Abstract
Retroviruses and retrotransposons assemble intracellular immature core particles around a RNA genome, and nascent particles collect in association with membranes or as intracellular clusters. How and where genomic RNA are identified for retrovirus and retrotransposon assembly, and how translation and assembly processes are coordinated is poorly understood. To understand this process, the subcellular localization of Ty3 RNA and capsid proteins and virus-like particles was investigated. We demonstrate that mRNAs, proteins, and virus-like particles of the yeast Ty3 retrotransposon accumulate in association with cytoplasmic P-bodies, which are sites of mRNA translation repression, storage, and degradation. Deletions of genes encoding P-body proteins decreased Ty3 transposition and caused changes in the pattern of Ty3 foci, underscoring the biological significance of the association of Ty3 virus-like protein components and P-bodies. These results suggest the hypothesis that P-bodies may serve to segregate translation and assembly functions of the Ty3 genomic RNA to promote assembly of virus-like particles. Because Ty3 has features of a simple retrovirus and P-body functions are conserved between yeast and metazoan organisms, these findings may provide insights into host factors that facilitate retrovirus assembly.
Keywords: Ty3, virus assembly, retrotransposon, retrovirus, mRNA localization, P-body
INTRODUCTION
Retroviruses and retrotransposons assemble intracellular immature core particles around a RNA genome, and nascent particles collect in association with membranes or as intracellular clusters (Garfinkel et al. 1985; Hansen et al. 1992; Coffin et al. 1997; Linial and Eastman 2003; Sfakianos et al. 2003; Teysset et al. 2003). An important aspect of the retrovirus life cycle is how the RNA genome and particle proteins are brought together during assembly to allow orderly progression of the life cycle. For some viruses, there is evidence that packaged and translated RNAs are segregated from early times; for others, it appears that genomic RNAs are translated and subsequently packaged (Butsch and Boris-Lawrie 2002). Recent experiments with Moloney murine leukemia virus and Mason-Pfizer monkey virus indicate that nascent viral proteins can facilitate delivery of the RNA to an assembly site (Basyuk et al. 2003; Sfakianos et al. 2003).
To gain insight into the retrovirus-like assembly process, we examined localization of the protein and RNA components of the yeast retrovirus-like element Ty3 (Sandmeyer et al. 2002). Ty3 is 5.4 kbp in length and is comprised of long terminal repeats (LTRs) of 340 bp flanking an internal domain of two overlapping ORFs, GAG3 and POL3. GAG3 encodes the major structural proteins, capsid (CA) and nucleocapsid (NC); POL3 encodes the aspartyl protease (PR), reverse transcriptase (RT), and integrase (IN) of Ty3. Proteins are synthesized initially as Gag3 and Gag3-Pol3 precursor polyproteins, which associate through Gag3 interactions into virus-like particles (VLPs) of ~50 nm (Hansen et al. 1992). The zinc finger of Ty3 NC is critical for proper assembly; by analogy with retroviruses, it is required for packaging of the genomic RNA (Orlinsky and Sandmeyer 1994). Subsequent to assembly, the polyprotein precursors are processed by PR, resulting in mature VLPs, which are analogous to retrovirus cores. Reverse transcription of the RNA yields cDNA, which is translocated into the nucleus and integrated into chromosomal DNA. Thus, although Ty3 lacks an envelope and extracellular phase of the cell cycle, it is similar in structure and function to an animal retrovirus.
RESULTS AND DISCUSSION
To localize Ty3 proteins in living cells, we fused sequence encoding green fluorescent protein (GFP) or red fluorescent protein (RFP) to the downstream end of the POL3 gene in a Ty3 element under transcriptional control of the GAL1-10 upstream activating sequence (UAS) (Fig. 1A ▶), which is induced by growth in galactose. Western analysis showed that Ty3-GFP/RFP was expressed and that both Gag3 and Gag3-Pol3-GFP/RFP proteins were properly processed (Fig. 1B ▶). Because proper processing requires assembly into the VLP, these results suggest the fusion proteins are fully functional and able to assemble within a VLP, a conclusion also supported by localization of these fusion proteins to VLP by electron microscopy (EM) (see below).
FIGURE 1.
Ty3-GFP, similar to wild-type Ty3, produces clusters of VLPs. (A) Ty3-GFP/RFP in pNB2127/pNB2266. The GFP/RFP coding region was inserted at an XmaI site so that GFP/RFP was expressed fused to the POL3 reading frame at the downstream end. (B) Cells expressing Ty3-GFP/RFP show correct proteolytic maturation of Gag3 into p27 and CA and production of IN-GFP/RFP from Gag3-Pol3-GFP/RFP. Cultures of BY4741 transformed with pDLC201, pNB2127, or pNB2266 were induced for Ty3 or Ty3-GFP/RFP expression for 6 h. Cell extracts were prepared and fractionated by electrophoresis on SDS–10% polyacrylamide gels. Proteins were transferred to Immobilon and analyzed by reacting with anti-GFP, anti-IN, and anti-CA IgG. Uninduced cell extracts are indicated by “U.” Positions of IN-GFP/RFP (88 kDa), IN (61 kDa), Gag3 (34 kDa), and CA (24 kDa) are indicated. (C) BY4741 cells transformed with pNB2127 and induced to express Ty3-GFP for different periods of time. Ty3-GFP is indicated by green; DAPI-stained DNA, pseudo-colored red. Scale bar, 5.0 μm. (D) BY4741 cells transformed with pNB2127 and induced for 6 h to express Ty3-GFP observed by EM. Scale bar, 0.5 μm; n indicates nucleus; v, vacuole. (E) Cells induced for Ty3 expression as described in D, except processed by reacting sequentially with rabbit polyclonal anti-GFP and goat anti-rabbit IgG conjugated to 15-nm gold particles. Scale bar, 0.24 μm.
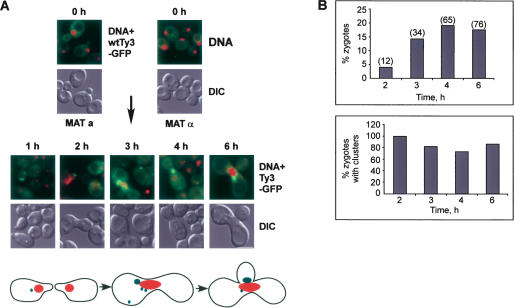
Examination of cells expressing the Ty3-GPF fusion protein revealed that the Ty3-GFP protein was distributed in a punctate pattern after 1 h of induction and in fewer, but larger, foci at later times (Fig. 1C ▶). Moreover, by indirect immunofluorescence, the CA and NC proteins colocalized with the Ty3-GFP, indicating that the accumulation of Ty3-GFP in foci was not restricted just to the fusion protein (data not shown). In addition, the pattern of Ty3-GFP fluorescence was similar in mating populations of cells in which Ty3 was expressed under its native pheromone-inducible promoter from a low-copy plasmid (Fig. 2 ▶). This latter result demonstrates that the accumulation of the Ty3-GFP protein in foci is not an artifact of expression of the Ty3 element from the GAL1-10 UAS. These results indicate that Ty3 proteins accumulate in specific cytoplasmic foci during the Ty3 life cycle, which could be sites of VLP assembly.
FIGURE 2.
Ty3 proteins occur in foci when Ty3 is expressed under its native promoter in mating cells. Sequences containing the coding region of Ty3-GFP were cloned under the native pheromone-inducible Ty3 LTR in the low-copy vector pRS316 (pNB2176). Ty3 BY4741 (MATa) cells transformed with pNB2176 and BY4742 (MATα) cells transformed with the high-copy vector pYES2.0 were mixed on solid SD-ura and allowed to undergo mating to induce Ty3-GFP expression. (A) Ty3-GFP clusters form in mating cells. Formation of zygotes at 0, 1, 2, 3, 4, and 6 h after mixing. (Top, upper panels) Nuclei and Ty3 clusters (DAPI, red; Ty3-GFP, green). (Top, lower panels) Differential interference contrast (DIC) micrographs of whole cells. Scale bar, 5.0 μm. (Bottom) Cartoon of mating cells. Cells form shmoo projections that fuse to give a zygote dumbbell which then buds. (B) Mating populations were monitored for percentage of cells in zygotes and percentage of zygotes with clusters.
If the Ty3 protein foci are sites of virion assembly, the Ty3 mRNA should also be present. To determine if Ty3 mRNA is concentrated in Ty3 protein foci, we inserted two copies of the binding sites for MS2 bacteriophage coat protein just downstream of the Ty3 POL3 ORF (Ty3-MS2) and coexpressed a MS2-GFP fusion protein. The interaction of the MS2-GFP protein with the MS2 sites on the RNA allows the tagged RNA to be specifically localized in cells (Bertrand et al. 1998). Cells expressing MS2-GFP and Ty3-MS2 showed multiple fluorescent foci, first faintly visible at 1 h after induction, that continued to enlarge with a concomitant reduction in number up to 6 h (Fig. 3A ▶, upper panel; 3B ▶). In contrast, in cells expressing only MS2-GFP, fluorescence was diffuse throughout this time course (Fig. 3A ▶, lower panel). Moreover, Ty3-MS2 and MS2-GFP fluorescence analyzed together with indirect immunofluorescence using antibodies specific for Ty3 CA showed that the Ty3 mRNA and CA colocalized (Fig. 3C ▶). Thus, Ty3 proteins and RNA are localized together to specific cytoplasmic foci.
FIGURE 3.
Ty3 mRNA occurs in foci with Ty3 protein. (A) Ty3-MS2 expression causes MS2-GFP to form foci. Yeast strain yTM443 was transformed with pMS2-GFP and pTy3-MS2 and was grown to logarithmic phase in SR-ura, -his. Ty3-MS2 was induced by addition of galactose (upper panel) or left uninduced (lower panel). Cells were transferred to -ura, -his, -met medium 30 min prior to sampling to induce the MS2-GFP MET25 promoter. As observed previously for expression from this promoter, some cells did not efficiently express MS2-GFP (Beach et al. 1999). Scale bar, 5.0 μm. (Right) Cartoon shows MS2-GFP binding to MS2 sites in Ty3-MS2 RNA. (B) Cells from cultures imaged in A (upper panel), were evaluated for number of Ty3-MS2 foci. Cells with background fluorescence or with diffuse fluorescence are combined in “no Ty3 clusters.” Over 300 cells were classified per time point. These trends were representative of other experiments. (C) Ty3-MS2 foci coincide with Ty3 CA clusters. Cells expressing MS2-GFP and Ty3-MS2 were induced as described above for 6 h. Ty3 CA was visualized by immunofluorescence using rabbit polyclonal anti-CA and goat anti-rabbit IgG conjugated to TRITC (red), and Ty3-MS2 mRNA was visualized by direct fluorescence (GFP, green). Cells were stained with DAPI to visualize DNA (blue). Cells in C were imaged by confocal fluorescence microscopy. Scale bar, 5.0 μm.
The presence of both Ty3 mRNA and proteins in the cytoplasmic foci and their continued accumulation over time were consistent with the idea that these were sites of VLP assembly. To determine if these foci contained VLPs, we examined their structure by using EM. Inspection of cells expressing Ty3-GFP showed accumulation of clusters of VLPs similar to what was previously reported for wild-type Ty3 (Hansen et al. 1992; Fig. 1D ▶), and immunoEM using anti-GFP showed that GFP was associated with VLPs (Fig. 1E ▶). In cells expressing Ty3 for 1 or 1.5 h, individual VLPs or clusters were rarely observed by EM (data not shown). After 3 h, however, consistent with observations using Ty3-GFP, small clusters were observed and larger clusters were apparent at 6 h. In combination, these results indicate that Ty3 proteins, mRNA, and VLPs all accumulate in cytoplasmic foci, which might be specific sites of VLP assembly.
Several observations led us to hypothesize that Ty3 foci might be associated with cytoplasmic P-bodies. First, P-bodies are sites of accumulation of nontranslating mRNAs, which can then be subject to degradation, recycling to translation, or storage for later reuse (Sheth and Parker 2003; Cougot et al. 2004; Brengues et al. 2005; Coller and Parker 2005). Thus, if VLP assembly initiated or occurred within P-bodies, competition between assembly and translation would be avoided. Moreover, P-body components were identified in genomic screens for mutations affecting retrotransposition of both Ty3 (dhh1Δ and xrn1/kem1Δ) (Irwin et al. 2005) and Ty1 (pat1Δ and lsm1Δ) (Griffith et al. 2003). In isolated testing, lsm1Δ and pat1Δ strains also show reduced Ty3 retrotransposition (data not shown).
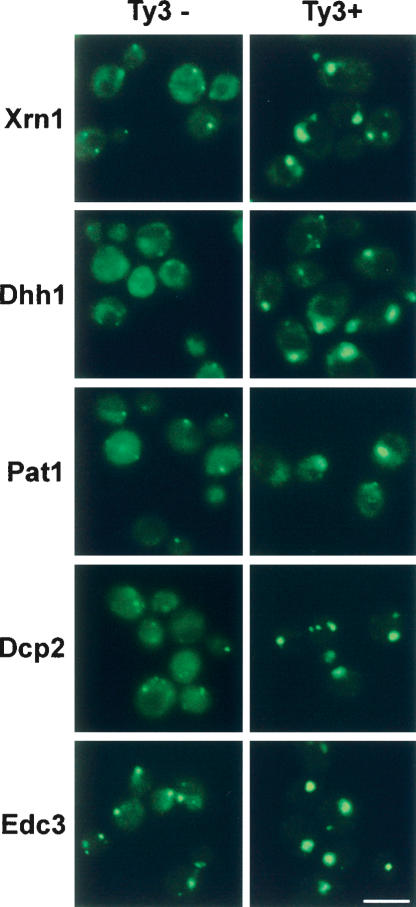
To test whether Ty3 VLP clusters were physically associated with P-bodies, we expressed the Ty3-RFP fusion protein in strains in which the individual P-body proteins Xrn1/Kem1, Dhh1, Pat1, Dcp2, and Edc3 were fused to GFP (Huh et al. 2003). Strikingly, we observed that in each case, Ty3-RFP colocalized with or near the P-body protein (Fig. 4 ▶). Careful inspection of images suggested that the individual RFP and GFP foci were overlapping but not perfectly congruent. Deconvolution of Z stack images for Dhh1-GFP and Xrn1/Kem1-GFP supported this interpretation (data not shown). Furthermore, immunoEM using anti-GFP in strains expressing Ty3 and Dhh1-GFP or Xrn1/Kem1-GFP showed colloidal gold labeling of GFP overlapping or adjacent to the assemblies of VLPs (Fig. 4B ▶; data not shown). Inspection of the GFP fluorescence showed that individual P-body proteins appear in differing numbers of foci (Fig. 4A ▶, cf. Dhh1p and Dcp2p). Moreover, for Dcp2p, which occurred in multiple foci, Ty3-RFP colocalized with only a subset of foci (Fig. 4A ▶). These results indicate that Ty3 proteins, RNAs, and VLPs accumulate in association with P-bodies, although it may be that only a subset of P-bodies is associated with Ty3 assembly.
FIGURE 4.
Ty3 VLPs associate with P-body proteins. (A) Ty3-RFP colocalizes with P-body proteins fused to GFP. Yeast strain BY4741 (wild type) and strains derived from it by fusion of the GFP coding region in frame downstream of genes encoding P-body proteins (labeled Xrn1, etc.) and for the gene encoding Nvj1, a nuclear-vacuolar junction protein, were transformed with the Ty3-RFP or RFP expression plasmids and grown in the presence of galactose for 4 h. No Ty3 refers to the strain containing RFP expressed from the galactose-inducible promoter in pYES2.0. RFP alone expressed at high levels does not form foci, and Nvj1-GFP does not colocalize with Ty3-RFP. Unmarked cells (wild type) do not show bleed-through of Ty3-RFP into the green channel. Scale bar, 5.0 μm. (B) Dhh1-GFP and Xrn1/Kem1-GFP are associated with Ty3 VLP clusters. Cells induced for expression of Ty3 for 4 h were processed for EM and reacted with anti-GFP and secondary antibodies conjugated to colloidal gold. Solid arrows indicate colloidal gold particles. Scale bar, 0.1 μm.
Additional evidence that Ty3 mRNAs interact with components of P-bodies comes from the observation that expression of the Ty3 RNA in cells growing in mid-to-late log leads to the hyperaccumulation of the P-body components in P-bodies compared with cells without Ty3 mRNA being expressed (Fig. 5 ▶), where P-body components are generally more distributed throughout the cytoplasm (Teixeira et al. 2005).
FIGURE 5.
Expression of Ty3 leads to hyper-accumulation of P-body components. Cells expressing P-body components fused to GFP (e.g., Xrn1-GFP) were transformed with pNB2289 (pYES2.0-RFP) (Ty3−) or with Ty3 plasmid pNB2266 (Ty3+). Cells were induced for Ty3 expression by growth in galactose and visualized as described in Figure 4 ▶ legend.
Evidence that the association of Ty3 components with P-bodies is functionally important is that lesions in components of P-bodies affect retrotransposition and alter the morphology of Ty3 foci. Specifically, Dhh1, a key protein for translation repression and targeting mRNAs to P-bodies, and Xrn1/Kem1, the 5′ to 3′ exonuclease found in P-bodies, are required for Ty3 retrotransposition (Irwin et al. 2005). Moreover, dhh1Δ strains showed more diffuse fluorescence of Ty3 proteins, and twice as many cells displayed multiple, small foci compared with wild-type cells (Fig. 6A,B ▶; data not shown). Xrn1/kem1Δ cells contain increased P-bodies (Sheth and Parker 2003), and the Ty3 protein in this mutant accumulates in association with large P-bodies (Fig. 6A ▶; data not shown).
FIGURE 6.
Ty3 VLP localization and morphogenesis is affected in P-body mutants. (A) Ty3-GFP clusters are disrupted in cells with deletions of genes encoding P-body proteins. BY4741 wild-type and dhh1Δ and xrn1/kem1Δ derivatives were transformed with pNB2127, grown to logarithmic phase in SR-ura, and induced to express Ty3-GFP for 4 h in the presence of galactose. Scale bar, 5.0 μm. (B) DhhΔ cells expressing Ty3-GFP from the culture shown in A were classified as described in Figure 2 ▶ legend. Over 250 mutants and >500 wild types were classified. This pattern is representative of that observed in other experiments.
Several observations now indicate that an important aspect of the Ty3 life cycle involves the association of Ty3 components with P-bodies. First, we observe that Ty3 proteins, mRNA, and VLPs all accumulate in association with P-bodies. Second, the expression of the Ty3 mRNA leads to increased P-body size. Third, dhh1Δ and xrn1/kem1Δ strains, which have defects in P-body function, show alterations in retrotrans-position (Irwin et al. 2005) and the morphology of Ty3 foci (Fig. 6 ▶). The simplest explanation for these observations is that Ty3 VLP assembly occurs in association with P-bodies. We cannot rigorously rule out the formal possibility that Ty3 VLPs assemble elsewhere in the cell and only subsequently aggregate in association with P-bodies. However, the absence of dispersed VLPs by EM and the nascent accumulation of Ty3-GFP in foci at 1 h, which is before we observe significant numbers of VLPs, suggests this is an unlikely possibility.
Interestingly, because translation is restricted from yeast P-bodies (Teixeira et al. 2005), these observations suggest that translation of the Ty3 proteins could occur in a diffuse manner followed by accumulation of the mRNA and Ty3 proteins in association with P-bodies. Assembly of VLPs in P-bodies might be advantageous because it would increase the local concentration of Ty3 RNA for packaging. Sequestering a nontranslating pool of mRNAs would also serve to restrict the disruption of packaging by translating ribosomes. Nonetheless, the specific molecular mechanistic linkage between the single wild-type P-body/Ty3 VLP cluster in most cells and the efficiency of transposition remains to be determined. Indeed, it is even possible that association of Ty3 VLPs with P-bodies affects steps in the life cycle after assembly.
Retroviruses assemble at characteristic cytoplasmic or membranous sites (Coffin et al. 1997). For example, in the case of Mason-Pfizer monkey virus, a β retrovirus, which assembles in pericentriolar clusters, specific chaperones have been implicated in the protein assembly process (Hong et al. 2001; Sfakianos et al. 2003) and in the case of human immunodeficiency virus, RNase L inhibitor is associated with protein assembly intermediates (Zimmerman et al. 2002). Nonetheless, relatively little is known about which, if any, host factors might play a specific role in directing genomic RNA into the assembly rather than translation pathway. Because P-bodies in mammalian cells are similar in composition and function to yeast P-bodies (Bashkirov et al. 1997; Ingelfinger et al. 2002; Lykke-Andersen 2002; Cougot et al. 2004; Andrei et al. 2005; Ferraiuolo et al. 2005; Kedersha et al. 2005), and in some cases P-bodies can associate with membranes (C. Beckham and R. Parker, unpubl.), a clear implication of our results is that P-body functions might also participate in core particle assembly for at least some other retroviruses/retrotransposons in eukaryotes, including mammals.
MATERIALS AND METHODS
Yeast and bacterial culture conditions
Yeast and bacterial culture methods were as previously described except where noted (Ausubel et al. 1999; Burke et al. 2000). Bacterial strain HB101 (F− hsd-20 [rB−mB−] recA13 leuB6 ara14 proA2 lacY1 galK2 rpsL20 [Smr] xyl-5 myl-1 supE44 λ) was used for all plasmid preparations. Ty3-GFP/RFP expression was in the BY4741 background (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (Open Biosystems) (Winzeler et al. 1999). Expression of Ty3-MS2 was in strain YTM443 (MATa trp1-H3 ura3-52 his3-200 ade2-101 lys2-1 leu1-12 can1-100 bar1::hisG GAL+ ΔTy3) (Menees and Sandmeyer 1994). P-body–GFP-tagged strains (Huh et al. 2003) and deletion strains (Winzeler et al. 1999) were in the BY4741 background. Deletion strains were purchased from Open Biosystems. The plasmid from which MS2-GFP was expressed under control of the MET25 promoter was described previously (Beach et al. 1999). For Ty3 expression, cells were grown to logarithmic phase in synthetic raffinose (SR)–ura medium and induced by the addition of galactose to 2% (SG). Cultures were maintained at 30°C, except for those used for EM, which were maintained at 25°C.
Recombinant DNA constructions
The Ty3-GFP plasmid was produced by first introducing the XmaI site downstream of Ty3 POL3 at nucleotide 5057 in pDLC201 (Hansen et al. 1988) (pNB2114). A PCR fragment containing the coding region for GFP (S65T) was ligated into a XmaI site so that GFP was expressed in frame with POL3 (pNB2127). The Ty3-RFP expression plasmid was created by cloning yeast codon-usage and context-optimized DsRed Monomer sequence (Clontech) into this XmaI site of pNB2114 (pNB2266). For the negative control, the same DsRed sequence was expressed from the HindIII/BamHI site of pYES2.0 (pNB2289). The DsRed Monomer DNA was assembled with a CODA Express kit (CODA Genomics Inc.) The Ty3-MS2 expression plasmid (pTy3-MS2) was created by ligating a duplex oligonucleotide containing a tandem repeat of the MS2 CA protein recognition sequence into the XmaI site of pNB2114.
Fluorescence microscopy
Yeast fluorescence microscopy was performed essentially as previously described (Oakes et al. 1998). The IgG fraction of rabbit polyclonal anti-Ty3 CA (Menees and Sandmeyer 1994) was affinity purified on a protein A–Sepharose CL-4B column (Amersham Biosciences). The resulting anti-CA (6.2 mg/mL) IgG stock was diluted 1:1000 before use in phosphate-buffered saline (PBS) containing BSA (20 mg/mL). Rabbit IgG was detected with TRITC-conjugated goat anti-rabbit IgG fraction (14 mg/mL) (Sigma) in PBS-BSA at a 1:5000 dilution. In order to detect DNA, living cells were grown in DAPI at a concentration of 1.25 μg/mL for several hours; fixed cells were stained with 1 μg/mL of DAPI in PBS for 5 min and washed with PBS three times.
Fluorescence microscopy was performed by using a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss Inc.) equipped with a 100× NA 1.3 objective. Fluors were detected by using Chroma filter sets: RFP and TRITC, 41004 (exciter, 560/555; emitter, 645/675); GFP, 41001 (exciter, 480/440; emitter, 535/550); DAPI, CZ 902 wide UV excitation with long-pass emission (exciter, 340/360; beam splitter, 400). Images were captured by using an AxioCam MRm camera with Axiovision 3.1 software. Images were processed and pseudocolored by using Adobe Photoshop 6.0 (Adobe Systems Inc.).
Confocal microscopy was performed with a Zeiss LSM50 Meta confocal fluorescence microscope equipped with Argon/2 laser (wavelength, 488 nm), He-Ne laser (wavelength, 543 nm), and two photon Ti:Sapphire femtosecond laser (wavelength 790 nm) using a 100× 1.3 NA objective and LSM510 Software. The multitrack option was used for image acquisition. Images were processed as described for conventional fluorescence microscopy.
Electron microscopy
Cells for EM were prepared as described previously (Giddings et al. 2001). Thin sections were viewed on a Philips CM10 electron microscope (Philips Electronic Instruments Co.) and recorded by using a Gatan digital camera and Digital Micrograph Software package (Gatan Inc.). For immunoEM, thin sections were similarly prepared and floated for 2 h on rabbit anti-CA diluted 1:800 or affinity purified rabbit anti-GFP (provided by J. Kahana and P. Silver, Harvard University, Cambridge, MA) diluted in 2% nonfat dry milk in PBS-Tween, followed by floating for 1 h on 15-nm gold-conjugated anti-rabbit IgG diluted 1:20 in the same blocking solution.
Immunoblot analysis
For immunoblot analysis of Gag3, IN, and IN-GFP/RFP fusion proteins from whole cell extracts, cultures of BY4741 and dhh1Δ and xrn1/kem1Δ derivatives were transformed with pDLC201, pNB2127, or pNB2266, induced as described above for microscopy. Immunoblot analysis was performed as described previously (Irwin et al. 2005).
Acknowledgments
We thank M. Oakes (University of California, Irvine) for advice on fluorescence microscopy and J. Meehl and A. Bednarz (University of Colorado, Boulder) and Z. Wang (University of California, Irvine) for technical assistance. This work was supported by grants from the NIH (to S.B.S. and R.P.) and from the Howard Hughes Medical Institute (to R.P.).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2264806.
REFERENCES
- Andrei, M.A., Ingelfinger, D., Heintzmann, R., Achsel, T., Rivera-Pomar, R., and Luhrmann, R. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 1999. Current protocols in molecular biology. Greene Publishing Associates/Wiley-Interscience, New York.
- Bashkirov, V.I., Scherthan, H., Solinger, J.A., Buerstedde, J.M., and Heyer, W.D. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basyuk, E., Galli, T., Mougel, M., Blanchard, J.M., Sitbon, M., and Bertrand, E. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5: 161–174. [DOI] [PubMed] [Google Scholar]
- Beach, D.L., Salmon, E.D., and Bloom, K. 1999. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9: 569–578. [DOI] [PubMed] [Google Scholar]
- Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S.M., Singer, R.H., and Long, R.M. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2: 437–445. [DOI] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., and Parker, R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D., Dawson, D., and Stearns, T. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Butsch, M. and Boris-Lawrie, K. 2002. Destiny of unspliced retroviral RNA: Ribosome and/or virion? J. Virol. 76: 3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin, J.M., Hughes, S.H., and Varmus, H.E., eds. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Coller, J. and Parker, R. 2005. General translation repression by activators of mRNA uncapping. Cell 122: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., and Seraphin, B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo, M.A., Basak, S., Dostie, J., Murray, E.L., and Schoer, R.A. 2005. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 170: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel, D.J., Boeke, J.D., and Fink, G.R. 1985. Ty element transposition: Reverse transcriptase and virus-like particles. Cell 42: 507–517. [DOI] [PubMed] [Google Scholar]
- Giddings, T.H., O’Toole, E.T., Morphew, M., Mastronarde, D.N., McIntosh, J.R., and Winey, M. 2001. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 67: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, J.L., Coleman, L.E., Raymond, A.S., Goodson, S.G., Pittard, W.S., Tsui, C., and Devine, S.E. 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, L.J., Chalker, D.L., and Sandmeyer, S.B. 1988. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol. Cell. Biol. 8: 5245–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, L.J., Chalker, D.L., Orlinsky, K.J., and Sandmeyer, S.B. 1992. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J. Virol. 66: 1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S., Choi, G., Park, S., Chung, A.S., Hunter, E., and Rhee, S.S. 2001. Type-D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J. Virol. 75: 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O’Shea, E.K. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Ingelfinger, D., Arndt-Jovin, D.J., Luhrmann, R., and Achsel, T. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrn1 in distinct cytoplasmic foci. RNA 8: 1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Irwin, B., Aye, M.S., Baldi, P., Beliakova-Bethell, N., Cheng, H., Dou, Y., Liou, W., and Sandmeyer, S. 2005. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 15: 641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fitzler, M.J., Scheuner, D., Kaufman, R.J., Golan, D.E., and Anderson, P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial, M.L. and Eastman, S.W. 2003. Particle assembly and genome packaging. Curr. Top. Microbiol. Immunol. 277: 89–110. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menees, T.M. and Sandmeyer, S.B. 1994. Transposition of the yeast retroviruslike element Ty3 is dependent on the cell cycle. Mol. Cell. Biol. 14: 8229–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes, M., Aris, J.P., Brockenbrough, J.S., Wai, H., Vu, L., and Nomura, M. 1998. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 143: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlinsky, K.J. and Sandmeyer, S.B. 1994. The Cys-His motif of Ty3 NC can be contributed by Gag3 or Gag3-Pol3 polyproteins. J. Virol. 68: 4152–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer, S.B., Aye, M., and Menees, T.M. 2002. Ty3: A position-specific, gypsylike element in Saccharomyces cerevisiae. In Mobile DNA II (eds. N.L. Craig et al.), pp. 663–682. ASM Press, Washington, DC.
- Sfakianos, J.N., LaCasse, R.A., and Hunter, E. 2003. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag poly-peptides to a pericentriolar region of the cell. Traffic 4: 660–670. [DOI] [PubMed] [Google Scholar]
- Sheth, U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., Sheth, U., Valencia-Sanchez, M.A., Brengues, M., and Parker, R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teysset, L., Dang, V.D., Kim, M.K., and Levin, H.L. 2003. A long terminal repeat-containing retrotransposon of Schizosaccharomyces pombe expresses a Gag-like protein that assembles into virus-like particles which mediate reverse transcription. J. Virol. 77: 5451–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al.1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Zimmerman, C., Klein, K.C., Kiser, P.K., Singh, A.R., Firestein, B.L., Riba, S.C., and Lingappa, J.R. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415: 88–92. [DOI] [PubMed] [Google Scholar]