Abstract
Polynucleotide kinase-phosphatase (Pnkp) from Clostridium thermocellum catalyzes ATP-dependent phosphorylation of 5′-OH termini of DNA or RNA polynucleotides and Ni2+/Mn2+-dependent dephosphorylation of 2′,3′ cyclic phosphate, 2′-phosphate, and 3′-phosphate ribonucleotides. CthPnkp is an 870-amino-acid polypeptide composed of three domains: an N-terminal module similar to bacteriophage T4 polynucleotide kinase, a central module that resembles the dinuclear metallo-phosphoesterase superfamily, and a C-terminal ligase-like adenylyltransferase domain. Here we conducted a mutational analysis of CthPnkp that identified 11 residues required for Ni2+-dependent phosphatase activity with 2′-AMP and 3′-AMP. Eight of the 11 CthPnkp side chains were also required for Ni2+-dependent hydrolysis of p-nitrophenyl phosphate. The ensemble of essential side chains includes the conserved counterparts (Asp187, His189, Asp233, Arg237, Asn263, His264, His323, His376, and Asp392 in CthPnkp) of all of the amino acids that form the dinuclear metal-binding site and the phosphate-binding site of bacteriophage λ phosphatase. Three residues (Asp236, His264, and Arg237) required for activity with 2′-AMP or 3′-AMP were dispensable for Ni2+-dependent hydrolysis of p-nitrophenyl phosphate. Our findings, together with available structural information, provide fresh insights to the metallophosphoesterase mechanism, including the roles of His264 and Asp236 in proton donation to the leaving group. Deletion analysis defined an autonomous phosphatase domain, CthPnkp-(171–424).
Keywords: RNA/DNA end-healing, phosphoesterase, dinuclear metal cluster, λ phosphatase
INTRODUCTION
When breakage of a 3′-5′ phosphodiester in DNA or RNA results in the formation of 5′-OH and 3′-PO4 (or 2′,3′ cyclic PO4) termini, the broken ends must be “healed” before they can be sealed by polynucleotide ligase. Healing occurs via hydrolysis of the 3′-PO4 (or 2′,3′ cyclic phosphate) to form a 3′-OH and phosphorylation of the 5′-OH to form a 5′-PO4 end. Bacteriophage T4 polynucleotide 5′-kinase/3′-phosphatase (Pnkp) is the prototypal end-healing enzyme (Richardson 1965; Novogrodsky and Hurwitz 1966; Novogrodsky et al. 1966; Cameron and Uhlenbeck 1977; Amitsur et al. 1987; Wang and Shuman 2001 Wang and Shuman 2002; Wang et al. 2002; Galburt et al. 2002). Additional Pnkp enzymes have been identified and characterized from other bacteriophages, eukarya, and a eukaryotic virus (Martins and Shuman 2004; Zhu et al. 2004; Bernstein et al. 2005; Blondal et al. 2005), but only recently has an end-healing activity been discovered in a bacterial proteome—that of the anaerobic thermophile Clostridium thermocellum (Martins and Shuman 2005).
CthPnkp catalyzes the phosphorylation of 5′-OH termini of DNA or RNA polynucleotides and the dephosphorylation of 2′,3′ cyclic phosphate, 2′-phosphate, and 3′-phosphate ribonucleotides. CthPnkp also catalyzes an auto-adenylylation reaction via a polynucleotide ligase-type mechanism. These characteristics are consistent with a role in end-healing during RNA or DNA repair. CthPnkp is a homodimer of an 870-amino-acid polypeptide composed of three catalytic domains: an N-terminal module that resembles the polynucleotide kinase domain of bacteriophage T4 Pnkp, a central metal-dependent phosphoesterase module, and a C-terminal module that resembles the nucleotidyl transferase domain of polynucleotide ligases (Fig. 1 ▶). Putative trifunctional homologs of CthPnkp are encoded by several other bacterial genera, including Kinetococcus, Nostoc, Streptomyces, Fusobacterium, Bacillus, and Helicobacter (Martins and Shuman 2005). Putative bifunctional Pnkp homologs in Deinococcus and Thermobifida are composed only of the kinase and the phosphoesterase domain equivalents of CthPnkp, with no equivalent of the adenylyltransferase domain.
FIGURE 1.
The phosphoesterase domain of CthPnkp. (Left) The amino acid sequence of CthPnkp(172–406) is aligned to the sequences of homologous proteins of Kineococcus radiotolerans, Nostoc sp. PCC 7120, Streptomyces coelicolor, Thermobifida fusca, Bacillus anthracis, Deinococcus radiodurans, and bacteriophage λ. Gaps in the alignment are indicated by dashes. The amino acids subjected to mutational analysis in the present study are indicated by |. The side chains that bind a dinuclear metal cluster and coordinate the phosphate in the active site in the crystal structure of λ phosphatase are highlighted in shaded boxes. Other positions of amino acid side chain identity or similarity in all proteins are indicated by dots above the alignment. (Right) The mutated amino acids of CthPnkp are listed next to their counterparts in λ phosphatase and the atomic contacts of these side chains in the crystal structure of λ phosphatase.
The distinctive feature of CthPnkp and its relatives visa ‘vis known RNA/DNA repair enzymes is that its 3′ end-modification component belongs to the calcineurin-type metallophosphoesterase superfamily (Goldberg et al. 1995; Rusnak and Mertz 2000). It contains putative counterparts of the amino acids that form the dinuclear metal-binding site and the phosphate-binding site of bacteriophage λ phosphatase (Voegtli et al. 2000). As with λ phosphatase (Zhuo et al. 1993), the 2′,3′ cAMP phosphatase activity of CthPnkp is specifically dependent on either nickel (Ni) or manganese. Here we address the following questions: (1) Is the primary structure similarity between CthPnkp and λ phosphatase functionally relevant? (2) If so, what features of the conserved side chains are essential for activity? (3) How do structure–function relationships illuminate the reaction mechanism? (4) Is phosphohydrolase activity restricted to nucleotide substrates? (5) Are the kinase and adenylyltransferase domains required for phosphatase activity, or does the phosphatase function reside within an autonomous domain of the CthPnkp protein?
RESULTS
Alanine scanning identifies the phosphoesterase active site of CthPnkp
The amino acid sequence of the central phosphatase-like domain of C. thermocellum Pnkp is shown in Figure 1 ▶, aligned to the sequences of homologous proteins of Kineococcus radiotolerans, Nostoc sp. PCC 7120, Streptomyces coelicolor, Thermobifida fusca, Bacillus anthracis, and Deinococcus radiodurans, and to the phosphoprotein phosphatase of bacteriophage λ. The alignment highlights the fact that CthPnkp and its bacterial homologs contain the full constellation of conserved residues that bind a dinuclear metal cluster and coordinate a sulfate in the active site in the crystal structure of λ phosphatase (Fig. 2 ▶; Voegtli et al. 2000). To probe the mechanistic and structural relatedness of CthPnkp and λ phosphatase, we introduced single alanine mutations at 12 positions within the full-length CthPnkp polypeptide, 11 of which (Asp187, His189, Asp233, Asp236, Arg237, Asn263, His264, Asp265, His323, His376, Asp392) are conserved in the bacterial homologs and λ phosphatase. The other targeted residue, Arg359, is conserved in the bacterial proteins but is apparently missing from λ phosphatase (Fig. 1 ▶).
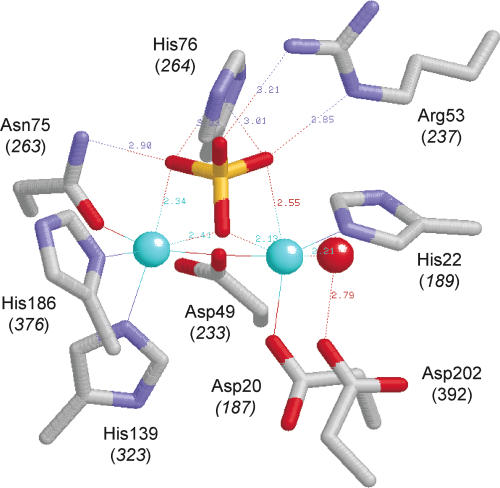
FIGURE 2.
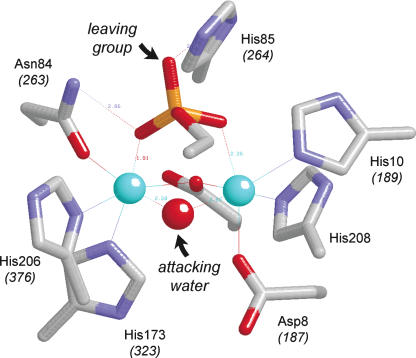
The active site of λ phosphatase. The figure depicts the active site of the manganese- and sulfate-bound λ phosphatase (molecule C) from the crystal structure of Voegtli et al. (2000; Protein Data Bank 1G5B). The amino acid side chains coordinating the binuclear metal cluster and the sulfate ion are shown. The corresponding amino acids of CthPnkp are indicated in parentheses. The manganese ions are colored cyan. Water is colored red. The structure is predicted to mimic the product (Pi) complex of the enzyme.
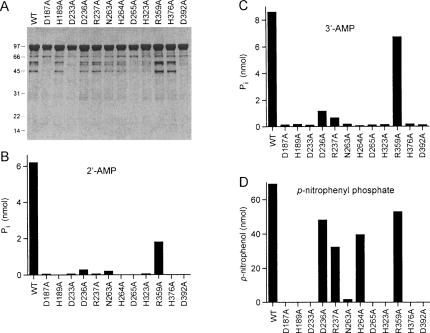
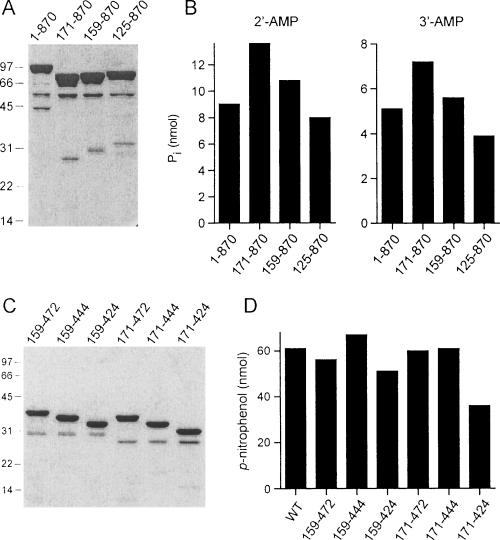
The CthPnkp-Ala mutants were produced in Escherichia coli as His10-tagged fusions in parallel with the wild-type protein and then purified from soluble bacterial extracts by adsorption to Ni-agarose and elution with buffer containing imidazole. SDS-PAGE analysis showed that the protein preparations consisted principally of a ~100-kDa polypeptide corresponding to His10-CthPnkp (Fig. 3A ▶). Minor polypeptides of 45 and 55 kDa likely corresponded to proteolytic fragments of CthPnkp (see below). The recombinant wild-type Pnkp catalyzed phosphate release from either 2′-AMP or 3′-AMP in the presence of NiCl2 (Fig. 3B,C ▶). Eleven of the alanine mutations either abolished or severely diminished phosphatase activity with 2′-AMP and 3′-AMP substrates. Our operational definition of a significant mutational effect is one that reduces activity to <20% of the wild-type value. The 11 residues defined by the alanine scan as essential or important for phosphatase function correspond to those that are conserved in λ phosphatase (Fig. 1 ▶). In contrast, the R359A mutation had little effect on hydrolysis of 3′-AMP (Fig. 3C ▶) and only a modest impact on hydrolysis on 2′-AMP (Fig. 3B ▶) that did not meet our criterion for functional importance. All of the Pnkp-Ala proteins retained polynucleotide kinase activity, measured by transfer of 32Pi from [γ-32P]ATP to the 5′-OH terminus of an 36-mer oligodeoxyribonucleotide to form a 5′ 32P-labeled oligonucleotide product (data not shown), signifying that the observed mutational effects on phosphatase function were probably not caused by global effects on protein folding. These results suggest that the phosphatase component of CthPnkp has an active site similar to that of λ phosphatase.
FIGURE 3.
Effects of alanine mutations on CthPnkp phosphatase activity. (A) Aliquots (5 μg) of the Ni-agarose preparations of the wild-type (WT) and Ala-mutant CthPnkp were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Reaction mixtures (25 μL) containing 50 mM Tris-HCl (pH 7.5), 5 mM NiCl2, 5 mM 2′-AMP, and 0.5 μg wild-type or mutant CthPnkp were incubated for 30 min at 45°C. (C) Reactions mixture (25 μL) containing 50 mM Tris-HCl (pH 7.5), 5 mM NiCl2, 5 mM 3′-AMP, and 1 μg CthPnkp were incubated for 30 min at 45°C. (D) Reaction mixtures (25 μL) containing 50 mM Tris-HCl (pH 7.5), 10 mM NiCl2, 10 mM p-nitrophenyl phosphate, and 0.5 μg CthPnkp were incubated for 30 min at 45°C.
Hydrolysis of a non-nucleotide phosphomonoester substrate
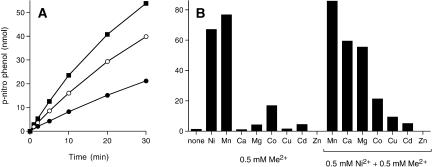
Although the linkage of the Clostridium 2′,3′-phosphatase to a polynucleotide kinase domain and a ligase-like adenylyl-transferase domain suggested a role in nucleic acid repair, the reports that λ-like phosphatases are able to hydrolyze non-nucleotide and nonprotein phosphate-containing substrates (Zhuo et al. 1993; Mertz et al. 1997; Chen et al. 2004; Schenk et al. 2005) prompted us to test the activity of CthPnkp with the generic substrate p-nitrophenyl phosphate. We found that the enzyme readily converted p-nitrophenyl phosphate (10 mM) to p-nitrophenol in the presence of 10 mM NiCl2. The rate and extent of product formation was proportional to input CthPnkp (Fig. 4A ▶). Hydrolysis of 10 mM p-nitro-phenyl phosphate was optimal at 2–10 mM Ni; activity at 0.5 mM Ni was 77% of the optimum value (data not shown). Magnesium, copper, cadmium calcium, or zinc (at 10 mM concentration) could not satisfy the divalent cation requirement (not shown). A mixing experiment was performed in which reactions containing 10 mM Ni were supplemented with 10 mM of each of the other divalent cations. Whereas magnesium and calcium neither significantly stimulated nor inhibited the hydrolysis of p-nitrophenyl phosphate, activity was abolished in the presence of copper, cadmium, or zinc (data not shown).
FIGURE 4.
Hydrolysis of p-nitrophenyl phosphate. (A) Kinetics. Reaction mixtures (200 μL) containing 50 mM Tris-HCl (pH 7.5), 10 mM NiCl2, 10 mM p-nitrophenyl phosphate, and either 1 μg (•), 2 μg (○), or 4 μg (▪) of CthPnkp were incubated at 45°C. Aliquots (25 μL) were withdrawn at the times specified and quenched immediately with sodium carbonate. p-Nitrophenol formation is plotted as a function of time. (B) Metal-specificity. Reaction mixtures (25 μL) contained 50 mM Tris-HCl (pH 7.5), 10 mM p-nitrophenyl phosphate, 1 μg CthPnkp, and either no divalent cation (none), 0.5 mM of the indicated divalent metal (Me2+) alone, or 0.5 mM NiCl2 plus 0.5 mM of the indicated divalent metal. All metals were added as the chloride salt. Reactions were incubated for 30 min at 45°C.
Our initial tests of manganese as a cofactor indicated that mixtures containing 10 mM p-nitrophenyl phosphate and manganese resulted in enzyme-independent generation of a brown-colored solution after the reactions were quenched with sodium carbonate, which caused a significant enzyme-independent background of absorbance at 410 nm. The background was absent at 0.5 mM manganese, increased linearly from 1–3 mM manganese, and plateaued at ≥5 mM. Whereas mixing 10 mM cobalt with 10 mM p-nitro-phenyl phosphate also generated a light brown color, this background was absent at 0.5 mM cobalt. To avoid these background complications, we reassayed the hydrolysis of p-nitrophenyl phosphate in the presence of 0.5 mM divalent cations. Ni and manganese supported similar levels of phosphatase activity, whereas cobalt was ~20% as effective (Fig. 4B ▶). Magnesium, calcium copper, cadmium, and zinc were ineffective. The selectivity for Ni and manganese in hydrolysis of p-nitrophenyl phosphate mirrors the requirements for hydrolysis of 2′,3′ cyclic AMP, 2′-AMP, and 3′-AMP substrates reported previously (Martins and Shuman 2005). When the metal mixing experiment was performed at 0.5 mM Ni and 0.5 mM of the second metal, we found that: (1) manganese did not synergize with Ni; (2) magnesium and calcium had little effect; (3) cobalt reduced activity to a level comparable to that of cobalt alone; and (4) copper, cadmium, and zinc were more strongly inhibitory. A simple interpretation of these results is that copper, cobalt, cadmium, and zinc compete with Ni for one or both sites in the binuclear metal cluster to generate a catalytically inert or impaired metalloenzyme, whereas magnesium and calcium do not compete well with Ni for the metal-binding sites.
Distinctive effects of alanine mutations on hydrolysis of p-nitrophenyl phosphate
Eight of the alanine mutations of Cth-Pnkp either abolished or severely diminished phosphatase activity with p-nitrophenyl phosphate as the substrate (Fig. 3D ▶). The residues thereby defined as essential for production of p-nitrophenol include all of the predicted direct or water-mediated ligands of the binuclear metal cluster (Asp187, His189, Asp233, Asn263, His323, His376, Asp392) (Figs. 1 ▶, 2 ▶), signifying that the same mechanism of metal-dependent catalysis is employed by CthPnkp with nucleotide and non-nucleotide substrates. Mutation of Arg359 had little impact on hydrolysis of p-nitrophenyl phosphate (Fig. 3D ▶), which was consistent with the results for the nucleotide substrates. The interesting findings were that Asp236, Arg237, and His264 were not required for hydrolysis of p-nitrophenyl phosphate, although they clearly were critical for activity with 2′-AMP and 3′-AMP (Fig. 3 ▶). We discuss these results below in the context of a model of general acid catalysis involving Asp236 and His264, whereby the requirement for proton donation is alleviated for hydrolysis of p-nitrophenyl phosphate by the low pKa of the p-nitrophenol leaving group.
Structure–function relationships at essential residues
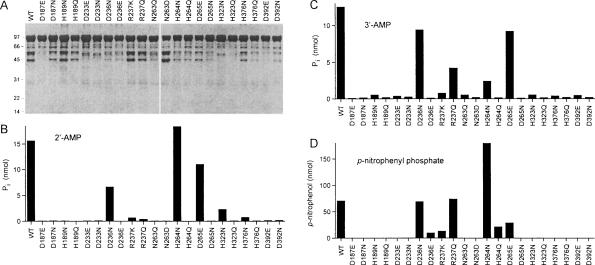
Conservative substitutions were introduced at the 11 positions defined by the alanine scan as essential/important for phosphatase activity with the 2′-AMP or 3′-AMP substrates. Aspartate was replaced by glutamate and asparagine, histidine by asparagine and glutamine, arginine by lysine and glutamine, and asparagine by aspartate and glutamine. The 22 conservative mutants were produced in E. coli as His10-tagged fusions and purified from soluble extracts by Ni-agarose chromatography, in parallel with the wild-type CthPnkp (Fig. 5A ▶). The effects of the mutations on the release of Pi from 2′-AMP and 3′-AMP substrates are shown in Figure 5, B and C ▶, respectively. Effects on the release of p-nitrophenol from p-nitro-phenyl phosphate are shown in Figure 5D ▶. The notable finding was that all seven side chains predicted to make direct or water-mediated contacts to the binuclear metal cluster (Asp187, His189, Asp233, Asn263, His323, His376, Asp392) were strictly essential for phosphatase activity with all three substrates; i.e., both of the conservative substitutions tested at each position either abolished or significantly suppressed product formation. These results attest to the lack of plasticity in the metal-binding site of CthPnkp.
FIGURE 5.
Effects of conservative mutations on CthPnkp phosphatase activity. (A) Aliquots (5 μg) of the Ni-agarose preparations of wild-type (WT) CthPnkp and conservative mutants were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Reaction mixtures (25 μL) containing 50 mM Tris-HCl (pH 7.5), 5 mM NiCl2, 5 mM 2′-AMP, and 1 μg CthPnkp as specified were incubated for 30 min at 45°C. (C) Reactions mixture (25 μL) containing 50 mM Tris-HCl (pH 7.5), 5 mM NiCl2, 5 mM 3′-AMP, and 1 μg CthPnkp as specified were incubated for 30 min at 45°C. (D) Reaction mixtures (25 μL) containing 50 mM Tris-HCl (pH 7.5), 10 mM NiCl2, 10 mM p-nitrophenyl phosphate, and 0.5 μg CthPnkp as specified were incubated for 30 min at 45°C.
Conservative substitutions at other positions resulted in significant gains of function compared with an alanine mutant. For example, whereas D265A was virtually inert with all substrates, introduction of a glutamate restored activity to 70%, 74%, and 39% of the wild-type value with 2′-AMP, 3′-AMP, and p-nitrophenyl phosphate, respectively (Fig. 5 ▶). In contrast, the conservative D265N change had no salutary effect. These results signify that a carboxylate functional group is critical at position 265. Note that this position is naturally occupied by glutamate (Glu77) in λ phosphatase and several bacterial homologs of CthPnkp (Fig. 1 ▶). The Glu77 side chain in λ phosphatase makes no contacts with either the metal cluster or the sulfate ion (a mimetic of phosphate) but rather appears to play a structural role in ensuring the proper conformation of the active site, via hydrogen bonds from the glutamate to three backbone amide nitrogens, including that of Asp49 (Asp233 in CthPnkp) (Fig. 1 ▶), which coordinates both of the divalent cations (Fig. 2 ▶).
The R237A change suppressed hydrolysis of 2′-AMP and 3′-AMP, but not p-nitrophenyl phosphate. Whereas the conservative R237Q mutant was as active as the wild-type enzyme with p-nitrophenyl phosphate (Fig. 5D ▶) and 34% as active as the wild-type with 3′-AMP (Fig. 5C ▶), there was no gain of function in hydrolysis of 2′-AMP (Fig. 5B ▶). Note that the conservative R237K mutant was as defective as R237A with 2′-AMP or 3′-AMP and was even more defective than was R237A with p-nitrophenyl phosphate (Fig. 5 ▶). These results show that (1) the requirement for Arg237 is substrate dependent; (2) a positive charge is not sufficient for optimal activity with any substrate tested, i.e., lysine is detrimental in lieu of arginine; and (3) hydrogen bonding capacity is probably the critical property for activity with 3′-AMP, insofar as glutamine elicited a gain of function. The crystal structure of λ phosphatase shows that the equivalent of Arg237 makes a bidentate contact with a sulfate ion that likely mimics the phosphate of the product complex (Fig. 2 ▶).
H264A also blocked hydrolysis of 2′-AMP and 3′-AMP, but not p-nitrophenyl phosphate. Replacement by asparagine restored hydrolysis of 2′-AMP and 3′-AMP to 118% and 19% of the wild-type levels, respectively, but glutamine had no salutary effect (Fig. 5B,C ▶). It is notable that the activity of the H264N protein with p-nitrophenyl phosphate was several fold higher than that of wild-type CthPnkp, whereas H264Q was 31% as active as wild type (Fig. 5D ▶). The crystal structure of λ phosphatase shows that the histidine equivalent to His264 of CthPnkp coordinates the sulfate ion via Nɛ (Fig. 2 ▶), while Nδ makes a hydrogen bond to the aspartate side chain corresponding to Asp236 of CthPnkp (Fig. 1 ▶). It is therefore surprising that replacing His264 with asparagine, which mimics the Nδ functional group of histidine, results in much higher activity than does glutamine, which mimics the Nɛ functional group. As discussed below, these results have mechanistic implications concerning models of general acid catalysis by members of the calcineurin-like superfamily (Rusnak and Mertz 2000; Hopfner et al. 2001).
D236A suppressed activity with 2′-AMP and 3′-AMP, but not with p-nitrophenyl phosphate. Asparagine restored activity to 42% and 75% of the wild-type levels with 2′-AMP, 3′-AMP, respectively, and supported full activity with p-nitrophenyl phosphate. In contrast, glutamate abolished activity with 2′-AMP and 3′-AMP and reduced activity with p-nitrophenyl phosphate to 14% of the wild-type level. The finding that an isosteric amide is functional in lieu of the carboxylate implies that hydrogen bonding capacity (presumably to Nδ of His264; see above) is the relevant characteristic of this side chain in promoting hydrolysis of nucleotide phosphomonoesters. We infer a tight steric constraint on this side chain, insofar as the extra methylene group of glutamate was detrimental to catalysis.
Effects of selected mutations on Km and kcat
We determined steady-state kinetic parameters for hydrolysis of p-nitrophenyl phosphate at pH 7.5 in the presence of 10 mM Ni. Formation of p-nitrophenol by CthPnkp displayed a hyperbolic dependence on the concentration of the p-nitro-phenyl phosphate substrate (data not shown). From a double reciprocal plot, we calculated a Km value of 2.5 mM and a kcat of 818 min−1 (Table 1 ▶). Kinetic parameters were then determined for the set of nine proteins mutated at residues Asp236, Arg237, and His264 (Table 1 ▶). The kcat values for D236A (512 min−1), R237A (352 min−1) and H264A (402 min−1) confirmed the inferences from single-point assays that none of the three side chains is important for the chemistry of p-nitrophenyl phosphate hydrolysis. The Km values of the three alanine mutants were similar (within a factor of two) to that of wild-type CthPnkp. Notable findings were that the conservative H264N mutation resulted in a eightfold increase in kcat (to 6770 min−1) relative to wild-type enzyme, without significantly affecting the Km for p-nitrophenyl phosphate, whereas the turnover number of the H264Q protein was about half that of H264A (Table 1 ▶). Grossly disparate effects of conservative replacements on kinetic parameters were also seen at Arg237, where lysine preserved substrate binding but reduced kcat to 108 min−1, whereas glutamine lowered the substrate affinity several-fold (Km 8.6 mM) but increased kcat slightly (to 1090 min−1 compared with wild-type CthPnkp.
TABLE 1.
Kinetic parameters for hydrolysis of p-nitrophenyl phosphate
| Protein | Km (mM) | kcat (min−1) |
| Wild-type | 2.5 ± 0.7 | 818 ± 77 |
| D236A | 1.3 ± 0.4 | 512 ± 81 |
| D236N | 0.85 ± 0.05 | 762 ± 21 |
| D236E | 2.5 ± 0.6 | 146 ± 2 |
| R237A | 4.0 ± 0.05 | 352 ± 30 |
| R237K | 3.2 ± 0.05 | 108 ± 7 |
| R237Q | 8.6 ± 1.1 | 1090 ± 48 |
| H264A | 1.7 ± 0.1 | 402 ± 42 |
| H264N | 3.8 ±1.2 | 6770 ± 1430 |
| H264Q | 2.8 ± 0.4 | 228 ± 39 |
Reaction mixtures (25 μL) containing 50 mM Tris-HCl (pH 7.5); 10 mM NiCl2; 0, 0.625, 1.25, 2.5, 5, 10, or 15 mM p-nitrophenyl phosphate; and a fixed amount (between 0.25 and 1 μg) of one of the indicated CthPnkp proteins were incubated for 15 min at 45°C. p-Nitrophenol production was plotted as a function of substrate concentration, and then Km and kcat values were calculated from double-reciprocal plots of the data. The values in the table are averages from two independent substrate titration experiments.
An autonomous phosphatase domain of CthPnkp
We produced recombinant His10-tagged versions of three N-terminal deletion mutants: CthPnkp-(125–870), CthPnkp-(159–870), and CthPnkp-(171–870). The truncated proteins were purified from soluble E. coli extracts by Ni-agarose chromatography and displayed the expected decrements in electrophoretic mobility compared with the full-length protein (Fig. 6A ▶). Whereas the ~55-kDa contaminant was present in all of the preparations, the ~45-kDa contaminant seen in the wild-type enzyme was absent in the truncated protein preparations, being replaced by a set of discrete lower-molecular-weight contaminants with mobility differences that correspond to the differences in their respective N termini (Fig. 6A ▶). This pattern suggests that the ~55-kDa contaminant is a C-terminal proteolytic fragment of CthPnkp, whereas the smaller polypeptides are proteolytic fragments derived from the upstream segment of the recombinant protein. All three N-terminal deletion mutants retained phosphatase activity with either 2′-AMP or 3′-AMP substrates (Fig. 6B ▶) but were inert with respect to 5′ polynucleotide kinase activity (data not shown). Thus, an upstream margin of the phosphatase domain (amino acid 171) is positioned only 17 amino acids upstream of essential residue Asp187 identified in the alanine scan.
FIGURE 6.
N-terminal and C-terminal deletions. Aliquots (5 μg) of the Ni-agarose preparations of the wild-type (WT) CthPnkp and the indicated N-terminal deletion mutants (A) or C-terminal truncation mutants (C) were analyzed by SDS-PAGE. The Coomassie blue-stained gels are shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Reactions mixtures (25 μL) containing 50 mM Tris-HCl (pH 7.5), 10 mM NiCl2, 1 μg CthPnkp as specified, and 5 mM 2′-AMP or 3′-AMP were incubated for 30 min at 45°C. (D) Reactions mixtures (25 μL) containing 50 mM Tris-HCl (pH 7.5), 10 mM NiCl2, 10 mM p-nitrophenyl phosphate, and 10 pmol of enzyme as specified were incubated for 30 min at 45°C.
To test if the C-terminal adenylyltransferase domain of CthPnkp plays a role in the phosphatase reaction, we produced two other series of deletion mutants with N termini fixed at either residue 159 or 171 (based on the results in Fig. 6B ▶) and C termini at residues 472, 444, or 424. The six truncated proteins were purified from soluble E. coli extracts by Ni-agarose chromatography and displayed the expected variations in electrophoretic mobility (Fig. 6C ▶). The pattern of minor polypeptide fragments—comprising a 30-kDa species when the N terminus was at residue 159 and a 28-kDa species when the N terminus was at residue 171—suggested that the proteins underwent proteolytic cleavage at a single discrete site located near the C termini of all of the proteins. The truncated proteins were tested for hydrolysis of p-nitrophenyl phosphate in parallel with wild-type CthPnkp; because the wild-type and truncated proteins differed in size, the input protein in the assay was adjusted to attain approximately equimolar enzyme. The deletion of protein distal to residues 472 or 444 had no apparent impact on activity (Fig. 6D ▶), whereas further truncation of the C terminus to residue 424 (located just 32 amino acids downstream of essential residue Asp392) had only a modest effect (Fig. 6D ▶). As the proteins in this series lacked the adenylyltransferase domain, we surmise that it plays no role in the phosphatase activity. We conclude that the phosphat-ase resides within an autonomous central domain of CthPnkp located between residues 171 and 424. Shorter truncation mutants were not tested.
DISCUSSION
Here we conducted a structure–function analysis of the phosphatase domain of CthPnkp, guided by its amino acid sequence similarity to λ phosphatase and other members of the metallophosphoesterase superfamily. The alanine scan pinpointed 11 residues required for hydrolysis of 2′-AMP and 3′-AMP that are conserved in several bacterial homologs of CthPnkp and in λ phosphatase. Eight of these side chains are also essential for hydrolysis of the non-nucleotide substrate p-nitrophenyl phosphate. Structure–activity relationships were clarified by conservative substitutions. Our results can be interpreted in light of structural information available for λ phosphatase (Voegtli et al. 2000), which shares with CthPnkp a requirement for either Ni or manganese, and for Pyrococcus furiosus Mre11 (Hopfner et al. 2001), a manganese-dependent DNA phosphodiesterase. The findings provide fresh insights to the metallophosphoesterase catalytic mechanism.
The active sites of λ phosphatase (Fig. 2 ▶) and PfuMre11 (Fig. 7 ▶) contain two manganese ions coordinated directly or via water by aspartate, histidine, and asparagine side chains. The metal ligands in λ phosphatase are strictly conserved in CthPnkp. In PfuMre11, the water-to-Asp constituent of the octahedral complex of one of the manganese ions is replaced by a direct contact to a histidine side chain (Fig. 7 ▶). The λ phosphatase structure contains a sulfate ion in a position that is proposed to mimic the product complex of the enzyme with phosphate. The PfuMre11 structure contains 5′-dAMP and can be viewed as a substrate complex for reasons discussed below. The mechanism of the phosphatase reaction is somewhat controversial, insofar as there are divergent models in the literature concerning the identity of the nucleophilic water, the expulsion of the leaving group, and the nature of the transition state (Rusnak and Mertz 2000; Voegtli et al. 2000; Hopfner et al. 2001). We favor the view that the bridging water between the two manganese ions seen in the PfuMre11 structure is the attacking nucleophile, because it is located 3.1 Å from the phosphorus atom and is almost perfectly apical to the phosphate oxygen atom of the leaving group, which is a nucleoside 3′-O in the case of Mre11 (Fig. 7 ▶). As discussed by Voegtli et al. (2000), the bridging water ligand between the manganese ions should have a low pKa and thereby be activated by the metals for attack on the phosphate. Comparison of the PfuMre11 substrate complex in Figure 7 ▶ with the λ phosphatase product complex in Figure 2 ▶ suggests that the attacking hydroxyl nucleophile becomes incorporated into the phosphate product and continues to occupy the bridging position between the two metal ions; moreover, there is an apparent stereochemical inversion of the phosphorus center, as expected for the in-line single displacement mechanism proposed for the metallophosphoesterases (Mueller et al. 1993). In addition to activating the nucleophile, the metals directly coordinate two nonbridging phosphate oxygens.
FIGURE 7.
The active site of PfuMre11. The figure depicts the active site of the manganese- and 5′-dAMP-bound P. furiosus phosphodiesterase Mre11 from the crystal structure of Hopfner et al. (2001) (Protein Data Bank 1II7). The amino acid side chains coordinating the binuclear metal cluster and the nucleotide 5′ phosphate are shown. The corresponding amino acids of CthPnkp are indicated in parentheses. The manganese ions are colored cyan. Water is colored red. The structure is predicted to mimic the substrate complex of the enzyme. The proposed nucleophilic water and the phosphate oxygen corresponding to the 3′-O leaving group are indicated. For simplicity, only the phosphate and the ribose O5 and C5 atoms of the nucleotide are shown.
We show here that every one of the seven side chains that comprise the metal-binding site of λ phosphatase is strictly essential for CthPnkp phosphatase activity with all substrates tested; not even conservative substitutions are tolerated at Asp187, His189, Asp233, Asn263, His323, His376, and Asp392. Mutational studies of λ phosphatase had established essential catalytic roles for five metal-binding residues: Asp20, His22, Asp49, Asn75, and His186 (Zhuo et al. 1994; White et al. 2001), which are equivalent to Asp187, His189, Asp233, Asn263, and His376 of CthPnkp. Of particular interest is the essential Asn263 side chain of CthPnkp, which is consistently seen to make contacts to both manganese (via Oδ) and one of the phosphate oxygens (via Nδ) (Figs. 2 ▶, 7 ▶). Our results indicate that both atomic contacts are required for activity.
We obtained additional functional insights to the reaction mechanism via the substrate-specific effects of mutations at Asp236, His264, and Arg237 of CthPnkp, whereby these residues are required for activity with 2′-AMP and 3′-AMP but not with p-nitrophenyl phosphate. His264 coordinates two phosphate oxygens in the λ phosphatase and PfuMre11 crystal structures; in the PfuMre11 substrate complex, it donates a hydrogen bond from Nɛ to the leaving group oxygen (Fig. 7 ▶). It has been suggested that this histidine could serve as a general acid catalyst, by donating a proton to the leaving group (Rusnak and Mertz 2000; Hopfner et al. 2001). The preservation of function of the H264A mutant with p-nitrophenyl phosphate compared with the nucleotide substrates can be rationalized by the fact that the pKa of p-nitrophenol (7.2) is ~7 pH units lower than that of the 2′-OH and 3′-OH leaving groups of 2′-AMP and 3′-AMP. Previous studies from this laboratory have shown that severe defects incurred by alanine mutations of a general acid catalyst of phosphoryl transfer reactions of DNA topoisomerase IB can be alleviated by simple chemical substitutions in the substrate that lower the pKa of the leaving group (Krogh and Shuman 2000, 2002). Whereas the substrate-specific effects of the CthPnkp H264A mutation are consistent with a mechanism of general acid catalysis, the findings that the H264N change restores activity with 2′-AMP and 3′-AMP substrates and significantly stimulates activity with p-nitrophenyl phosphate (Table 1 ▶) complicate the story. As mentioned above, the fact that asparagine but not glutamine is beneficial is inconsistent with a model whereby the gain of function is imparted by restoring a direct hydrogen bond between the amide group and the leaving phosphate oxygen. Rather, we propose that asparagine-264 is able to coordinate a water molecule, which in turn donates a proton to the leaving group. This scenario preserves the requirement for proton donation to expel the leaving group when its pKa is high but allows for use of either a protein side chain as a general acid or an ordered water molecule as a specific acid.
The plasticity of the CthPnkp active site in accommodating asparagine instead of His264 contrasts with the intolerance of λ phosphatase for asparagine in lieu of the corresponding His76 side chain (Zhuo et al. 1994; Mertz et al. 1997). The finding that the H76N mutant of λ phosphatase exerted quantitatively similar effects on activity with p-nitrophenyl phosphate (pKa 7.2) and phenyl phosphate (pKa 10) can be interpreted as signifying that the histidine is not required for protonation of the leaving group (Mertz et al. 1997), although it remains possible that the histidine does play a critical role as a general acid in reactions with phosphoprotein substrates that have leaving groups with much higher pKa values (Rusnak and Mertz 2000). Members of the metallophosphoesterase family may differ in the dependence of their catalytic parameters on the pKa of the leaving group (Schenk et al. 2005).
Asp236 of CthPnkp is equivalent to Asp52 of λ phosphatase, which contacts Nδ of the proposed histidine general acid. The Asp–His pair could comprise a potential proton relay to the leaving group. The fact that the D236A mutant retains activity with p-nitrophenyl phosphate is consistent with a role in leaving group expulsion. Our mutational analysis of CthPnkp shows that an asparagine suffices for activity with all substrates; i.e., hydrogen-bonding, rather than full proton transfer between the Asp and His, is the relevant factor for catalysis.
Arg237 of CthPnkp corresponds to Arg53 in λ phosphatase, which makes multiple contacts with the sulfate (phosphate) oxygens in the crystal structure, suggestive of a role in transition state stabilization (Voegtli et al. 2000). Consistent with this idea, mutating Arg53 to alanine in λ phosphatase results in a 10−3 decrement in Mn2+-dependent hydrolysis of p-nitrophenyl phosphate with no effect on the Km for p-nitrophenyl phosphate (Zhuo et al. 1994). The R237A mutation of CthPnkp affects activity with 2′-AMP and 3′-AMP but has relatively little impact on either Km or kcat for hydrolysis of p-nitrophenyl phosphate, a finding that contrasts sharply with the result for λ phosphatase. It seems unlikely that the lack of requirement for Arg237 in hydrolysis of p-nitrophenyl phosphate is related to the pKa of the leaving group, insofar as arginine is a less plausible candidate than is histidine as a proton donor, although we cannot completely exclude this possibility at present. The fact that there are no contacts between arginine or lysine residues and the phosphate of 5′-dAMP in the crystal structure of PfuMre11 (Hopfner et al. 2001) attests to the variability of the requirements for phosphate contacts above and beyond those provided by the two metals and the catalytic asparagine (Asn263 in CthPnkp). Indeed, the amino acid sequence of PfuMre11 lacks an arginine at the position corresponding to Arg237 and Arg53 of CthPnkp and λ phosphatase, respectively (Hopfner et al. 2001). If the essential role of the arginine is exerted via transition state stabilization, then the differential requirement for this residue might reflect differences in the nature of the transition state. For example, in an associative mechanism, the phosphorane transition state develops an additional negative charge compared with the ground state, in which case the arginine could help neutralize this extra charge. In a dissociative mechanism, the metaphosphate transition state has a net loss of one negative charge compared with the ground state, in which case electrostatic effects of the arginine contacts might not be as critical for catalysis. With this line of reasoning, we speculate that CthPnkp hydrolysis of p-nitrophenyl phosphate (where leaving group departure is potentially easier) might have relatively more dissociative character, while hydrolysis of 2′-AMP and 3′-AMP might have a more associative character. Kinetic studies have suggested that λ phosphatase catalyzes manganese-dependent hydrolysis of p-nitrophenyl phosphate via a dissociative transition state (Hoff et al. 1999). The fact that the R237Q change in CthPnkp restores activity, relative to the alanine mutant, with 3′-AMP (and p-nitrophenyl phosphate) (Table 1 ▶) but not with 2′-AMP hints that there may be additional mechanistic differences in the way the enzyme reacts with various nucleotide substrates.
A complete description of the mechanism of the CthPnkp phosphatase will hinge on obtaining atomic structures of the enzyme at various steps along the reaction pathway. The delineation here of an autonomous phosphatase domain fragment of the trifunctional CthPnkp protein will facilitate efforts toward structure determination via crystallography.
MATERIALS AND METHODS
Materials
2′-AMP, 3′-AMP, p-nitrophenyl phosphate, and p-nitrophenol were purchased from Sigma. Malachite green reagent was purchased from BIOMOL Research Laboratories.
Recombinant CthPnkp
Alanine substitution mutations were introduced into the pET16-CthPnkp expression plasmid by PCR using the two-stage overlap extension method (Martins and Shuman 2005). N-terminal truncation alleles CthPnkp-(125–870), CthPnkp-(159–870), and CthPnkp-(171–870) were generated by PCR amplification using sense primers that introduced an NdeI site and a methionine codon in lieu of the codons for Gln124, Pro158, or Pro170. The C-terminal truncation alleles CthPnkp-(159–472), CthPnkp-(159–444), CthPnkp-(159–424), CthPnkp-(171–472), CthPnkp-(171–444), and CthPnkp-(171–424) were constructed by PCR amplification using an anti-sense primer that introduced a stop codon in lieu of the codons for Ile473, Ile445, or Lys425. All plasmid inserts were sequenced to confirm the presence of the intended mutations and verify that no other coding changes had been introduced during amplification and cloning. Wild-type and mutant pET16-CthPnkp plasmids were transformed into E. coli BL21(DE3). Cultures (500 mL) were induced to express CthPnkp as described previously (Martins and Shuman 2005); the recombinant His10-tagged CthPnkp proteins were purified from soluble bacterial extracts by Ni-agarose affinity chromatography (Martins and Shuman 2005).
Nucleotide 2′, 3′ phosphatase assay
Phosphate release was measured colorimetrically by using the malachite green method (Lanzetta et al. 1979). Reaction mixtures (25 μL) containing 50 mM Tris-HCl (pH 7.5), 5 mM 2′-AMP or 3′-AMP, 5 mM NiCl2, and CthPnkp as specified were incubated for 30 min at 45°C. The reactions were quenched by adding 1 mL of malachite green reagent. Release of phosphate was determined by measuring A620 and interpolating the value to a phosphate standard curve.
Hydrolysis of p-nitrophenyl phosphate
Reaction mixtures (25 μL) containing 50 mM Tris-HCl (pH 7.5), 10 mM p-nitrophenyl phosphate, 10 mM NiCl2, and CthPnkp as specified were incubated for 30 min at 45°C. The reactions were quenched by adding 0.9 ml of 1 M Na2CO3. Release of p-nitro-phenol was determined by measuring A410 and interpolating the value to a p-nitrophenol standard curve.
Acknowledgments
This work was supported by NIH grant GM42498. S.S. is an American Cancer Society Research Professor.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2196406.
REFERENCES
- Amitsur, M., Levitz, R., and Kaufman, G. 1987. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 6: 2499–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, N.K., Williams, R.S., Rakovszky, M.L., Cui, D., Green, R., Karimi-Busheri, F., Mani, R.S., Galicia, S., Koch, C.A., Cass, C.E., et al. 2005. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell 17: 657–670. [DOI] [PubMed] [Google Scholar]
- Blondal, T., Hjorleifsdottir, S., Aevarsson, A., Fridjonsson, O.H., Skirnisdottir, S., Wheat, J.O., Hermannsdottir, A.G., Hreggvidsson, Bacterial polynucleotide kinase-phosphatase G.O., Smith, A.V., and Kristjansson, J.K. 2005. Characterization of a 5′-polynucleotide kinase/3′-phosphatase from bacteriophage RM378. J. Biol. Chem. 280: 5188–5194. [DOI] [PubMed] [Google Scholar]
- Cameron, V. and Uhlenbeck, O.C. 1977. 3′-Phosphatase activity in T4 polynucleotide kinase. Biochemistry 16: 5120–5126. [DOI] [PubMed] [Google Scholar]
- Chen, S., Yakunin, A.F., Kuznetsova, E., Busso, D., Pufan, R., Proudfoot, M., Kim, R., and Kim, S.H. 2004. Structural and functional characterization of a novel phosphodiesterase from Methanococcus janaschii. J. Biol. Chem. 279: 31854–31862. [DOI] [PubMed] [Google Scholar]
- Galburt, E.A., Pelletier, J., Wilson, G., and Stoddard, B.L. 2002. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure 10: 1249–1260. [DOI] [PubMed] [Google Scholar]
- Goldberg, J., Huang, H., Kwon, Y., Greengard, P., Nairn, A.C., and Kuriyan, J. 1995. Three-dimensional structure of the catalytic sub-unit of protein serine/threonine phosphatase-1. Nature 376: 745–753. [DOI] [PubMed] [Google Scholar]
- Hoff, R.H., Mertz, P., Rusnak, F., and Hengge, A.C. 1999. The transition state of the phosphoryl transfer reaction catalyzed by the λ Ser/Thr protein phosphatase. J. Am. Chem. Soc. 121: 6382–6390. [Google Scholar]
- Hopfner, K.P., Karcher, A., Craig, L., Woo, T.T., Carney, J.P., and Tainer, J.A. 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105: 473–485. [DOI] [PubMed] [Google Scholar]
- Krogh, B.O. and Shuman, S. 2000. Catalytic mechanism of DNA topoisomerase IB. Mol. Cell 5: 1035–1041. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Proton relay mechanism of general acid catalysis by DNA topoisomerase IB. J. Biol. Chem. 277: 5711–5714. [DOI] [PubMed] [Google Scholar]
- Lanzetta, P.A., Alvarez, L.J., Reinach, P.S., and Candia, O.A. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100: 95–97. [DOI] [PubMed] [Google Scholar]
- Martins, A. and Shuman, S. 2004. Characterization of a baculovirus enzyme with RNA ligase, polynucleotide 5′ kinase and polynucleotide 3′ phosphatase activities J. Biol. Chem. 279: 18220–18231. [DOI] [PubMed] [Google Scholar]
- ———. 2005. An end-healing enzyme from Clostridium thermocellum with 5′ kinase, 2′,3′ phosphatase, and adenylyltransferase activities. RNA 11: 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz, P., Yu, L., Sikkink, R., and Rusnak, F. 1997. Kinetic and spectroscopic analysis of mutants of a conserved histidine in the metallophoshatases calcineurin and λ protein phosphatase. J. Biol. Chem. 272: 21296–21302. [DOI] [PubMed] [Google Scholar]
- Mueller, E.G., Crowder, M.W., Averill, B.A., and Knowles, J.R. 1993. Purple acid phosphatase: a diiron enzyme that catalyzes a direct phospho group transfer to water. J. Am. Chem. Soc. 115: 2974–2975. [Google Scholar]
- Novogrodsky, A. and Hurwitz, J. 1966. The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid: Phosphorylation at 5′-hydroxyl termini. J. Biol. Chem. 241: 2923–2932. [PubMed] [Google Scholar]
- Novogrodsky, A., Tal., M., Traub, A., and Hurwitz, J. 1966. The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid: Further properties of the 5′-hydroxyl polynucleotide kinase. J. Biol. Chem. 241: 2933–2943. [PubMed] [Google Scholar]
- Richardson, C.C. 1965. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc. Natl. Acad. Sci. 54: 158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnak, F. and Mertz, P. 2000. Calcineurin: Form and function. Physiol. Rev. 80: 1483–1521. [DOI] [PubMed] [Google Scholar]
- Schenk, G., Gahan, L.R., Carrington, L.E., Mitic, N., Valizadeh, M., Hamilton, S.E., de Jersey, J., and Guddat, L.W. 2005. Phosphate forms an unusual tripodal complex with the Fe-Mn center of sweet potato purple acid phosphatase. Proc. Natl. Acad. Sci. 102: 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegtli, W.C., White, D.J., Reiter, N.J., Rusnak, F., and Rosenzweig, A.C. 2000. Structure of the bacteriophage λ Ser/Thr protein phosphatase with sulfate ion bound in two coordination modes. Biochemistry 39: 15365–15374. [DOI] [PubMed] [Google Scholar]
- Wang, L.K. and Shuman, S. 2001. Domain structure and mutational analysis of T4 polynucleotide kinase. J. Biol. Chem. 276: 26868–26874. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Mutational analysis defines the 5′-kinase and 3′-phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids. Res. 30: 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.K., Lima, C.D., and Shuman, S. 2002. Structure and mechanism of T4 polynucleotide kinase: An RNA repair enzyme. EMBO J. 21: 3873–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, D.J., Reiter, N.J., Sikkink, R.A., Yu, L., and Rusnak, F. 2001. Identification of the high affinity Mn2+ binding site of bacteriophage λ phosphoprotein phosphatase: Effects of metal ligand mutations on electron paramagnetic resonance spectra and phosphatase activities. Biochemistry 40: 8918–8929. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Yin, S., and Shuman, S. 2004. Characterization of polynucleotide kinase/phosphatase enzymes from mycobacteriophages Omega and Cjw1 and vibriophage KVP40. J. Biol. Chem. 279: 26358–26369. [DOI] [PubMed] [Google Scholar]
- Zhuo, S., Clemens, J.C., Hakes, D.J., Barford, D., and Dixon, J.E. 1993. Expression, purification, crystallization, and biochemical characterization of a recombinant protein phosphatase. J. Biol. Chem. 268: 17754–17761. [PubMed] [Google Scholar]
- Zhuo, S., Clemens, J.C., Stone, R.L., and Dixon, J.E. 1994. Mutational analysis of a Ser/Thr phosphatase: Identification of residues important in phosphoesterase substrate binding and catalysis. J. Biol. Chem. 269: 26234–26238. [PubMed] [Google Scholar]