Abstract
RNase P complexed with external guide sequence (EGS) represents a novel nucleic-acid-based gene interference approach to modulate gene expression. In this study, a functional EGS RNA was constructed to target the overlapping mRNA region of two human cytomegalovirus (HCMV) capsid proteins, the capsid scaffolding protein (CSP) and assemblin. The EGS RNA was shown to be able to direct human RNase P to cleave the target mRNA sequence efficiently in vitro. A reduction of ~75%–80% in the mRNA and protein expression levels of both CSP and assemblin and a reduction of 800-fold in viral growth were observed in human cells that expressed the functional EGS, but not in cells that either did not express the EGS or produced a “disabled” EGS that carried nucleotide mutations that precluded RNase P recognition. The action of the EGS is specific as the RNase P-mediated cleavage only reduces the expression of the CSP and assemblin but not other viral genes examined. Further studies of the antiviral effects of the EGS indicate that the expression of the functional EGS has no effect on HCMV genome replication but blocks viral capsid maturation, consistent with the notion that CSP and assemblin play essential roles in HCMV capsid formation. Our study provides the first direct evidence that EGS RNAs effectively inhibit HCMV gene expression and growth. Moreover, these results demonstrate the utility of EGS RNAs in gene therapy applications, including the treatment of HCMV infection by inhibiting the expression of virus-encoded essential proteins.
Keywords: RNase P, cytomegalovirus, external guide sequence, gene targeting, gene therapy
INTRODUCTION
Nucleic-acid-based gene interference strategies, such as anti-sense oligonucleotides, ribozymes or DNAzymes, and RNA interference (RNAi), represent powerful research tools and promising therapeutic agents for human diseases (Stein and Cheng 1993; Santoro and Joyce 1997; Wong-Staal et al. 1998; Scherer and Rossi 2003). The gene-targeting agents used can be a conventional antisense oligonucleotide, an antisense catalytic molecule (ribozyme or DNA enzyme), or an anti-sense molecule with an additional (guide) sequence that targets the mRNA for degradation by endogenous RNases such as RNase L and the RNA-induced silencing complexes (RISC) (Stein and Cheng 1993; Maran et al. 1994; Yuan and Altman 1994; Santoro and Joyce 1997; Wong-Staal et al. 1998; Scherer and Rossi 2003). Each of these approaches has its own advantages and limitations in term of targeting efficacy, sequence specificity, toxicity, and delivery efficiency in vivo. Improving these current technologies and developing new nucleic-acid-based strategies will provide exciting tools and reagents for basic research and clinical applications including therapeutic interventions.
Ribonuclease P (RNase P) has been proposed as a novel RNA-based gene interference strategy for knocking down gene expression (Gopalan et al. 2002; Raj and Liu 2003). This enzyme, which is a ribonucleoprotein complex found in all organisms examined, is responsible for the maturation of 5′ termini of all tRNAs, which account for ~2% of total cellular RNA (Frank and Pace 1998; Altman and Kirsebom 1999). It catalyzes a hydrolysis reaction to remove the leader sequence of precursor tRNA. Human RNase P has at least 10 polypeptides and an RNA subunit (H1 RNA) (Altman and Kirsebom 1999; Gopalan et al. 2002). One of the unique features of RNase P is its ability to recognize the structures, rather than the sequences, of the substrates, which allows the enzyme to hydrolyze different natural substrates in vivo or in vitro. Accordingly, any complex of two RNA molecules that resembles a tRNA molecule can be recognized and cleaved by RNase P (Fig. 1A–C ▶; Forster and Altman 1990; Yuan et al. 1992). One of the RNA molecules is called the external guide sequence (EGS). In principle, an mRNA sequence can be targeted for RNase P cleavage by using EGSs to hybridize with the target RNA and direct RNase P to the site of cleavage. The EGSs used to direct human RNase P for targeted cleavage resemble three-quarters of a tRNA molecule and consist of two sequence elements: a targeting sequence complementary to the mRNA sequence and a RNase P recognition sequence, which is a portion of the natural tRNA sequence and is required for interacting with RNase P (Fig. 1B,C ▶; Yuan et al. 1992). Subsequent studies have shown that expression of EGSs can modulate gene expression in bacteria and in mammalian cells (Yuan et al. 1992; Guerrier-Takada et al. 1995; Ma et al. 2000; Zhu et al. 2004). We have previously shown that EGS RNAs derived from a natural tRNA efficiently directed human RNase P to cleave the mRNA sequence encoding the thymidine kinase (TK) of herpes simplex virus 1 (HSV-1) in vitro (Kawa et al. 1998; Zhou et al. 2002). A reduction of 75%–80% in the TK mRNA and protein expression was observed in HSV-1 infected cells expressing the EGS RNAs.
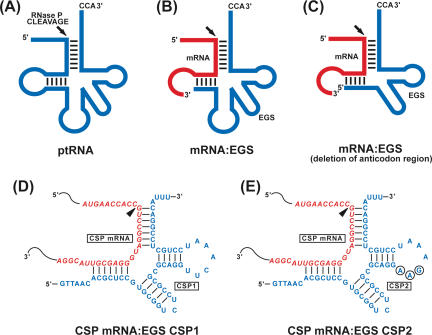
FIGURE 1.
(A) Schematic representation of a natural substrate (ptRNA) for RNase P. (B,C) A hybridized complex of a target RNA (e.g., mRNA) and an EGS that resembles the structure of a tRNA and can be cleaved by RNase P. C results from B by deleting the anticodon domain of the EGS, which is dispensable for EGS targeting activity (Yuan and Altman 1994). The site of cleavage by RNase P is marked with an arrowhead. (D,E) Complexes between the CSP mRNA sequence and EGS CSP1 and CSP2, respectively. The sequence of these EGSs equivalent to the tRNA sequence was derived from tRNAser and resembled the T-stem and loop and variable region of the tRNA molecule. Binding of CSP1 to the target mRNA results in a helix structure of 7 bp that is equivalent to the D stem of a tRNA, which usually has 4 bp. Our choice of a D stem-equivalent structure of 7 bp instead of 4 bp is to increase the targeting sequence specificity of the EGS.
Compared to other nucleic-acid-based gene interference strategies, such as the RNAi approach, which induces the cellular RISC RNase to cleave a target mRNA (Scherer and Rossi 2003), targeted cleavage of mRNA by RNase P using EGSs provides a unique approach to inactivate any RNA of known sequence expressed in vivo. To date, extensive studies exploiting the potential of antisense, ribozymes, and RNAi molecules have been carried out, including several clinical trials using these technologies for treating certain human diseases (Wong-Staal et al. 1998; Scherer and Rossi 2003). However, there are only a few studies evaluating the utility of the RNase P-based technology in targeting disease-associated mRNAs in human cells. In the present study, EGS RNAs were constructed to target the mRNA encoding the capsid scaffolding protein (CSP) of human cytomegalo-virus (HCMV), and their activity in blocking viral gene expression and growth in HCMV-infected cells was examined.
HCMV, a human herpesvirus, causes serious clinical manifestations in newborns and immunocompromised populations such as AIDS patients (Mocarski and Courcelle 2001). For example, this virus is the leading cause of congenital infections associated with mental retardation in newborns (Pass 2001). Continued development of effective antiviral compounds and approaches is central for the treatment and prevention of infections by HCMV as well as other herpesviruses. HCMV CSP completely overlaps with and is within the 3′ coding sequence of another viral capsid protein, assemblin (Welch et al. 1991). Both CSP and assemblin are believed to be essential for HCMV capsid formation and viral replication (Mocarski and Courcelle 2001). Thus, CSP and assemblin may serve as targets for novel drug development to combat HCMV infection.
In this study, we showed that a constructed EGS induced RNase P to cleave the target mRNA sequence in vitro. Moreover, intracellular expression of the EGS using retro-viral expression vectors leads to a significant inhibition of the expression of viral CSP and assemblin. An 800-fold reduction in viral growth was observed in the EGS-expressing cells. Our study provides the direct evidence that the RNase P-based approach is highly effective in inhibiting HCMV gene expression and growth by targeting the CSP mRNA. These results also demonstrate the feasibility of developing highly active EGSs as a novel class of antiviral agents for treatment of human viral diseases.
RESULTS
In vitro RNase P-mediated cleavage of the HCMV CSP mRNA sequence induced by the constructed EGSs
To achieve optimal cleavage, it is critical to choose the target regions of the CSP mRNA that are accessible to binding by the EGS because most intracellular RNAs are usually associated with proteins and are present in highly organized and folded conformations. In vivo mapping with dimethyl sulphate (DMS) has been extensively used to determine the accessibility of mRNA and structure of RNAs in cells (Liu and Altman 1995; Zaug and Cech 1995). Using this method, we examined the accessibility of the 5′ region around the translation initiation site of the CSP mRNA. Cells were infected with HCMV and then incubated with culture media that contained DMS. DMS entered the cells and modified the nucleotides of the mRNA regions that were accessible. Total mRNAs were subsequently isolated from the infected cells. Those regions of the CSP mRNA that were modified by DMS were mapped by primer extension assays in the presence of reverse transcriptase. A position, 359 nt downstream from the translational initiation codon, was chosen as the cleavage site for EGSs. This site appeared to be one of the regions most accessible to DMS modification (data not shown). Moreover, its flanking sequence exhibited several sequence features that need to be present in order to interact with an EGS and RNase P to achieve efficient cleavage. These features include that (1) the nucleotides 3′ and 5′ adjacent to the site of cleavage are a guanosine and a pyrimidine, respectively, and (2) a U is 8 nt downstream from this cleavage site. The interactions of these sequence elements with the EGS facilitate the formation of the mRNA–EGS complex into a tRNA-like structure while those with RNase P are critical for recognition and cleavage by the enzyme (Yuan and Altman 1994).
Two EGSs were constructed (Fig. 1D,E ▶): CSP1, which resembles a part of the tRNAser structure, contains a T-loop and stem, and a variable region but not the anticodon region that is dispensable for EGS activity (Fig. 1C,D ▶; Yuan and Altman 1994). Binding of CSP1 to the target mRNA results in a helix structure of 7 bp that is equivalent to the D stem of a tRNA, which usually has 4 bp. Our choice of a D stem-equivalent structure of 7 bp instead of 4 bp is to increase the targeting sequence specificity of the EGS. CSP2 was derived from CSP1 by introducing base-substitution mutations in three positions of the T-loop. The nucleotides in these three positions are highly conserved among tRNA molecules and are important for folding of the tRNA molecules and their recognition by RNase P (Frank and Pace 1998; Altman and Kirsebom 1999). The DNA sequences coding for the EGSs were generated by PCR using the DNA sequence for tRNAser as the template and primers that contained the sequences complementary to the targeted region of the CSP mRNA. These DNA sequences were under the control of the promoter for T7 RNA polymerase, and EGS RNAs were synthesized in vitro from the DNA sequences by T7 RNA polymerase.
The EGSs were subsequently incubated with human RNase P and substrate csp39, which contained the targeted CSP mRNA sequence of 39 nt. In the absence of any EGS, no cleavage of the CSP mRNA sequence was observed (Fig. 2 ▶, lane 4). Efficient cleavage of this substrate by RNase P was apparent in the presence of EGS CSP1 and >60% of the substrate was cleaved within 45 min (Fig. 2 ▶, lane 1). In contrast, cleavage of the same substrate by RNase P was barely detected in the presence of EGS CSP2 (Fig. 2 ▶, lane 2).
FIGURE 2.
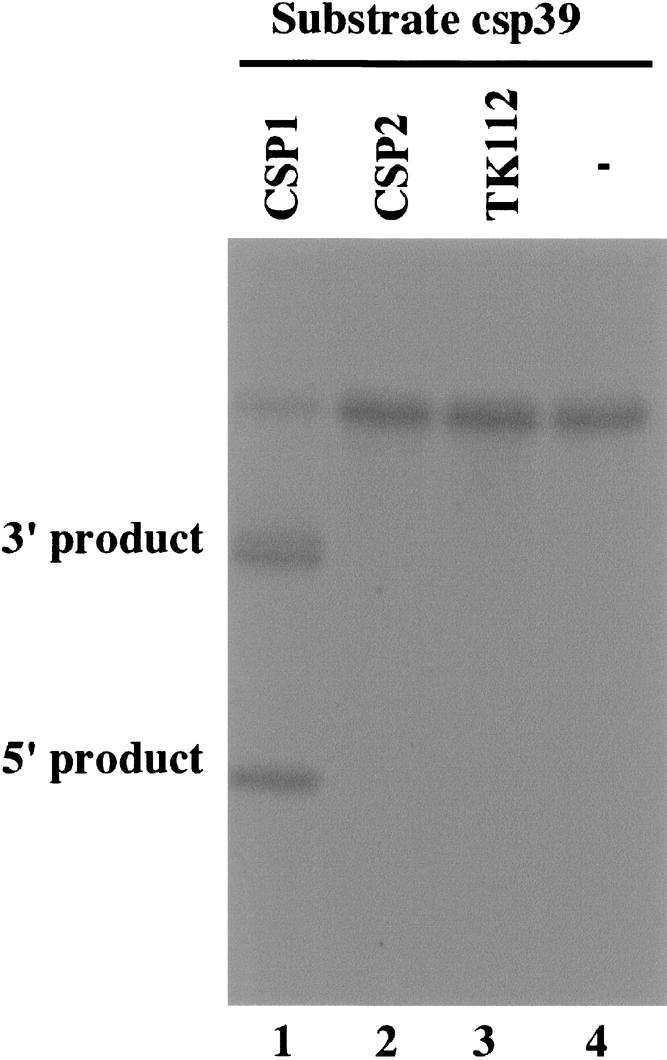
Cleavage of CSP mRNA sequence (substrate csp39) by human RNase P in the presence of different EGSs. No EGS was added to the reaction mixture in lane 4. Ten nanomolar of the EGS CSP1 (lane 1), CSP2 (lane 2), and TK112 (lane 3) were incubated with [32P]-labeled CSP RNA substrate (20 nM) and human RNase P (2 units) at 37°C in a volume of 10 μL for 45 min in buffer A (50 mM Tris at pH 7.4, 100 mM NH4Cl, and 10 mM MgCl2). More than 60% of the substrate was cleaved within 45 min. Experimental details can be found in Materials and Methods.
It is possible that the differential cleavage efficiencies observed with CSP1 and CSP2 were due to their different binding affinities to the CSP mRNA sequence. To investigate whether this was the case, the binding between the EGSs and substrate csp39 was assayed using a gel-shift assay. In this assay, the EGSs were incubated with the substrate to allow for binding, and the EGS-csp39 complexes were separated in polyacrylamide gels under nondenaturing conditions. Similar amounts of complexes formed between these two EGSs and the CSP mRNA sequence were observed when the same amount of EGSs was used (data not shown). Further detailed assays under different concentrations of the EGSs and csp39 indicated that the binding affinity of CSP2 to csp39 (Kd = 850 ± 150 nM) is similar to that of CSP1 (Kd = 900 ± 200 nM). Meanwhile, a very little amount of cleavage products was observed in the presence of CSP2 even under high concentrations of the EGS and RNase P and a prolonged incubation period (Fig. 2 ▶, lane 2; data not shown). These observations suggest that the mutations in CSP2 do not significantly affect the binding affinity of the EGS to the mRNA sequence but abolish its targeting activity to induce RNase P cleavage, possibly by disrupting the recognition of EGS–CSP mRNA complex by RNase P. Thus, CSP2 RNA may be used as a control for the antisense effect in our experiments in cultured cells (see below).
Efficient expression of the EGSs in human cells
The DNA sequences coding for CSP1 and CSP2 were cloned into retroviral vector LXSN and placed under the control of the small nuclear U6 RNA promoter, which has previously been shown to express EGS RNAs steadily (Miller and Rosman 1989; Yuan et al. 1992; Kawa et al. 1998). This promoter is transcribed by RNA polymerase III, and its transcripts are highly expressed and primarily localized in the nucleus (Yuan et al. 1992; Good et al. 1997; Kawa et al. 1998). To construct cell lines that express EGS RNAs, amphotropic packaging cells (PA317) were transfected with LXSN–EGS DNAs to produce retroviral vectors that contained the genes for EGS RNAs. Human U373MG cells were then infected with these vectors, and cells expressing the EGSs were cloned.
An additional cell line, which expressed EGS TK112 that targeted the mRNA for thymidine kinase (TK) of herpes simplex virus (HSV-1) (Kawa et al. 1998), was also constructed. No RNase P-mediated cleavage of substrate csp39 in the presence of TK112 was observed in vitro (Fig. 2 ▶, lane 3). We used this cell line to determine whether EGS RNA with an incorrect guide sequence could target CSP mRNA in tissue culture. The constructed lines and a control line in which cells were transfected with LXSN vector DNA alone were indistinguishable in terms of their growth and viability for up to 3 mo (data not shown), suggesting that the expression of the EGSs did not exhibit significant cytotoxicity.
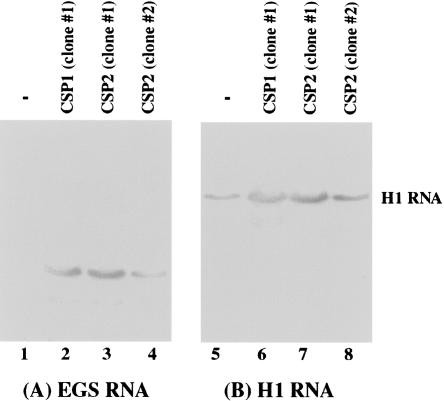
We determined the level of EGS RNA in each cell clone using Northern analysis with a DNA probe that is complementary to EGS RNA. The RNA samples isolated from cells were separated in gels and transferred to two identical membranes. One membrane was hybridized with a radio-labeled probe that contained the EGS RNA sequence while the other was probed with a labeled DNA sequence coding for H1 RNA, the RNA subunit of human RNase P and a nuclear RNA (Bartkiewicz et al. 1989), which was used as the internal control. The EGS RNAs were readily expressed in the nuclei as they were detected in the nuclear RNA fractions (Fig. 3 ▶; data not shown). This is consistent with previous observations in our laboratory as well as others that the transcripts expressed by the U6 promoter are primarily localized in the nuclei (Yuan et al. 1992; Good et al. 1997; Kawa et al. 1998). The different levels of EGS RNA expression between the two cloned cell lines (Fig. 3 ▶, cf. lanes 3 and 4) are presumably due to the incorporation of the LXSN–EGS sequence into different locations of the host chromosome, and its expression is influenced by the flanking sequence at the insertion site. We only used the cell lines that expressed similar levels of these EGS RNAs for further studies in tissue culture.
FIGURE 3.
The expression of EGS RNAs in cultured cells. Northern analyses were carried out using nuclear RNA fractions isolated from parental U373MG cells (−, lanes 1,5) and a cloned cell line that expressed CSP1 (lanes 2,6), and from two cloned cell lines expressing EGS CSP2 (lanes 3,4,7,8). Equal amounts of each RNA sample (30 μg) were separated on 2% agarose gels that contained formaldehyde and transferred to two identical nitrocellulose membranes. One membrane was hybridized to a [32P]-radiolabeled probe that contained the DNA sequence coding for EGS CSP1 (lanes 1–4) while the other was probed with a labeled DNA sequence coding for H1 RNA (lanes 5–8), the RNA subunit of human RNase P and a nuclear RNA (Bartkiewicz et al. 1989). The hybridized products corresponding to the full-length retro-viral transcripts (~6 kb), transcribed from the LTR promoter, are at the top of the gel and are not shown.
EGS RNA-directed inhibition of viral CSP and assemblin expression
To determine the efficacy of the EGS RNAs in inhibiting the expression of their viral targets, cells were infected with HCMV at a multiplicity of infection (MOI) of 0.05–1. Total RNAs were isolated from the infected cells at 8–72 h post-infection. The expression levels of CSP and assemblin mRNAs were determined by Northern analyses. The level of the 5-kb-long viral immediate-early transcript (5 kb RNA), whose expression is not regulated by CSP or assemblin under the assay conditions (Mocarski and Courcelle 2001), was used as an internal control for the quantitation of expression of CSP and assemblin mRNAs (Fig. 4 ▶). A reduction of ~80% ± 8% and 81% ± 7% (average of three experiments) in the expression level of CSP and assemblin mRNA was observed in cells that expressed EGS CSP1, respectively (Table 1 ▶). In contrast, a reduction of <10% in the expression level of these two mRNAs was observed in cells that expressed CSP2 or TK112. These results suggest that the significant reduction of the target mRNA expression in cells that expressed CSP1 was due to the RNase P-mediated cleavage directed by the EGS. The low level of inhibition found in cells that expressed CSP2 was presumably due to an antisense effect because CSP2 exhibits binding affinity to the target sequence similar to CSP1 but does not direct RNase P for targeted cleavage.
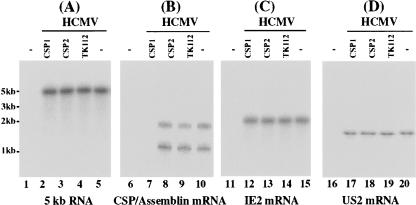
FIGURE 4.
Levels of HCMV mRNAs as determined by Northern analysis. Cells (1 × 106) were either mock-infected (lanes 1,6,11,16) or infected with HCMV (MOI = 1) (lanes 2–5,7–10,12–15,17–20) and were harvested at 24 h post-infection. Northern analyses were carried out using RNA isolated from parental U373MG cells (−, lanes 1,5,6,10,11,15,16,20) and cell lines that expressed CSP1 (lanes 2,7,12,17), CSP2 (lanes 3,8,13,18), and TK112 (lanes 4,9,14,19). Equal amounts of each RNA sample (30 μg) were separated on agarose gels that contained formaldehyde, transferred to a nitrocellulose membrane, and hybridized to a [32P]-radiolabeled probe that contained the cDNA sequence of the HCMV 5-kb transcript (lanes 1–5), CSP mRNA (lanes 6–10), IE2 mRNA (lanes 11–15), and US2 mRNA (lanes 16–20). The hybridized products corresponding to the 5-kb RNA, CSP, assemblin, IE2, and US2 mRNAs were ~5, 1.1, 2.1, 2.2, and 1.5 kb in size, respectively (Mocarski and Courcelle 2001).
TABLE 1.
Levels of inhibition of the expression of viral genes in cells that expressed CSP1, CSP2, and TK112, as compared to the levels of inhibition in cells that did not express an EGS (U373MG)
| EGS RNA | |||||
| Viral gene class | U373MG | TK112 | CSP1 | CSP2 | |
| IE2 mRNA | α | 0% | 1% | 1% | 2% |
| US2 mRNA | β | 0% | 0% | 3% | 2% |
| CSP mRNA | γ | 0% | 1% | 80 ± 8% | 7% |
| Assemblin mRNA | γ | 0% | 0% | 81 ± 7% | 7% |
| UL44 protein | β, γ | 0% | 0% | 3% | 1% |
| UL83 protein | γ | 0% | 1% | 2% | 1% |
| CSP protein | γ | 0% | 0% | 80 ± 8% | 6% |
| Assemblin | γ | 0% | 0% | 78 ± 7% | 6% |
The values shown are the means derived from triplicate experiments, and values for the standard deviation that were less than 5% are not shown.
The protein levels of CSP and assemblin in EGS-expressing cells are expected to decline due to decreased levels of their mRNAs. Proteins were isolated from cells at 24–72 h post-infection, separated in SDS-polyacrylamide gels, and transferred to identical membranes. Two of these membranes were stained with an anti-CSP and anti-assemblin antibody, respectively (Fig. 5B,C ▶). Another membrane was stained with a monoclonal antibody against human actin (anti-Actin) (Fig. 5A ▶). The latter serves as an internal control for the quantitation of CSP and assemblin protein expression. The expression of CSP, assemblin, and actin was quantitated using a chemiluminescent substrate for the antibody staining (Fig. 5 ▶). The results of three independent experiments are summarized in Table 1 ▶: A reduction of ~80% ± 8% and 78% ± 7% in the level of CSP and assemblin proteins was observed in cells that expressed EGS CSP1, respectively. In contrast, a reduction of <10% was found in cells that expressed CSP2 or TK112 RNAs. The low level of reduction in the expression level of CSP and assemblin proteins observed in cells that expressed CSP2 was probably due to the antisense effect of the EGS.
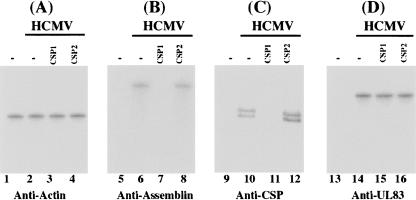
FIGURE 5.
Levels of human actin and HCMV proteins as determined by Western blot analysis. Protein samples were isolated from the parental U373MG cells or EGS-expressing cells that were either mock-infected (lanes 1,5,9,13) or infected with HCMV (MOI = 0.5–1) (lanes 2–4,6–8,10–12,14–16) for 48 h, separated in either SDS-polyacrylamide gels, and then transferred to membranes. One membrane was allowed to react with a monoclonal antibody (Anti-actin) against human actin (A) while the others were stained with the monoclonal antibody (Anti-assemblin, Anti-CSP, and Anti-UL83) against HCMV assemblin, CSP, or viral late protein UL83, respectively (B–D).
EGS-directed inhibition of HCMV growth by targeting CSP mRNA
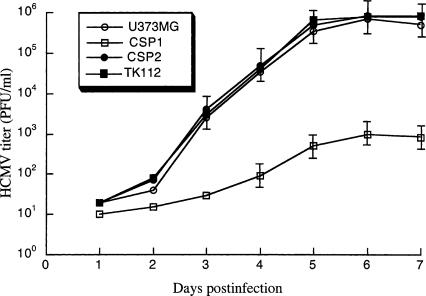
To determine whether the growth of HCMV is inhibited in the EGS-expressing cells, cells were infected by HCMV at an MOI of 1–5. We prepared virus stocks from the infected cultures (cells and culture medium together) at 1-d intervals through 7 d post-infection and determined the number of plaque-forming units (PFU) by measuring the viral titer on human foreskin fibroblasts. After 5 d post-infection, a reduction of at least 800-fold in viral yield was observed in cells that expressed EGS CSP1 (Fig. 6 ▶). No significant reduction was found in those that expressed the control EGSs CSP2 or TK112 (Fig. 6 ▶; data not shown). These results suggest that EGS-mediated targeting of viral CSP and assemblin mRNAs effectively inhibits HCMV growth.
FIGURE 6.
Growth of HCMV in U373MG cells and cell lines that expressed EGS RNAs. Cells (5 × 105) were infected with HCMV at a MOI of 2. Virus stocks were prepared from the infected cells at 1-d intervals through 7 d post-infection and the PFU count was determined by measurement of the viral titer on human foreskin fibroblasts. These values are derived from the means from triplicate experiments. The standard deviation is indicated by the error bars.
EGS RNA-directed targeting of CSP mRNA specifically blocks HCMV capsid formation but has no effects on the expression of other viral genes or viral genome replication
HCMV CSP is believed to be essential for viral capsid assembly (Mocarski and Courcelle 2001). Meanwhile, it is possible that the observed reduction of viral growth in the EGS-expressing cells is not necessarily due to specific EGS RNA-directed RNase P cleavage of CSP and assemblin mRNAs but is due to other effects of the EGS on viral lytic replication that are unrelated to the consequence of the EGS-directed cleavage or the inhibition of viral CSP or assemblin expression, such as blocking the expression of viral immediate-early genes. To exclude these possibilities and further determine the antiviral mechanism of the EGS-directed cleavage, two sets of experiments were carried out to investigate the step of the viral lytic cycles that is blocked in the cells that expressed CSP1.
In the first set of experiments, the expression of other viral genes in the cells is examined. Inhibition of CSP and assemblin expression is not expected to affect the expression of other viral genes, including immediate-early (α), early (β), and late (γ) genes, which are not regulated by the CSP or the assemblin (Mocarski and Courcelle 2001). Northern analyses were carried out to determine the expression levels of the IE2 (an α transcript) and US2 mRNA (a β transcript) (Fig. 4C,D ▶). Moreover, we performed Western analyses to determine the expression level of viral protein UL44, a viral late (β γ) protein and UL83, a viral late (γ) protein (Fig. 5D ▶; Table 1 ▶). No significant difference in the expression level of these genes was observed in cells that expressed CSP1, CSP2, or TK112 (Table 1 ▶). These results suggest that the expression of CSP1 specifically inhibits the expression of CSP and assemblin and does not affect overall viral gene expression. Moreover, our results are consistent with the notion that neither CSP nor assemblin is required for the expression of most viral genes.
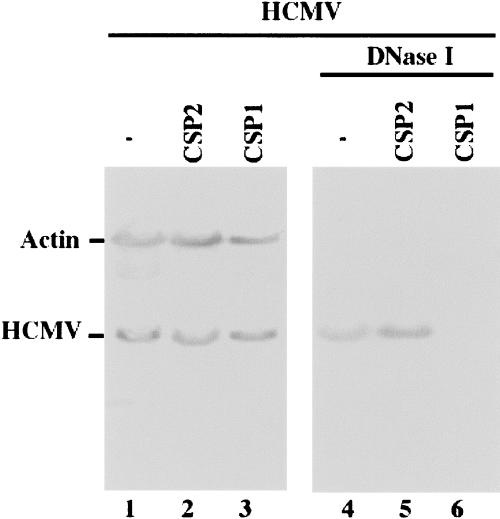
In the second set of experiments, we investigated whether viral genomic replication as well as capsid maturation is affected in the cells that expressed CSP1 RNA. Total DNA was isolated from HCMV-infected cell lysates. The level of intracellular viral DNA was determined by PCR detection of HCMV IE1 sequence, using the level of β-actin DNA as the internal control. Since HCMV only replicates in an episomal form and does not integrate its DNA into the host genome (Mocarski and Courcelle 2001), the amount of the intracellular viral DNA detected by the PCR assay represents the replication level of the viral genome. No significant difference in the level of viral DNA was found in cells that expressed CSP1, CSP2, and TK112, suggesting that a reduction of CSP and assemblin expression by the EGS does not affect the step of viral genome replication (Fig. 7 ▶, lanes 1–3). To examine viral capsid formation, the level of encapsidated viral DNA was assayed in order to determine the level of mature capsid formed in the infected cells. DNA samples were isolated from HCMV-infected cell lysates that were treated with DNase I. The encapsidated viral DNAs should be resistant to DNase I digestion while those that are not packaged in the capsid are susceptible to degradation. The level of intracellular “encapsidated” viral DNA was determined by PCR detection of HCMV IE1 sequence. When assaying the DNA samples from cell lysates that were not treated with DNase I, we found no significant difference in the level of total intracellular (both encapsidated and uncapsidated) viral DNA in the EGS-expressing cells (Fig. 7 ▶, lanes 1–3). When the DNase I-treated samples were assayed, however, the encapsidated DNA was hardly detected in the cells that expressed CSP1 (Fig. 7 ▶, lanes 4–6). These observations suggest that EGS-mediated inhibition of CSP and assemblin expression does not affect the replication of viral DNA but blocks the event(s) during capsid formation.
FIGURE 7.
Level of encapsidated (B) and total intracellular (A) viral DNA as determined by semiquantative PCR. Total DNA (lanes 1–3) or DNase I-treated DNA samples (lanes 4–6) were isolated from cells that either did not express an EGS (−, lanes 1,4) or express EGS CSP2 (lanes 2,5) or CSP1 (lanes 3,6). Cells were infected with HCMV at MOI of 1. We determined the levels of viral IE1 sequence by PCR using human actin DNA sequence as the internal controls. The amplification by PCR was within the linear range. The radiolabeled PCR products were separated in 4% nondenaturing polyacrylamide gels and quantitated with a Storm840 phosphorimager.
DISCUSSION
The EGS-based technology represents an attractive approach for gene inactivation since it utilizes endogenous RNase P to generate highly efficient and specific cleavage of the target RNA (Altman and Kirsebom 1999; Raj and Liu 2003). In particular, RNase P is among the most ubiquitous and essential enzymes found in nature, as it is responsible for processing all tRNA molecules. Moreover, RNase P-mediated cleavage directed by EGSs is highly specific and does not generate “irrelevant cleavage” that is usually associated with RNase H-mediated cleavage induced by conventional DNA-based oligonucleotides (Altman and Kirsebom 1999; Ma et al. 2000). Thus, EGS molecules represent promising general gene-targeting agents that can be used in both basic research and clinical applications.
Targeting activity and specificity of EGS RNAs
Several criteria must be satisfied if successful targeting with the EGS technology is to be achieved. Among these are high efficiency of cleavage, sequence specificity of the EGS, and efficient delivery of the reagents. We have constructed EGS RNAs that target the overlapping region of HCMV CSP and assemblin mRNAs and have shown that these EGSs directed human RNase P to cleave CSP mRNA sequence efficiently in vitro. Moreover, we showed that these EGSs were expressed stably in human cell culture, and a reduction of ~75%–80% in the expression level of CSP and assemblin and a reduction of >800-fold in viral growth were observed in cells that expressed a designed EGS (i.e., CSP1). In contrast, a reduction of <10% in the levels of CSP/assemblin expression and viral growth was observed in cells that expressed control EGS CSP2. CSP2 bound efficiently to the CSP mRNA sequence but contained point mutations that disrupted RNase P recognition. No reduction was observed in cells that expressed EGS TK112 that targeted another mRNA. These results suggest that the overall observed inhibition with CSP1 was primarily due to targeted cleavage by RNase P induced by this EGS as opposed to the antisense effect or other nonspecific effects of the EGSs.
The targeting activity of EGSs appears to be specific. First, expression of the EGSs did not exhibit significant cytotoxicity, as cells expressing EGSs are indistinguishable from the parental cells in terms of cell growth and viability for up to 3 mo (data not shown). Second, the inhibition of viral growth and capsid formation seems to be due to the reduction of CSP and assemblin, since the level of DNA encapsidation and the expression of CSP and assemblin were found to be greatly decreased in cells expressing CSP1 and not in the control cells expressing CSP2 or TK112 (Figs. 4 ▶, 5 ▶, 7 ▶; data not shown). Third, expression of the EGS only inhibits the expression of the CSP and assemblin mRNAs. We found no reduction in the expression levels of other viral genes examined (e.g., IE2, US2, UL44, and UL83) in EGS-expressing cells (Figs. 4 ▶, 5 ▶; Table 1 ▶). Fourth, the replication of viral genomic DNA does not appear to be affected by the expression of EGS CSP1 and the subsequent reduction of CSP and assemblin expression (Fig. 7 ▶). Together, these results support the belief that CSP and assemblin are essential for capsid assembly and do not involve in viral gene expression and genome replication.
Efficient cleavage of the CSP mRNA sequence by human RNase P was observed in vitro with the EGS (i.e., CSP1) that targets CSP mRNA but not with TK112. Moreover, a significant reduction of the expression was observed in cells that expressed CSP1 but not TK112. Two types of interaction between the EGS and target mRNA govern the specificity of the EGS targeting approach (Yuan and Altman 1994; Altman and Kirsebom 1999). One is the Watson–Crick base-pairing interaction between the antisense domain of the EGS and the target mRNA. The other type of interaction is between the RNase P recognition domain (e.g., T-loop and stem) and the mRNA. This interaction facilitates the folding of the EGS–mRNA complex into a tRNA-like molecule and stabilizes the mRNA–EGS complex. Manipulation of this latter interaction would make the EGS technology potentially more sequence specific than the conventional antisense approach.
Delivery of the EGS into the nuclear compartment is essential to the success of the EGS technology since RNase P is exclusively localized in the nuclei. The expression cassette we used to produce these EGSs is the promoter for small nuclear U6 RNA (Yuan et al. 1992; Good et al. 1997; Kawa et al. 1998). This promoter has been used extensively to express functional RNAs and ribozymes for gene targeting applications, and the transcript from this promoter is quite stable and primarily localized in the nuclei (Yuan et al. 1992; Good et al. 1997; Kawa et al. 1998). In addition, the efficient delivery and proper localization of the EGS may be mediated by cellular tRNA-binding proteins. These proteins could interact with the tRNA-like domains of the EGS and target the EGS to the nuclear compartment that contains RNase P. Further exploitation of these interactions will facilitate development of EGSs as novel gene-targeting agents for both basic research and clinical therapeutic applications.
Gene-targeting application of EGS RNAs for the inhibition of herpesvirus infection
HCMV is a member of the human herpesvirus family, which includes seven other different viruses such as HSV and Epstein–Barr virus (Mocarski and Courcelle 2001). All these viruses can engage in lytic replication as well as establish latent infections. HCMV CSP and assemblin are believed to be required for viral capsid formation. Their homologous proteins have been found in other herpes-viruses and are highly conserved among all herpesviruses (Mocarski and Courcelle 2001), suggesting that these proteins may serve as the ideal targets for drug development for the treatment and prevention of the infections by HCMV as well as other herpesviruses. Our results presented in this study indicate that EGS RNA-mediated inhibition of the expression of HCMV CSP and assemblin leads to a significant (~800-fold) reduction of viral growth, and they are consistent with the notion that blocking the expression of these two proteins should yield effective antiviral therapy. To further evaluate their anti-HCMV activity, these EGSs can be delivered into the monocyte/macrophage-lineage cells where HCMV is believed to establish latent infection (Mocarski and Courcelle 2001). These studies will determine whether the EGSs can abolish viral gene expression in these cells and prevent HCMV reactivation from latent infection into lytic replication. Meanwhile, our results have shown that EGSs can be used as a tool to study the function of HCMV essential genes. It has been technically difficult to construct complementing cell lines that support the replication of HCMV mutants with deletions of viral essential genes. Thus, genetic analyses of many HCMV essential genes have not been successful and little is currently known about the functional role of these genes in viral replication. The EGS-directed RNase P-mediated inhibition of the expression of viral essential genes represents an attractive approach to study the function of these genes in CMV infections. These studies will further facilitate the development of the EGS-based technology for general gene-targeting applications.
MATERIALS AND METHODS
Construction of plasmids, EGS RNAs, and RNA substrate for studies in vitro
The DNA sequence that encodes substrate csp39 was constructed by PCR using pGEM3zf(+) as a template and oligonucleotides AF25 (5′-GGAATTCTAATACGACTCACTATAG-3′) and sAP3 (5′-CGGGATCCGTAACGCTCCCATCCGGACGGTGGTTCATCCTATAGTGAGTCGTATTA-3′) as 5′ and 3′ primers, respectively. The DNA sequences coding for the EGSs were synthesized by the polymerase chain reaction (PCR), using construct pTK112 DNA (Kawa et al. 1998) as the template and were cloned under the control of the T7 RNA polymerase promoter. All oligonucleotides used as PCR primers were synthesized in a DNA synthesizer. To construct pCSP1, the 5′ and 3′ primers were oligoCSP11 (5′-GGGTTAACACGCUCCGTGCGGTCTCC-3′) and oligoCSP12 (5′-AAGCTTTAAATGTCCGGAGCAGGATTTG-3′), respectively. To construct pCSP2, the 5′ and 3′ primers were oligoCSP11 and the oligoCSP21 (5′-AAGCTTTAAATGTCCGGAGCAGGATTTCT TCCTGCGCGCG-3′), respectively.
RNase P assay and in vitro studies
Human RNase P was prepared from HeLa cellular extracts as described previously (Kawa et al. 1998; Zhu et al. 2004). EGS RNAs and the CSP mRNA substrate (i.e., csp39) were synthesized in vitro by T7 RNA polymerase (Promega Inc.) as per the manufacturer’s recommendations and further purified on 8% urea/polyacrylamide gels. Subsequently, the EGS RNAs and [32P]-labeled CSP RNA substrate were incubated with human RNase P. The cleavage reactions were carried out at 37°C in a volume of 10 μL for 45 min in buffer A (50 mM Tris at pH 7.4, 100 mM NH4Cl, and 10 mM MgCl2) (Kawa et al. 1998; Zhu et al. 2004). Cleavage products were separated in denaturing gels and quantitated with a Storm phosphorimager (Molecular Dynamics).
Assaying for the binding between EGSs and CSP mRNA sequence in vitro was performed as described previously (Kawa et al. 1998; Zhu et al. 2004). In brief, various concentrations of EGSs were preincubated in buffer A at 37°C for 15 min before mixing with an equal volume of different concentrations of substrate RNA preheated under identical conditions. The samples were incubated for 10–30 min to allow binding, then loaded on a 5% polyacrylamide gel, and run at 10 W. The electrophoresis running buffer contained 100 mM Tris-HEPES (pH 7.5) and 10 mM MgCl2. The value of Kd was then extrapolated from a graph plotting percent of product bound versus EGS concentration. The values were the average of three experiments.
Viruses, cells, and antibodies
Human primary foreskin fibroblasts (HFFs), astrocytoma U373MG cells, and PA317 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum. The propagation of HCMV (AD169) in HFFs and U373MG cells was carried out as described previously (Trang et al. 2000; Dunn et al. 2003). The anti-rabbit polyclonal antibodies against HCMV CSP and assemblin were kindly provided by Annette Meyer of Pfizer, Inc. The monoclonal antibody against human actin was purchased from Sigma, Inc. The monoclonal antibodies c1202, c1203, and c1205, which react with HCMV proteins UL44, IE1/IE2, and UL83, were purchased from Goodwin Institute for Cancer Research.
Construction of EGS-expressing cells
The protocols for the construction of U373MG cells expressing different EGSs were modified from Miller and Rosman and have been described previously (Miller and Rosman 1989; Trang et al. 2000). In brief, amphotropic PA317 cells were transfected with retroviral vector DNAs (LXSN-CSP1, LXSN-CSP2, and LXSN-TK112) with the aid of a mammalian transfection kit purchased from GIBCO BRL. Forty-eight hours post transfection, culture supernatants that contained retroviral vectors were collected and used to infect U373MG cells. At 48–72 h post-infection, cells were incubated in culture medium that contained 600 μg/mL neomycin. Cells were subsequently selected in the presence of neomycin for 2 wk and neomycin-resistant cells were cloned.
For Northern analyses of the expression of the EGSs, nuclear RNA fractions from EGS-expressing cells were isolated as described previously (Kawa et al. 1998; Trang et al. 2000). The RNA fractions were separated in 2.5% agarose gels that contained formaldehyde, transferred to nitrocellulose membranes, hybridized with the [32P]-radiolabeled DNA probes that contained the DNA sequences coding for CSP1, TK112, and H1 RNA, and finally analyzed with a Storm840 phosphor-imager. The radiolabeled DNA probe used to detect EGS RNAs was synthesized by using a random primed labeling kit (Boehringer Manheim Co.).
Viral infection and assays for viral gene expression and growth
T-25 flasks of cells (~106 cells) were either mock-infected or infected with HCMV as described previously (Trang et al. 2000; Dunn et al. 2003). The MOI is specified as that in the Results section. The infected cells were incubated for 8–72 h and viral mRNAs or proteins were isolated as described previously (Kawa et al. 1998; Zhu et al. 2004). To measure the levels of viral immediate-early (IE) transcripts, some of the cells were also treated with 100 μg/mL cycloheximide prior to and during infection.
The RNA fractions were separated in 1% agarose gels that contained formaldehyde, transferred to a nitrocellulose membrane, hybridized with the [32P]-radiolabeled DNA probes that contained the HCMV or human β-actin DNA sequences, and analyzed with a Storm840 phosphorimager. The DNA probes used to detect human β-actin mRNA, HCMV immediate-early 5-kb RNA transcript, IE2 mRNA, US2 mRNA, and CSP and assemblin mRNA were synthesized from plasmids pβ-actin RNA, pCig27, pIE2, pCig38, and pCSP, respectively.
For Western analyses, we separated the polypeptides from cell lysates on either SDS/7.5% polyacrylamide gels or SDS/9% poly-acrylamide gels cross-linked with N,N″ methylenebisacylamide. We then transferred the separated polypeptides electrically to nitrocellulose membranes. We stained the membranes using the antibodies against HCMV proteins and human actin in the presence of a chemiluminescent substrate (Amersham Inc.), and analyzed the stained membranes with a Storm840 phosphori-mager. Quantitation was performed in the linear range of RNA and protein detection.
To determine the level of the inhibition of viral growth, 5 × 105 cells were either mock-infected or infected with HCMV at an MOI of 1–5. The infection was carried out by incubating cells with DMEM in the absence or presence of viruses at 37°C for 90 min, and then with fresh DMEM for different periods of time (Trang et al. 2000; Dunn et al. 2003). The cells and medium were harvested at 1, 2, 3, 4, 5, 6, and 7 d post-infection and viral stocks were prepared by adding 10% skim milk followed by sonication. The titers of the viral stocks were determined by performing plaque assays on human foreskin fibroblasts (Trang et al. 2000; Dunn et al. 2003). The values obtained were the average from triplicate experiments.
Assaying the level of viral genome replication
Cells (5 × 105) grown on 6-well plates were mock-infected or infected with HCMV. After a 1.5-h incubation at 37°C, the inoculum was removed and the cells were further incubated and harvested at 48–96 h post-infection. Total and encapsidated (DNase I-treated) DNAs were isolated essentially as described (Gao et al. 1994; Matusick-Kumar et al. 1994) and used as the PCR DNA templates.
Viral DNA was detected by PCR amplification of the viral immediate-early IE1 sequence, using human β-actin sequence as the internal control. The PCR reaction consisted of 20 cycles with denaturation at 94°C for 1 min, followed by primer annealing at 47°C for 1 min and extension at 72°C for 1 min. The last cycle was again an extension at 72°C for 10 min. The 5′ and 3′ primers used to amplify the actin sequence were Actin5 (5′-TGACGGGGTCACCCA CACTGTGCCCATCTA-3′) and Actin3 (5′-CTAGAAGCATTGCG GTGGCAGATGGAGGG-3′), respectively (Daftarian et al. 1996) while those to amplify the HCMV IE1 sequence were CMV3 (5′-CCAAGCGGCCTCTGATAACCAAGCC-3′) and CMV4 (5′-CAGCACCATCCTCCTCTTCCTCTGG-3′), respectively (Demmler et al. 1988). PCR cycles and other conditions were optimized to assure that the amplification was within the linear range.
We carried out the PCR reactions in the presence of α-[32P]-dCTP. The radiolabeled DNA samples were separated on polyacrylamide gels and then scanned with a Storm840 phosphorimager. We also generated a standard (dilution) curve by amplifying different dilutions of the template DNA. The plot of counts for both HCMV and β-actin versus dilutions of DNA did not reach a plateau for the saturation curve (data not shown) under the conditions described above, indicating that quantitation of viral DNA could be accomplished. Moreover, we observed that the ratio of viral DNA to β-actin remained constant with respect to each DNA dilution in the standard curve, suggesting that the assay is adequately accurate and reproducible. The PCR results were derived from three independent experiments.
Acknowledgments
We thank Jiaming Zhu, Rong Hai, and Jarone Lee for technical assistance. P.T. was a recipient of a predoctoral fellowship from the American Heart Association (Western States Affiliate). K.K. acknowledges fellowship support from the University of California at Berkeley. F.L. is an American Heart Association Established Investigator and a Scholar of the Leukemia and Lymphoma Society. The research has been supported by grants from the March of Dimes Birth Defects Foundation and the NIH (DE14842).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2184706.
REFERENCES
- Altman, S. and Kirsebom, L.A. 1999. Ribonuclease P. In The RNA world (eds. R.F. Gesteland et al.), pp. 351–380. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bartkiewicz, M., Gold, H., and Altman, S. 1989. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes & Dev. 3: 488–499. [DOI] [PubMed] [Google Scholar]
- Daftarian, P.M., Kumar, A., Kryworuchko, M., and Diaz-Mitoma, F. 1996. IL-10 production is enhanced in human T cells by IL-12 and IL-6 and in monocytes by tumor necrosis factor-alpha. J. Immunol. 157: 12–20. [PubMed] [Google Scholar]
- Demmler, G.J., Buffone, G.J., Schimbor, C.M., and May, R.A. 1988. Detection of cytomegalovirus in urine from newborns by using polymerase chain reaction DNA amplification. J. Infect. Dis. 158: 1177–1184. [DOI] [PubMed] [Google Scholar]
- Dunn, W., Chou, C., Li, H., Hai, R., Patterson, D., Stolc, V., Zhu, H., and Liu, F. 2003. Functional profiling of human cytomegalovirus genome. Proc. Natl. Acad. Sci. 100: 14223–14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, A.C. and Altman, S. 1990. External guide sequences for an RNA enzyme. Science 249: 783–786. [DOI] [PubMed] [Google Scholar]
- Frank, D.N. and Pace, N.R. 1998. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67: 153–180. [DOI] [PubMed] [Google Scholar]
- Gao, M., Matusick-Kumar, L., Hurlburt, W., DiTusa, S.F., Newcomb, W.W., Brown, J.C., McCann 3rd, P.J., Deckman, I., and Colonno, R.J. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 68: 3702–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, P.D., Krikos, A.J., Li, S.X., Bertrand, E., Lee, N.S., Giver, L., Ellington, A., Zaia, J.A., Rossi, J.J., and Engelke, D.R. 1997. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 4: 45–54. [DOI] [PubMed] [Google Scholar]
- Gopalan, V., Vioque, A., and Altman, S. 2002. RNase P: Variations and uses. J. Biol. Chem. 277: 6759–6762. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Li, Y., and Altman, S. 1995. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc. Natl. Acad. Sci. 92: 11115–11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawa, D., Wang, J., Yuan, Y., and Liu, F. 1998. Inhibition of viral gene expression by human ribonuclease P. RNA 4: 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. and Altman, S. 1995. Inhibition of viral gene expression by the catalytic RNA subunit of RNase P from Escherichia coli. Genes & Dev. 9: 471–480. [DOI] [PubMed] [Google Scholar]
- Ma, M., Benimetskaya, L., Lebedeva, I., Dignam, J., Takle, G., and Stein, C.A. 2000. Intracellular mRNA cleavage induced through activation of RNase P by nuclease-resistant external guide sequences. Nat. Biotechnol. 18: 58–61. [DOI] [PubMed] [Google Scholar]
- Maran, A., Maitra, R.K., Kumar, A., Dong, B., Xiao, W., Li, G., Williams, B.R., Torrence, P.F., and Silverman, R.H. 1994. Blockage of NF-κB signaling by selective ablation of an mRNA target by 2-5A antisense chimeras. Science 265: 789–792. [DOI] [PubMed] [Google Scholar]
- Matusick-Kumar, L., Hurlburt, W., Weinheimer, S.P., Newcomb, W.W., Brown, J.C., and Gao, M. 1994. Phenotype of the herpes simplex virus type 1 protease substrate ICP35 mutant virus. J. Virol. 68: 5384–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A.D. and Rosman, G.J. 1989. Improved retroviral vectors for gene transfer and expression. Biotechniques 7: 980–990. [PMC free article] [PubMed] [Google Scholar]
- Mocarski, E.S. and Courcelle, C.T. 2001. Cytomegalovirus and their replication. In Fields virology (eds. D.M. Knipe and P.M. Howley), pp. 2629–2673. Lippincott-William & Wilkins, Philadelphia, PA.
- Pass, R.F. 2001. Cytomegalovirus. In Fields virology (eds. D.M. Knipe and P.M. Howley), pp. 2675–2706. Lippincott-William & Wilkins, Philadelphia, PA.
- Raj, S.M. and Liu, F. 2003. Engineering of RNase P ribozyme for gene-targeting applications. Gene 313: 59–69. [DOI] [PubMed] [Google Scholar]
- Santoro, S.W. and Joyce, G.F. 1997. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. 94: 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, L.J. and Rossi, J.J. 2003. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 21: 1457–1465. [DOI] [PubMed] [Google Scholar]
- Stein, C.A. and Cheng, Y.C. 1993. Antisense oligonucleotides as therapeutic agents—Is the bullet really magical? Science 261: 1004–1012. [DOI] [PubMed] [Google Scholar]
- Trang, P., Lee, M., Nepomuceno, E., Kim, J., Zhu, H., and Liu, F. 2000. Effective inhibition of human cytomegalovirus gene expression and replication by a ribozyme derived from the catalytic RNA subunit of RNase P from Escherichia coli. Proc. Natl. Acad. Sci. 97: 5812–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, A.R., Woods, A.S., McNally, L.M., Cotter R.J., and Gibson, W. 1991. A herpesvirus maturational proteinase, assemblin: Identification of its gene, putative active site domain, and cleavage site. Proc. Natl. Acad. Sci. 88: 10792–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Staal, F., Poeschla, E.M., and Looney, D.J. 1998. A controlled, Phase 1 clinical trial to evaluate the safety and effects in HIV-1 infected humans of autologous lymphocytes transduced with a ribozyme that cleaves HIV-1 RNA. Hum. Gene Ther. 9: 2407–2425. [DOI] [PubMed] [Google Scholar]
- Yuan, Y. and Altman, S. 1994. Selection of guide sequences that direct efficient cleavage of mRNA by human ribonuclease P. Science 263: 1269–1273. [DOI] [PubMed] [Google Scholar]
- Yuan, Y., Hwang, E.S., and Altman, S. 1992. Targeted cleavage of mRNA by human RNase P. Proc. Natl. Acad. Sci. 89: 8006–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug, A.J. and Cech, T.R. 1995. Analysis of the structure of Tetra-hymena nuclear RNAs in vivo: Telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA 1: 363–374. [PMC free article] [PubMed] [Google Scholar]
- Zhou, T., Kim, J., Kilani, A.F., Kim, K., Dunn, W., Jo, S., Nepomuceno, E., and Liu, F. 2002. In vitro selection of external guide sequences for directing RNase P-mediated inhibition of viral gene expression. J. Biol. Chem. 277: 30112–30120. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Trang, P., Kim, K., Zhou, T., Deng, H., and Liu, F. 2004. Effective inhibition of Rta expression and lytic replication of Kaposi’s sarcoma-associated herpesvirus by human RNase P. Proc. Natl. Acad. Sci. 101: 9073–9078. [DOI] [PMC free article] [PubMed] [Google Scholar]