3′ end processing of mouse histone pre-mRNA: Evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 22: 4023–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit, A., Koning, T.W., Soldati, D., Melin, L., and Schümperli, D. 1993. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 21: 1569–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.N., Wittop, K.T., Schümperli, D., and Muller, B. 1998. A 5′–3′ exonuclease activity involved in forming the 3′ products of histone pre-mRNA processing in vitro. RNA 4: 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.-F., Whitfield, M.L., Ingledue, T.I., Dominski, Z., and Marzluff, W.F. 1996. The protein which binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes & Dev. 10: 3028–3040. [DOI] [PubMed] [Google Scholar]