Abstract
We have encountered an unexpected property of rRNA secondary structures that may generalize to all RNAs. Analysis of 8892 ribosomal RNA sequences and structures from a wide range of species revealed unexpected universal compositional trends. First, different categories of rRNA secondary structure (stems, loops, bulges, and junctions) have distinct, characteristic base compositions. Second, the observed patterns of variation are similar among sequences from large and small rRNA subunits and all domains of life, despite extensive evolutionary divergence. Surprisingly, these differences do not seem to be related to selection for different compositions in different structural categories, but rather relate to the overall composition of the molecule: Randomized RNAs with no evolutionary history show the same structure-dependent compositional biases as rRNAs. These compositional trends may improve the accuracy of RNA secondary structure prediction, because they allow us to compare predicted structures against known compositional preferences. They also suggest caution in interpreting differences in the rate of change of the GC content in different parts of the molecule as evidence of differential selection.
Keywords: RNA secondary structure, base composition, selection, self-organization
INTRODUCTION
RNA molecules can be divided into secondary structure components, which may have distinct biological functions. For example, loops that terminate a single stem, such as the GNRA tetraloop, may engage in tertiary interactions, or junctions that link several helices together may be selected to orient the helices specifically. Recently, comparisons between crystal structures of homologous RNAs have revealed a surprising amount of flexibility in how a given structure is achieved (Westhof and Massire 2004), and within the context of highly conserved structural motifs, the ability for bases to substitute for one another can be predicted from first principles of structural similarity (Lescoute et al. 2005). Although these structural constraints are critical for understanding how highly conserved regions of RNA essential for biological function evolve, we expect that more general rules that constrain the less highly conserved features of RNA secondary structure also exist. These rules may help us improve algorithms for structure prediction and will inform our understanding of the vast diversity of RNAs that can perform a given catalytic task.
Because organisms vary widely in genome GC content in a manner consistent with directional mutation pressure (Sueoka 1962, 1988), we might expect the different parts of the RNA molecule to change in composition at different rates due to the different selective constraints in different regions, just as the three reading frames within mRNAs change in composition at different rates that reflect the average effect of mutations in each frame (Muto and Osawa 1987). Specifically, third position changes have the least effect because they are often synonymous, and second position changes have the greatest effect because they often substitute an amino acid that is chemically very different. This gives rise to substantially different slopes when regressing the GC content at a particular codon position against the overall GC content and also affects the amino acid composition of the protein correspondingly (Sueoka 1961; Muto and Osawa 1987; Lobry 1997; Sueoka 1999; Singer and Hickey 2000; Knight et al. 2001). Indeed, different RNA molecules such as tRNAs, rRNAs, and mRNAs also change in composition at different rates relative to overall GC content (Muto and Osawa 1987). Even within a single molecule, the paired and unpaired regions of 16S rRNA in bacteria and archaea have been shown to differ in slope substantially (Wang and Hickey 2002). In this article, we test whether these differences in response to overall genome GC content hold for finer-grained structural categories, within both large and small subunit rRNAs in all three domains of life. We also test whether the differences in response are due to differences in purifying selection in the different regions, or whether they are due to intrinsic differences in the amount of base-pairing expected in sequences of different composition (Schultes et al. 1999).
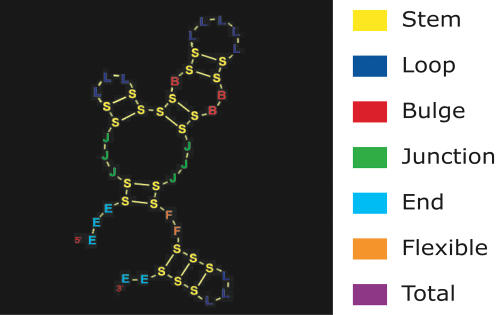
In RNA, each nucleotide can be assigned to one of six secondary structure categories: stem, loop, bulge, junction, or end, or a type of unpaired base that we provisionally call “flexible” (Fig. 1 ▶). Stems are the base-paired regions of the molecule. Loops, bulges, and junctions are unpaired regions enclosed by stems. Let the degree of an unpaired region be the number of stems attached to it. Then, loops have degree one, bulges have degree two, and junctions have a degree higher than two. The ends are all unpaired bases on the 5′ and 3′ end of the molecule. Flexible bases—also known as “freely rotating joints” (Schuster et al. 1994), although this may be a misnomer at the tertiary structure level—make up unpaired regions that connect two stems but that are not part of a closed RNA structure.
FIGURE 1.
Structural elements in RNA secondary structure. There are six different structural categories: stem, loop, bulge, junction, end, and flexible. Each element has been assigned a color that is used throughout this article to visualize data on that element. The seventh color (purple) is used to show data for the whole molecule (total).
We examined whether the four bases were differentially abundant in these different structural categories. We had three primary motivations for this analysis: First, we wanted to test whether a finer-grained analysis of the unpaired bases would reveal differences among bulges, loops, and junctions; second, we wanted to test whether any compositional patterns were specific to 16S rRNA, as previously observed (Wang and Hickey 2002), or were shared between subunits and domains of life; third, we wanted to test whether the compositional patterns resulted from selection on the biological sequences or would be obtained from any arbitrary sequence of the same composition. If there are consistent differences in the compositions of different structural elements that hold across many types of RNA molecule, we may be able to use these differences to refine the accuracy of secondary structure prediction programs such as BayesFold (Knight et al. 2004) by testing whether a computed secondary structure matches the known compositional preferences.
In this study we asked the following four questions about rRNA structure and composition:
Do the different categories of unpaired regions differ in composition from one another? There is a known bias toward purines in rRNAs and several other biological RNAs (Elson and Chargaff 1955; Schultes et al. 1997 Schultes et al. 1999; Lao and Forsdyke 2000), which can only come from the unpaired regions because the paired regions have a 1:1 ratio of purines to pyrimidines. We tested whether the different types of unpaired regions (especially loops, bulges, and junctions) contribute equally to this purine bias in the vast sample of rRNAs now available in the rRNA database (Wuyts et al. 2001, 2002) and, more generally, whether compositions in these unpaired regions are identical within each molecule.
Are patterns of composition conserved across different molecules and different domains of life? rRNA is often used to infer phylogenetic relationships between species, in part because it is seldom horizontally transferred. Because there has been no detectable recombination between the large and the small subunits, or between sequences from the three domains of life (bacteria, archaea, and eukaryotes), the six combinations of domains and subunits provide six independent evolutionary “experiments” about the extent to which a long RNA molecule can vary while maintaining its function. We tested whether the patterns of variation were similar in each subunit (the two subunits differ in function) and each domain of life, which could suggest that the processes of RNA evolution are generalizable across RNAs of different kinds and long time spans.
Are differences in composition between structural categories due to natural selection? Structural motifs that depend on specific sequences— including but not limited to UNCG tetraloops (Tuerk et al. 1988; Molinaro and Tinoco 1995), GNRA tetraloops (Woese et al. 1990), A platforms (Cate et al. 1996), and the A minor motif (Wimberly et al. 2000; Gutell et al. 2000; Doherty et al. 2001; Nissen et al. 2001)—have been proposed to play a major role in structuring rRNA and in causing the purine bias in unpaired regions. If this hypothesis were correct, we would expect that natural rRNA sequences, which are under strong natural selection to maintain their function and hence structure, might have substantially different compositions in each structural component than would randomized sequences with the same composition. Evolved RNAs would certainly be expected to have a much smaller range of composition. However, if the differences between structural categories arise automatically from differences in overall sequence composition, we would expect that the natural and randomized sequences would show similar compositional patterns and spans in different structural components.
Are differences in the strength of the response of each structural category to changes in genome GC content due to natural selection? The composition of the rRNA is known to correlate strongly with the composition of the surrounding genomic context (Muto and Osawa 1987; Guy and Roten 2004), except in hyperthermophiles (in which it correlates with optimal growth temperature) (Galtier and Lobry 1997). We tested whether these correlations hold true for each structural category independently, or whether the structural categories are negatively correlated such that directional changes in one category are counterbalanced by opposite changes in another structural category (this latter scenario would be expected if the composition of the overall rRNA sequence were under selection). Traditionally the correlations in GC content with other informational macromolecules have been explained by purifying selection (Sueoka 1961; Muto and Osawa 1987; Wang and Hickey 2002); we looked at the role of self-organization by examining these correlations in randomly generated RNA sequences.
RESULTS AND DISCUSSION
Are the different types of unpaired regions identical in composition?
Analyses of the base composition of RNA have typically focused on the differences between paired and unpaired regions (Schultes et al. 1997; Wang and Hickey 2002). Here, we address the differences among six separate elements of secondary structure. This separation is easily justified, because all structural elements are believed to have distinct functions in the molecule. For example, junctions affect the spatial orientation of the stems that they connect and certain kinds of loops and bulges are involved in docking interactions that hold the three-dimensional structure together (Tinoco 1996). There is thus no reason to believe a priori that different unpaired elements would have the same base composition. Similarly, ends and flexible regions are often combined into one category, “external elements” (Hofacker et al. 1994), but there are reasons to believe that they might have different compositions. The ends are not constrained in their conformation, but the flexible regions are bounded by helices and thus may appear more similar to junctions than to free ends.
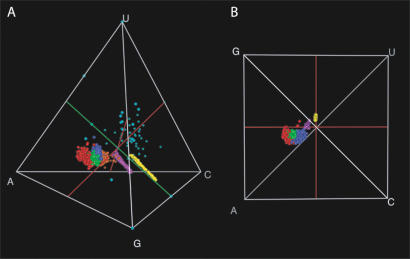
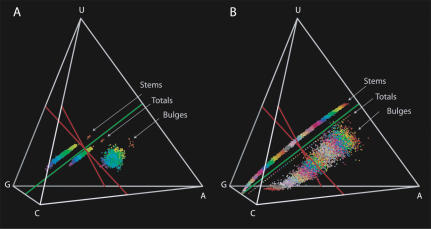
The base composition for each of the six structural elements and the overall base composition of the molecule are visualized in composition space (Fig. 2 ▶). An important feature for orientation in this space is Chargaff ’s axis. This axis, where the amounts of C and G are equal and the amounts of A and U are equal, indicates the line in composition space where Watson-Crick base-pairing holds exactly. Deviations from Chargaff ’s axis tell us about compositional differences due to processes other than changes in GC content, which can simply result from compensatory mutations in stems. Our results show that all structural elements have distinct compositions. The compositions of the whole molecules and the stems show linear distributions along Chargaff ’s axis, as expected (Schultes et al. 1997), with considerable variation in GC content but very little variation in the other directions (Fig. 2B ▶). Remarkably, the three unpaired regions that contain a substantial number of bases (loops, bulges, and junctions) have separate distributions. The ends and the flexible bases are scattered throughout composition space, because there are very few bases in these categories, so the sampling error is large. Therefore these latter two categories are excluded from the rest of the analysis.
FIGURE 2.
Compositional biases in SSU archaea. The composition space is visualized as a tetrahedron, which can be viewed from many different perspectives. All structural elements have a different color, as defined in Figure 1 ▶. In the oblique view (A), we show the linear distributions along Chargaff’s axis (green line) of the totals and stems. The loops, the bulges, and the junctions are clearly distinct. The ends and the flexible bases contain few bases and are therefore scattered by sampling error throughout composition space. In the other view, down Chargaff’s axis (B), in which the variation in GC content is not visible, we show how constrained all elements are in their variation in the two other directions. This view also emphasizes the purine bias in the totals and the unpaired regions.
In a first attempt to quantify the compositional patterns, we looked at the amount of bias and its direction for the mean composition of every structural element, except ends and flexible bases, for all annotated sequences. We calculated the amount of bias as the smallest distance between the mean of the sample and Chargaff ’s axis. Looking down Chargaff ’s axis, the bias can be in any of four directions: toward purines (AG), pyrimidines (UC), nucleotides with an amino group (AC), or nucleotides with a keto group (GU). Thus, we expressed the bias as the sum of the excess of either G or C and the excess of either A or U. For example, the sequence CUUAAAGGGG, which consists of 10% C, 20% U, 30% A, and 40% G, has an excess of 0.3 (0.4 − 0.1) in the direction of G and an excess of 0.1 (0.3 − 0.2) in the direction of A. The total bias in the example is therefore 0.4 (0.3 + 0.1), where 75% of the bias is toward G and 25% is toward A.
We present several general observations based on these calculations. First, looking at the composition of the total molecule, LSU sequences are more biased than are SSU sequences, and bacteria are more biased than are archaea, which are more biased than are the eukaryotes. Second, the molecules contain a purine bias, which consists of more G than A: ~60% G for the archaea and bacteria and up to 94% for eukaryotes (in the SSU eukaryotes, the other part is 6% U). Third, most of the variation in GC content of the total molecules can be explained by the stems that form very similar distributions along Chargaff ’s axis. The stems have an almost equal bias in all domains of life toward U and G, because of wobble base pairs. Interestingly, SSU sequences have a higher GU bias in their stems than LSU sequences do. Finally, the unpaired regions explain the purine bias in the molecules. For both archaea and bacteria, we find that the bulges are the most biased, that the loops are least biased, and that the junctions are between the loops and the bulges. The purine bias is on average 65% A in these domains. In eukaryotes, the bias in the unpaired regions is much smaller overall, and the order from least to most biased is as follows: loop, bulge, junction.
Because rRNA sequences are biased toward purines and because the overall composition is constrained by a sum, the paired regions and the unpaired regions will necessarily differ in composition. Specifically, a line drawn through points representing the composition of the paired and the unpaired parts of an RNA molecule will pass through the overall composition, showing that the compositions of the paired and the unpaired regions differ from the overall composition in precisely opposite directions. However, the magnitude of the change in composition can differ, because the paired and the unpaired regions can contain different numbers of bases, and the different types of unpaired regions, for example, loops, bulges and junctions, are not constrained to share the same composition. Thus, for example, if GC pairs were preferentially incorporated into stems, the compositional differences between paired and unpaired regions would be much greater than would be the case if the bases that participate in pairs were randomly chosen from the whole molecule. Similarly, the amount of the sequence contained in each structural category is potentially free to vary, affecting the extent to which each component can differ in composition from the overall sequence. Thus the compositions of individual structural components cannot be inferred from the number of base pairs and the overall composition of the molecule, and this compositional information may provide important clues about the assembly of RNA structures.
The unexpected differences in composition among the three different unpaired structural categories suggest that these categories should be considered separately in studies of RNA composition, and underscore the importance of the fine-grained approach.
Do the different structural categories have the same composition in the large and small subunit rRNA, and across all domains of life?
The three domains of life diverged billions of years ago and, although the rRNA molecule is conserved for function in the different domains, the nonfunctional parts presumably varied independently in each lineage. Since there is no known sequence homology between the two ribosomal subunits, these subunits have no apparent shared ancestry. It is therefore surprising to see the same patterns of variation across all domains and both subunits. These patterns of variation, or space in which the rRNA molecules can mutate freely without losing their function, are represented by the tight distributions in composition space.
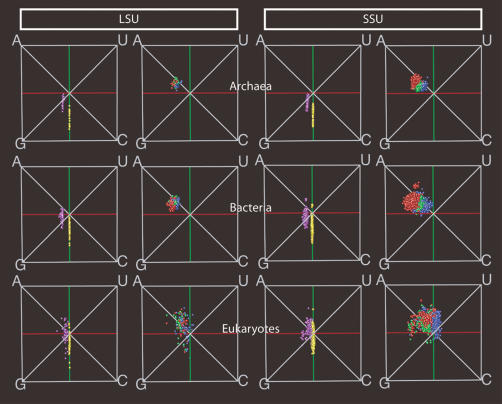
Figure 3 ▶ shows the compositions of the structural elements for large and small subunit sequences from archaea, bacteria, and eukaryotes. The distributions for LSU and SSU sequences within one domain are remarkably similar with respect to location and variation. The separation among the various structural elements is more pronounced in the SSU sequences, because many more SSU sequences than LSU sequences were available for analysis.
FIGURE 3.
Visual comparisons of sequences from two ribosomal subunits and three domains of life. There is one row for each domain, where we show left to right LSU totals and stems; LSU loops, bulges, and junctions; SSU totals and stems; SSU loops, bulges, and junctions (colors are defined in Fig. 1 ▶). (Top to bottom) The rows show archaea, bacteria, and eukaryotes. The similarities are striking, considering the long time of evolutionary divergence between the different domains.
Across domains, slightly more difference is visible. For example, the GC content differs, as shown by the positions of the totals along Chargaff’s axis, and there is less purine bias in eukaryotes than in the archaea and the bacteria. Also, there is more scatter in the eukaryotes, which may be an artifact of the sequence alignments. We have shown that removal of sequences with extreme overall base composition greatly reduces the scatter in all structural elements (data not shown). Despite these differences, the separation among the loops, the bulges, and the junctions is clear in all three domains, and similar patterns of variation are visible. In particular, one should note the similar relative positioning of the loops, the bulges, and the junctions in composition space for archaea and bacteria.
Samples that look distinct by eye need not be statistically different. Therefore, we applied Monte Carlo simulations to determine whether the differences between any combination of two samples (structural elements) within one species and domain (126 combinations in total) were significant. The calculations showed that the differences between all combinations but one were highly significant: The remaining P-values were <0.02 and for all SSU samples the P-values were <1/n, where n is the number of randomizations (10,000 in our experiment). Consequently, the different structural categories have significantly different compositions in the large and the small subunits and in the three domains of life, although these differences might be due to overall differences in the composition of the molecule (see below).
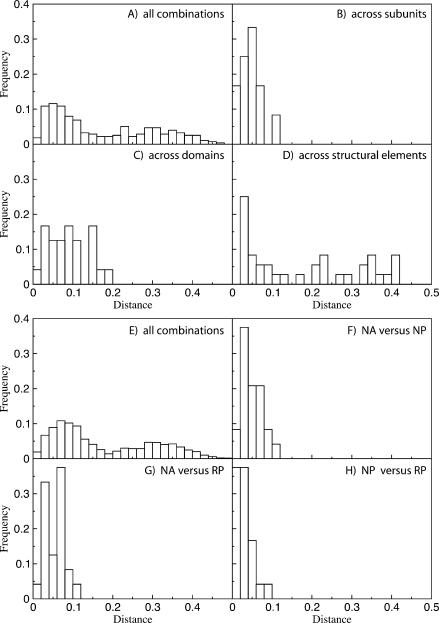
Despite the statistical significance of the differences, the observed compositional biases are visually strikingly similar across both subunits and all domains of life. Thus we tested whether the patterns were significantly similar using one-tailed two-sample t-tests on the distances between different groups. Looking separately at the differences in subunit, domain, and structural element tells us which variable causes the most difference in means. Figure 4 ▶ (top) shows the distance distributions of all possible combinations (Fig. 4A ▶) and matches across subunits (Fig. 4B ▶), domains (Fig. 4C ▶), and structural elements (Fig. 4D ▶). Comparing clusters within a domain and a structural element on subunit gave the highest significance (P = 0.00032, t = −3.5, df = 286). In other words, we found the greatest similarities between samples that came from the same structural category and the same domain but from different subunits. The visual similarities between clusters within a subunit and structural element, but across domains, were confirmed by the t-test (P = 0.00075, t = −3.2, df = 298). The data did not cluster by structural element (P = 0.68, t = 0.47, df = 310).
FIGURE 4.
Histograms of the distances between the means of different samples. The top half shows what factor, out of subunit, domain, or structural element, is most important for compositional similarity. (A) The distances for all possible combinations of samples. The three other histograms show the distances from subsets of all these combinations: combinations within the same domain and structural element, across subunit (B); combinations within subunit and structural element, and across domains (C); and combinations within subunit and domain, across structural elements (D). The bottom half addresses the similarities between annotated and predicted structures. (E) The distances between all possible combinations. The other three graphs show subsets within domain, subunit and structural category, but across sequence and structure type: NA vs. NP (F), NA vs. RP (G), NP vs. RP (H).
Thus, the compositions of stems, loops, bulges, and junctions differed significantly from one another in all samples tested. Although the composition of a given structural component (e.g., junctions) differed significantly between domains and subunits, overall the composition of a particular component was significantly more similar across domains and subunits than chance would predict. Additionally, the composition of stems varied much more than did the composition of the unpaired components, varying especially greatly in GC content. These results confirmed previous observations that the composition of unpaired regions in 16S rRNA are tightly constrained (Wang and Hickey 2002).
Are the constraints on rRNA composition due to natural selection?
Although the different structural components of rRNA are tightly constrained within characteristic regions of the space of possible compositions, these constraints might arise naturally from the process of RNA folding rather than because of purifying selection on the natural rRNA molecules. To test whether the differences between structural components within a molecule and the constraints on the composition of each structural component were due to selection, we compared the natural sequences to arbitrary, randomized sequences with the same composition. Any effects due to selection on the natural sequences should not be observed in the randomized sequences.
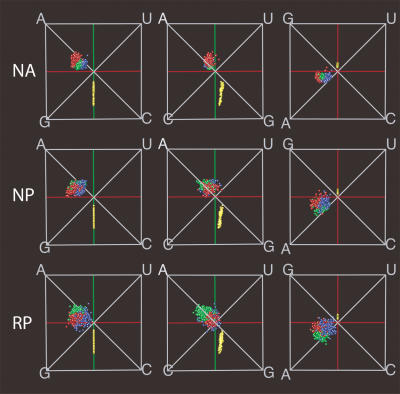
Because obtaining structures for long, arbitrary RNA sequences is prohibitively expensive, we estimated the secondary structures of the randomized sequences using the Vienna RNA folding package (Hofacker et al. 1994). Because the predicted structures are likely to contain errors, we also predicted the structures for the natural rRNA sequences using the same methods. This allowed us to separate effects due to inaccuracies in the structure prediction, which would be expected to be similar for natural and randomized sequences, from effects due to special properties of the natural sequences. We thus examined three types of data: annotated structures of natural sequences (NA), computer-predicted structures of the natural sequences (NP), and computer-predicted structures of randomized sequences (RP). We used the NP structures to test the effects of computer prediction, and we used the RP structures to test whether the compositional biases in structural components depended on the sequence, as opposed to the composition, of the natural rRNAs (the randomized sequences were constrained to have the same composition as the natural sequences).
The compositional biases observed in RP structures are much more similar to the annotated sequences than was expected (Fig. 5 ▶, top and bottom). Comparing the NP structures to the NA structures reveals some loss of information due to the computer predictions (Fig. 5 ▶, top and middle). Specifically, the variance of the samples increases in the NP structures, and some of the distinction among structural components is lost. Remarkably, however, the separation among loops, bulges, and junctions is still visible. The prediction of the composition of the stems is very good, probably because base-pairing dominates in the predictions.
FIGURE 5.
Comparison of the compositional biases found in annotated and predicted structures. The first row shows the annotated structures of natural sequences (NA) in three different perspectives. The second row, used as a benchmark, shows the predicted structures of the natural sequences (NP). The predicted structures of randomized sequences (RP) are shown in the third row. (Colors are defined in Fig. 1 ▶.)
Finally, the compositional biases in the RP structures are almost identical to those in the NP structures (Fig. 5 ▶, middle and bottom). We observe slightly more variation in the samples from randomized sequences, because these sequences are completely unrelated to each other. In contrast, the natural sequences are all recognizably homologous.
We also tested whether the compositions of each structural category in the NA, NP, and RP structures were significantly similar to one another by using the same test as for similarities between domains and subunits. On the lower half of Figure 4 ▶ are the graphs associated with the comparison of natural annotated (NA) structures with the computer predictions of the natural sequences (NP), and the predictions of the randomized sequences (RP). The statistics confirm the visual observations discussed above. Results of t-tests between the subsets and the distribution of all combinations (Fig. 4E ▶) show that matches across the computer predicted structures (NP vs. RP) (Fig. 4H ▶) are most significant (P = 2.1×10−9, t = 5.9, df = 2578). The matches both across NA and NP structures (Fig. 4F ▶), and across NA and RP structures (Fig. 4G ▶) are still highly significant (P = 1.6 ×10−7, t = −5.1 and P = 5.9 ×10−7, t = −4.9, respectively), despite the observed shifts of the unpaired regions in composition space.
Consequently, the different compositions of paired and unpaired regions, and of the different types of unpaired regions, do not depend on the sequence (to the limits of our ability to predict the structure with RNAfold) but only on the overall composition of the molecule. This suggests that differential selection for composition in the different structural categories does not cause the differences in composition, but rather that they arise automatically from the process of RNA folding.
Are the different responses to overall GC content in paired and unpaired regions due to natural selection?
If the constraints on the composition of the bases in each structural component are not due to selection, the different responses of each category to overall changes in genomic GC content might not depend on selection either. Accordingly, we tested whether the slope of the regression line relating GC content in each structural component and in the rRNA molecule overall or in the coding sequences in the genome differed between the natural rRNA sequences and randomized sequences with the same composition.
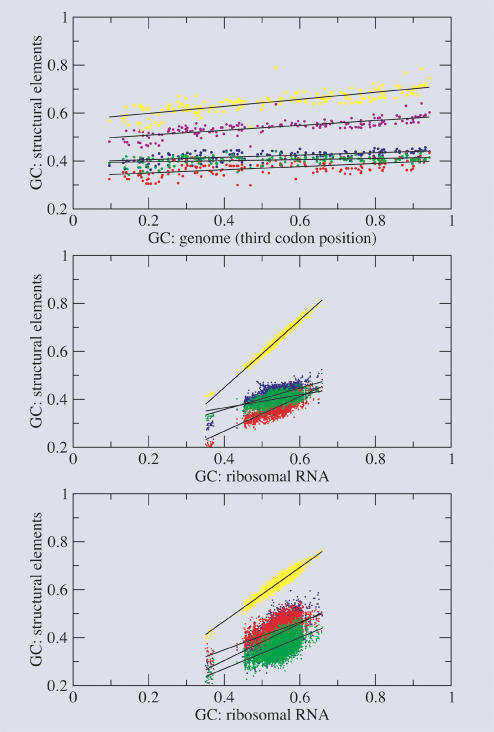
Figure 6 ▶ (top) shows the known correlation between genomic GC content at the selectively neutral third codon position and GC content of the total ribosomal RNA (Muto and Osawa 1987). Positive correlations between the GC content of the third codon position in protein-coding regions and the GC content of paired and unpaired regions in rRNA have been observed in bacteria (N. Sueoka, pers. comm.). The same positive correlation holds true for each structural category individually, and the major difference in slope is between paired and unpaired elements. Graphs of the GC content of the ribosomal RNA versus the GC content in the different structural elements magnify these differences (Fig. 6 ▶, middle), since the values are now constrained by a sum, and, at low overall GC content, the composition of the stems is thus much closer to the composition of the unpaired regions than at high overall GC content. The slopes of the stems are much steeper than the slopes of the unpaired regions. There is no systematic distinction in slopes among loops, bulges, and junctions. We find that the correlations are positive for all structural elements (i.e., stem, loop, bulge, and junction) for both subunits and all domains. This means that there is no compensation in base composition across different structural elements.
FIGURE 6.
Correlations with GC content in bacterial SSU rRNA. First, we show the correlation between GC content in all structural elements of the ribosomal RNA (including the total ribosomal RNA) and the GC content in the third codon position in the genome (top). The lower two graphs show the correlations between all structural elements and the GC content of the ribosomal RNA for annotated structures of real rRNA sequences (middle) and for predicted structures of randomized sequences (bottom). (Colors are defined in Fig. 1 ▶.)
Surprisingly, we see very similar correlations in unevolved (or randomized) sequences (Fig. 6 ▶, bottom). This raises the question of whether the stems are functionally less important and thus not (as strongly) restricted in their mutations, or whether the compositional variation in the stems can be explained by the different overall amount of pairing in RNA sequences of different composition (Schultes et al. 1999). In general, the slope of the stems is more shallow, and the slopes of the unpaired regions are steeper than observed in the annotated sequences.
We visualized these correlations in the tetrahedron by grouping sequences by GC content and color-coding them accordingly (Fig. 7A ▶). A given color in each structural element thus refers to the same set of sequences, which are grouped by the GC content of the total molecule. The simplex gives us more information than the previously shown graphs in the sense that we can see the relative positioning of the sets with similar GC content in the different structural elements. The clusters of sequences with similar GC content are still distinct clusters in all structural elements.
FIGURE 7.
Variation in GC content in natural SSU bacterial sequences (A) and arbitrary sequences with the same composition as the bacteria (B). Sequences with the same color have a similar composition (specifically GC content). In the real SSU bacteria (A), the clusters of sequences with similar overall base composition are recovered as tight clusters in all structural elements (only stems and bulges are shown). The same phenomenon is visible in the arbitrary sequences (B). The totals are tiny dots in the graph, because all 100 random sequences in each interval have the same composition. The GC content in all structural elements is strongly correlated with the GC content in the overall molecule.
This clustering might be due to either of two factors: sequence homology or compositional similarity. In the annotated sequences, we cannot separate these two potential causes, because homologous rRNA sequences have both similar sequences and similar compositions due to evolutionary conservation. To address this issue, we can use randomized sequences, which have strong compositional similarities, but no sequence homology whatsoever. If we find the same clustering behavior in these randomized sequences, nucleotide composition, rather than sequence homology, must be the driving force behind the characteristic compositions in the different structural elements.
Randomized sequences show strikingly similar patterns to the natural sequences (Fig. 7B ▶). These sequences are constructed by calculating the base composition on 2% intervals on a line through the mean of the SSU bacteria, parallel to Chargaff’s axis, creating 100 random sequences of length 1500 in each interval, folding the sequences with RNAfold, and applying the same classification as used throughout the analysis. The randomized sequences form very smooth distributions through composition space with seemingly mathematical precision. The clusters of sequences with the same GC content in their total molecule (dots in the graph) are visible as tight clusters in each structural category. This pattern (Fig. 7 ▶) has two implications: First, the base composition of structural categories is consistent at a given sequence composition, and second, similar base composition in the whole molecule implies similar base composition in each structural category.
Several mechanisms might influence these structure-dependent compositional biases. The first is purifying selection, which would cause the nucleotide composition of the whole sequence (and thus of all elements of the structure) to change in one direction by mutation, limited by the rate at which deleterious mutations are filtered out by selection. Purifying selection would explain the difference in slope between paired and unpaired elements of related and functional sequences in terms of different functional constraints for each structural element (Wang and Hickey 2002). For comparison, in coding sequences, the three codon positions have different rates of change in response to changes in genome GC content, which can be interpreted in terms of purifying selection (Muto and Osawa 1987; Sueoka 1988; Lobry and Sueoka 2002). However, the purifying selection model would predict that randomized sequences would show no difference in slopes between paired and unpaired regions, because they have no functions that need to be conserved and, in any case, share no evolutionary history.
Contrary to this prediction, we found that even randomized sequences have different rates of response to change in composition in each of the structural elements. Although it is possible that purifying selection accentuates these differences, much of the observed pattern can be attributed to the effects of folding, and claims about the extent of purifying selection based on these slopes (Wang and Hickey 2002) should be treated with caution. Purifying selection is not required to explain the compositional differences among stems, loops and bulges, although it may affect details of the slopes.
The second mechanism is adaptive (or positive) selection, which means selection in favor of a particular composition, presumably because the composition is required for function, such as GNRA tetraloops (Woese et al. 1990) and the other motifs described above. Selection for a particular sequence could in principle generate any possible composition, divided in any way among the structural components. In other words, the function of the ribosomal RNA might require more of certain bases in certain structural components, and this positive selection might generate the compositional differences (Lao and Forsdyke 2000; N. Sueoka, pers. comm.). We need adaptive selection to explain the existence of functional RNAs and many ubiquitous structural motifs, but we do not need it to explain the compositional biases, because they also occur in nonevolved, nonfunctional sequences as an effect of RNA folding. However, positive selection might explain the subtle deviations in real rRNAs from what is expected by chance based on the randomized sequences.
Although rRNA sequences are highly selected and conserved, the compositional biases are consistent with those in randomized sequences, suggesting that the compositional biases in all structural elements are inherent to any sequence with the same base composition. Thus, the major force behind the formation of structural biases appears to be what we call “self-organization,” the intrinsic factors such as base-pairing and stacking that drive secondary structure formation.
What explains the trends in the composition of the structural elements?
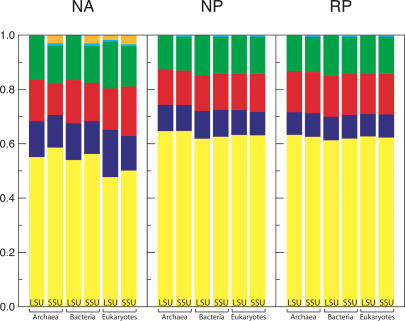
Having demonstrated that the different structural components of rRNA differ in composition from one another in both subunits and all three domains of life and that these differences appear to be driven by the overall composition of the molecule, we tested which parameters affect the result. First, we investigated the accuracy of the RNA folding in terms of its ability to assign bases to the correct structural categories. In addition to the base composition of structural elements, we examined the fraction of bases in all categories. We analyzed the NA, NP, and RP structures. We found that the fraction of bases ending up in each structural feature is approximately the same for all domains of life and that there are consistent differences between large and small subunit sequences: SSU rRNA has a higher percentage of base pairs than LSU sequences do (Fig. 8 ▶, left). It seems that the amount of base-pairing differs between LSU and SSU sequences but that the remaining bases are divided almost equally over the loops, the bulges, and the junctions. On average, <4% of bases appear in ends and flexible regions. Figure 8 ▶ (middle and right) shows that computer predictions systematically result in too many base pairs and thus too few bases in the unpaired regions, which might account for the observed increase in variation for the NP structures. In addition, the predictions are similar for sequences with the lengths of either typical LSU or SSU sequences. There is no visible difference between the predictions of the natural and the randomized sequences, suggesting that the bias is due to the folding procedure rather than being sequence-specific. Although covariation methods, with which the annotated structures are predicted, can systematically underpredict base-pairing because they cannot detect pairing involving absolutely conserved positions, the magnitude of the change (>10% of the sequence is incorrectly predicted to be paired) is much greater than the error in the covariation structures.
FIGURE 8.
Comparison of the fractions of bases in all structural elements between natural annotated (NA) structures, predicted structures of natural sequences (NP), and predicted structures of randomized sequences (RP). In each graph we show (from left to right) the average fractions in each element for LSU and SSU archaea, bacteria, and eukaryotes. (Colors are the same as in Fig. 1 ▶.)
Second, we tested whether some of the effects were due to the annotations in the databases. We identified 174 rRNAs that were derived from the same original GenBank record in the rRNA database and the CRW database, of which only 66 had identical sequences in the two databases. We redid the analysis by using only this subset of rRNAs and the predictions to each database. Although the structures differed in some detail, there was no meaningful difference between the compositions of the structural components calculated from each set, which differed by <2% on average (ranging from 0.01% to 4.5%) and were very similar visually. We also verified that the structures of the sequences for which high-resolution crystal structures were available were correctly represented in the databases, and we found that these structures were 97% identical to those in the CRW database (when considering only Watson-Crick and wobble pairing), consistent with previous reports (Gutell et al. 2002).
Third, we tested whether the thermodynamic parameters affected the result. The energies for tetraloops and certain other “special” sequences used by RNAfold are calculated by using sequence databases that include rRNA sequences and might unfairly bias the structures for arbitrary sequences to resemble the structures for natural sequences in composition. However, repeating the analysis with the “−4” option in RNAfold, which eliminates the contribution of tetraloop energies, did not affect the compositions of the different structural components significantly.
We next tested whether the differences between the unpaired structural components arose simply from the difference in pairing strength between AU and GC base pairs. The RNAfold program provides an option to fold sequences by using the abstract “ABCD” alphabet, in which A pairs with B and C pairs with D and in which all kinds of base pairs have the same energy parameters for pairing and stacking. Repeating the analysis by translating the sequences into the ABCD alphabet and folding with the thermodynamic parameters for AU or GC pairs (i.e., all pairs were treated as AU, or all were treated as GC) gave strikingly different compositions from normal folding, in part because GU pairs could not be incorporated in this model. However, in all cases, the loops, the bulges, and the junctions differed from each other in composition. Reassuringly, sequences in which the meanings of the bases were permuted (e.g., U might be exchanged with C) gave symmetric patterns, indicating that whichever bases are in excess over the 1:1 purine:pyrimidine ratio required for stems will be found in the unpaired regions to a similar degree to the bases that were in excess in the original composition. In other words, when all bases have the same energies, any bases in excess will be found more frequently in the unpaired regions; however, when the thermodynamic parameters are taken into account, the identity of the bases matters because of differences in pairing and stacking energies.
These results suggest that the causes of differences among bulges, loops, and junctions are not related to their properties as parts of nucleic acid sequences per se but are rather a general property of the class of formal grammars that includes non-pseudo-knotted structures when applied to arbitrary character strings. The results also indicate that the null hypothesis for studies of composition should not be that all unpaired structural components are identical in composition.
Conclusions
We have demonstrated several important features of nucleotide composition patterns within ribosomal RNA. First, there are striking similarities in the composition of the different structural categories across both ribosomal subunits and the three domains of life, despite much evolutionary divergence. Second, randomized sequences appear almost identical to natural sequences in the composition of each structural component; furthermore, they show the same patterns of variation, even though these randomized sequences are not evolved and do not have biological functions. Third, the GC content in all structural categories is positively correlated with the GC content of the ribosomal RNA overall, and randomized sequences show similar correlations to the annotated sequences. Finally, the nucleotide composition of individual structural features proves robust over multiple randomizations, since clusters of sequences with similar base compositions yield consistent clusters for each structural element.
These results for randomized sequences emerged solely from the inherent features of RNA folding, as reproduced by the dynamic programming method and thermodynamic parameters used for energy minimization in RNAfold. Our conclusions thus depend on the ability of these algorithms to provide information about arbitrary sequences: Although the predictions are far from perfect, there is no reason to believe that they are biased in ways that would give the observed patterns as an artifact. The thermodynamic parameters are derived from melting experiments on oligonucleotides (Mathews et al. 1999), which are short sequences that are neither evolved nor biologically active. There is thus no reason to believe that the rules derived from experiments on them would apply only to biologically active sequences and not to arbitrary RNA sequences. The predictions also use special bonus energies for particular loop sequences, which are based on experimental data and supported by statistics on known RNA structures. These energies improve the predictions for natural RNA sequences that were not themselves used to derive the parameters (Mathews et al. 1999), and are thus likely to provide the best available estimate of the structures of arbitrary sequences. Changing details of the parameters, such as eliminating the bonuses for tetraloops (which are inferred from a database of structures) did not affect our results.
The computer predictions are sufficiently accurate to capture the features we examined: The predictions of the natural sequences closely resemble the patterns observed from annotated sequences. The predictions are very accurate at specifying whether bases are paired or unpaired (Mathews et al. 1999), suggesting that the composition of the stems is probably most accurate, although there is less accuracy in predicting the overall topology of the molecule (data not shown). The predictions are good enough to show the separation between the unpaired regions. However, this distinction is less sharp than in the annotated sequences, which might be due to some mixing of the unpaired categories.
The discovery of general rules that determine the amount of base-pairing and the nucleotide composition of a molecule will have important consequences for the accuracy of secondary structure prediction programs, such as BayesFold (Knight et al. 2004). If the compositional preferences we have demonstrated for rRNA generalize to other molecules, we may be able to assess the plausibility of a structure by asking whether the compositional patterns comply with the specific compositional statistics, thus improving the predictions. Specifically, a structure that reproduces typical compositional biases in the different structural elements is more likely to be correct. However, the similarities between the compositions in each component of the true structure and the structures predicted by current methods suggest that the power of this approach may be limited to eliminating the more egregious mispredictions. A more promising difference is in the amount of the sequence that is assigned to each structural category, which shows clear differences between the natural and the predicted structures. We should be able to compensate for the systematic deviations in current computer predictions, especially the excess of base pairs.
Because the constraints on the compositions of each structural component and the slopes of the compositional responses of each structural component to changes in overall and genome GC content are very similar, the null model for evolutionary studies of rRNA should not be that these components behave identically but rather that compositional differences would be expected even in random sequences. Our results suggest that only parts of the rRNA are under strong selection and that most of the molecule is able to change neutrally. Testing whether other classes of RNA that are under stronger selection, such as the 5S rRNA, may reveal cases where the change in each structural component does differ from what would be observed in random sequences of the same composition (and hence the action of selection), but we see no evidence for these effects in rRNA.
MATERIALS AND METHODS
Data collection and processing
We downloaded all large subunit (LSU) and small subunit (SSU) rRNA sequences from the European Ribosomal RNA database (Wuyts et al. 2001, 2002) at http://www.psb.ugent.be/rRNA on November 11, 2003, which had not been subsequently updated as of July 2005. We retrieved the sequences in distribution format, which contains gaps and secondary structure information interleaved with the sequence. Additionally, we downloaded the helix numbering for all domains. In principle, the helix numbering provides the locations of the upstream and downstream parts of each helix in the alignment, although not all sequences comply with this numbering. As a control for the effects of the alignment in the database, we also used rRNA sequences and structures from the Gutell Comparative RNA Web (Cannone et al. 2002) and from the Protein Data Bank (Bernstein et al. 1977).
We obtained natural rRNA sequences, which could be used for computer predictions, by stripping out all gaps and secondary structure information from our annotated data. We created randomized versions of our annotated data by shuffling the natural sequences completely, using the Fisher-Yates shuffle algorithm as implemented in the random module of the Python standard library. In this way, all structural motifs are broken, but the overall base composition of the molecule is unaltered.
The structures associated with the rRNA sequences in the database are predicted by comparative sequence analysis. We refer to these structures as “annotated” because they are based on experimental evidence and have been compared to crystal structures. For randomized sequences there are no secondary structure models available. Because experimentally determining structures for these sequences is impossible, we used RNAfold from the Vienna RNA folding package (Hofacker et al. 1994), which implements the Zuker folding algorithm (Zuker and Stiegler 1981) to estimate an optimal secondary structure both for each natural sequence and for each permuted sequence.
RNAfold returns the optimal structures in dot-bracket (or Vienna) format. In order to compare the annotated structures and the computer-predicted structures, we developed an algorithm to convert the distribution format from the database into the Vienna format. Based on the helix numbering, it finds the most likely pairs of upstream and downstream helix parts. We verify the actual base-pairing and solve the matching for helix parts that are incorrectly annotated or unannotated. Pseudo-knots are discarded because the Vienna format cannot denote them, but because they comprise <2% of all base pairs in rRNA (Mathews et al. 1999), this limitation has little effect on our results.
The database contained 21,782 sequences. About 50% of these sequences were unusable: They contained too many undetermined positions (>50), had an odd number of helix parts, contained pairing helix parts of different lengths, etc. From the remaining 50% with good data, our conversion algorithm could reliably convert 10,254 structures into dot-bracket format, which corresponded to a data loss of 0.86% of the total number of sequences (Table 1 ▶). In our analysis, we focused on RNA from nuclear genomes: We included archaea, bacteria, and eukaryotes (263, 5530, and 3099 sequences, respectively; 8892 sequences in total).
TABLE 1.
Number of sequences analyzed; we focused on archaea, bacteria, and eukaryotes (8892 sequences total)
| In database | Unusable | Good | Analyzed | Data loss (%) | |
| LSU | |||||
| Archaea | 37 | 6 | 31 | 31 | 0.00 |
| Bacteria | 399 | 129 | 270 | 270 | 0.00 |
| Eukaryotes | 157 | 79 | 78 | 78 | 0.00 |
| Mitochondria | 659 | 225 | 434 | 430 | 0.92 |
| Plastid | 70 | 7 | 63 | 63 | 0.00 |
| Total | 1322 | 446 | 876 | 872 | 0.11 |
| SSU | |||||
| Archaea | 590 | 358 | 232 | 232 | 0.00 |
| Bacteria | 12,107 | 6839 | 5268 | 5260 | 0.15 |
| Eukaryotes | 6590 | 3493 | 3097 | 3021 | 2.45 |
| Mitochondria | 1039 | 274 | 765 | 764 | 0.13 |
| Plastid | 134 | 29 | 105 | 105 | 0.00 |
| Total | 20,460 | 10,993 | 9467 | 9382 | 0.89 |
Decomposing secondary structure into structural categories
We identified secondary structure elements in two steps. First, by using the dot-bracket notation of the structure, we built an ordered rooted tree (Hofacker et al. 1994; Schuster et al. 1994), a tree representation of the structure in which the nodes correspond to bases or base pairs, ordered from the 5′ to the 3′ end. Next, we assigned each base to a structural category during a tree traversal, ignoring the virtual root. Bases associated with internal nodes (i.e., base pairs) are assigned to a stem. Leaf nodes that are children of the root are either “ends” or “flexible bases,” depending on their position relative to the outgoing stems. All other leaves are assigned to loops, bulges, and junctions based on the number of stems going out of their parent node. The result of this process is a string of labels representing the structural elements of each base, which correspond exactly to the dot-bracket notation.
Calculating and visualizing base composition
We calculated the base composition for each structural element by grouping all bases within a particular element together and counting the number of each of the four bases: U, C, A, and G. We normalized this composition vector by the number of residues in the element (N) in order to compare elements containing different numbers of bases. The base composition of any RNA sequence can be visualized in a tetrahedral unit simplex (Schultes et al. 1997; Fig. 2 ▶). In this unit simplex, the three pairwise combinations of bases define three orthogonal axes. For example, the amount of G + C defines a position along Chargaff’s axis, where G = C and A = U. The two other axes are the purine–pyrimidine axis, plotting the amount of A + G versus C + U, and the amino–keto axis, plotting the amount of A + C versus G + U. The four bases form the four vertices; sequences containing more of a particular base lie closer to the vertex for that base.
For our particular analysis, we plotted seven dots for each sequence: six for the structural elements (stem, loop, bulge, junction, end, and flexible) and one for the overall base composition of the molecule. Plotting the base compositions for many RNA sequences allowed us to see the similarities or the differences among species, structural elements, ribosomal subunits, or domains of life.
We used the program MAGE (Richardson and Richardson 1992) to visualize the composition simplex. This program treats the three dimensions (A/N, C/N, G/N) as orthogonal axes and applies a distortion matrix to make them look like a tetrahedron. However, we could not use these distorted coordinates to calculate distances between points or samples. Therefore, we converted the coordinates by using combinations of the four bases as axes that form the orthogonal right-handed Cartesian coordinate system described above.
Testing whether samples are different
To test whether the difference in location between two samples was significant, we used Monte Carlo simulations. We compared the observed distance between two samples, i.e., the Euclidean distance between the means of the two samples, to the distribution of distances between many pairs of random samples resampled from the original data points. This technique does not depend on assumptions about the shape or variance of the underlying distributions.
To apply this technique, we first pooled the points in the two samples. Next, we randomly permuted the list of samples and divided the list into two groups that contained the same number of points as the original samples. Finally, we compared the distance between the means of the randomized samples to the distance between the means of the original samples. We repeated this 10,000 times, except when a small preliminary sample was sufficient to show that the difference was not significant. The P-value is the number of times the observed distance was greater than or equal to the benchmark divided by the number of randomizations. Any P-value ≤0.05 was considered significant.
Testing whether samples are similar
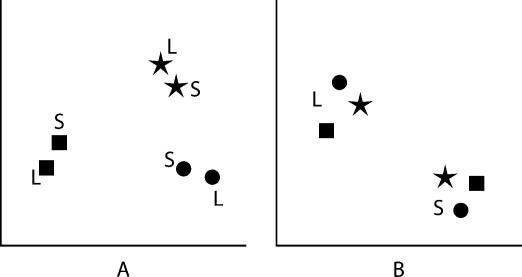
The Monte Carlo simulations sensitively reveal whether samples differ but cannot directly tell us which samples are similar. We needed to test whether patterns were more similar within structural categories, domains, or ribosomal subunits. For example, looking at this problem in only two dimensions, the data might be clustered as in Figure 9A ▶. This figure shows a situation in which the strongest similarities are within each domain rather than within each subunit, suggesting that the domain is more important in determining composition. Alternatively, the data might be clustered as in Figure 9B ▶, where subunit identity dominates the clustering. In the first case, the distances between points within a domain will on average be smaller than the distances between all combinations of two points. In the second case, the distances between points within a subunit will be smaller than the distances between all combinations of points. To generalize, points within a cluster will on average be closer than points chosen at random. We can compare these two populations of distances (within and between putative clusters) by using a one-tailed two-sample t-test: The lower the P-value, the greater the significance of the relationship represented by the clustering.
FIGURE 9.
Possible outcomes of data clustering. Symbols represent the base compositions for archaea (squares), bacteria (circles), and eukaryotes (stars) in two dimensions. Letters indicate the ribosomal subunit: large (L) and small (S). Data might be clustered by domain of life (A), where most similarity is within a particular domain and across subunits. In contrast, the subunit might be the most important factor for base composition (B), in which case sequences from the same subunit would be most similar, independent of the domain of life to which the sequences belong.
We applied this procedure in three dimensions to all annotated data. We looked for similarities among two subunits, three domains, and four structural elements. We considered only stems, loops, bulges, and junctions. The number of possible combinations between n samples is n(n − 1)/2, so in case of 24 (2 × 3 × 4) samples, the number of distances between (the means of) any two samples is 276 (24 × 23/2). Within clusters of equal domain and structural category (across subunits), we had 12 distances; within subunits and structural elements (across domains), 24 distances; and across structural elements, 36 distances.
We also applied this method to confirm the visual similarities between annotated and computer-predicted structures. This gives three times as many samples as above, thus 2556 distances in the full sample. We made three subsets, each time within a subunit, domain, and structural category, but across structure type (NA, NP, and RP). Each of the subsets contained 24 distances. The distributions of distances are visualized with histograms and compared with a one-tailed two-sample t-test.
Acknowledgments
Our thanks goes to Erik Schultes, Noboru Sueoka, Lionel Guy, and Catherine Lozupone for their careful reading and discussion of the manuscript. We also thank Eric Westhof and two anonymous reviewers for many helpful suggestions that improved the manuscript. Some parts of this work were supported by NIH research grant no. GM 30881 and NASA Center for Astrobiology grant no. NCC2-1052.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2183806.
REFERENCES
- Bernstein, F., Koetzle, T., Williams, G., Meyer Jr., E., Brice, M., Rodgers, J., Kennard, O., Shimanouchi, T., and Tasumi, M. 1977. The Protein Data Bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 112: 535–542. [DOI] [PubMed] [Google Scholar]
- Cannone, J., Subramanian, S., Schnare, M., Collett, J., D’Souza, L., Du, Y., Feng, B., Lin, N., Madabusi, L., Muller, K., et al. 2002. The Comparative RNA Web (CRW) Site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J., Gooding, A., Podell, E., Zhou, K., Golden, B., Szewczak, A., Kundrot, C., Cech, T., and Doudna, J. 1996. RNA tertiary structure mediation by adenosine platforms. Science 273: 1696– 1699. [DOI] [PubMed] [Google Scholar]
- Doherty, E.A., Batey, R.T., Masquida, B., and Doudna, J.A. 2001. A universal mode of helix packing in RNA. Nat. Struct. Biol. 8: 339– 343. [DOI] [PubMed] [Google Scholar]
- Elson, D. and Chargaff, E. 1955. Evidence of common regularities in the composition of pentose nucleic acids. Biochim. Biophys. Acta 17: 367–376. [DOI] [PubMed] [Google Scholar]
- Galtier, N. and Lobry, J. 1997. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 44: 632–636. [DOI] [PubMed] [Google Scholar]
- Gutell, R.R., Cannone, J.J., Shang, Z., Du, Y., and Serra, M.J. 2000. A story: Unpaired adenosine bases in ribosomal RNAs. J. Mol. Biol. 304: 335–354. [DOI] [PubMed] [Google Scholar]
- Gutell, R., Lee, J., and Cannone, J. 2002. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 12: 301–310. [DOI] [PubMed] [Google Scholar]
- Guy, L. and Roten, C. 2004. Genometric analyses of the organization of circular chromosomes: A universal pressure determines the direction of ribosomal RNA genes transcription relative to chromosome replication. Gene 340: 45–52. [DOI] [PubMed] [Google Scholar]
- Hofacker, I., Fontana, W., Stadler, P., Bonhoeffer, L., Tacker, M., and Schuster, P. 1994. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 125: 167–188. [Google Scholar]
- Knight, R., Freeland, S., and Landweber, L. 2001. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2: http://genomebiology.com. [DOI] [PMC free article] [PubMed]
- Knight, R., Birmingham, A., and Yarus, M. 2004. Bayesfold: Rational 2° folds that combine thermodynamic, covariation, and chemical data for aligned RNA sequences. RNA 10: 1323–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao, P. and Forsdyke, D. 2000. Thermophilic bacteria strictly obey Szybalski’s transcription direction rule and politely purine-load RNAs with both adenine and guanine. Genome Res. 10: 228– 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute, A., Leontis, N.B., Massire, C., and Westhof, E. 2005. Recurrent structural RNA motifs, isostericity matrices, and sequence alignments. Nucleic Acids Res. 33: 2395–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry, J. 1997. Influence of genomic G+C content on average aminoacid composition of proteins from 59 bacterial species. Gene 205: 309–316. [DOI] [PubMed] [Google Scholar]
- Lobry, J. and Sueoka, N. 2002. Asymmetric directional mutation pressures in bacteria. Genome Biol. 3: http://genomebiology.com. [DOI] [PMC free article] [PubMed]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Molinaro, M. and Tinoco Jr., I. 1995. Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: Thermodynamic and spectroscopic applications. Nucleic Acids Res. 23: 3056–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto,A. and Osawa, S. 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. 84: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J., Ban, N., Moore, P., and Steitz, T. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 98: 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, D. and Richardson, J. 1992. The kinemage: A tool for scientific communication. Protein Sci. 1: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes, E., Hraber, P., and LaBean, T. 1997. Global similarities in nucleotide base composition among disparate functional classes of single-stranded RNA imply adaptive evolutionary convergence. RNA 3: 792–806. [PMC free article] [PubMed] [Google Scholar]
- ———. 1999. Estimating the contributions of selection and self-organization in RNA secondary structure. J. Mol. Evol. 49: 76– 83. [DOI] [PubMed] [Google Scholar]
- Schuster, P., Fontana, W., Stadler, P., and Hofacker, I. 1994. From sequences to shapes and back: A case study in RNA secondary structures. Proc. R. Soc. Lond. B Biol. Sci. 255: 279–284. [DOI] [PubMed] [Google Scholar]
- Singer, G. and Hickey, D. 2000. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 17: 1581–1588. [DOI] [PubMed] [Google Scholar]
- Sueoka, N. 1961. Compositional correlation between deoxyribonucleic acid and protein. Cold Spring Harb. Symp. Quant. Biol. 26: 35–43. [DOI] [PubMed] [Google Scholar]
- ———. 1962. On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl. Acad. Sci. 48: 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1988. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. 85: 2653–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1999. Two aspects of DNA base composition: G+C content and translation-coupled deviation from intra-strand rule of AT and GC. J. Mol. Evol. 49: 49–62. [DOI] [PubMed] [Google Scholar]
- Tinoco Jr., I. 1996. RNA enzymes: Putting together a large ribozyme. Curr. Biol. 6: 1374–1376. [DOI] [PubMed] [Google Scholar]
- Tuerk, C., Gauss, P., Thermes, C., Groebe, D., Gayle, M., Guild, N., Stormo, G., d’Aubenton-Carafa, Y., Uhlenbeck, O., Tinoco Jr., I., et al. 1988. CUUCGG hairpins: Extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc. Natl. Acad. Sci. 85: 1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. and Hickey, D. 2002. Evidence for strong selective constraint acting on the nucleotide composition of 16S ribosomal RNA genes. Nucleic Acids Res. 30: 2501–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof, E. and Massire, C. 2004. Evolution of RNA architecture. Science 306: 62–63. [DOI] [PubMed] [Google Scholar]
- Wimberly, B., Brodersen, D., Clemons Jr., W., Morgan-Warren, R., Carter, A., Vonrhein, C., Hartsch, R., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Woese, C.R., Winker, S., and Gutell, R.R. 1990. Architecture of ribosomal RNA: Constraints on the sequence of “tetra-loops.” Proc. Natl. Acad. Sci. 87: 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts, J., De Rijk, P., Van de Peer, Y., Winkelmans, T., and De Wachter, R. 2001. The European large subunit ribosomal RNA database. Nucleic Acids Res. 29: 175–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts, J., Van de Peer, Y., Winkelmans, T., and De Wachter, R. 2002. The European database on small subunit ribosomal RNA. Nucleic Acids Res. 30: 183–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. and Stiegler, P. 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 9: 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]