Abstract
Successful investigation of common diseases requires advances in our understanding of the organization of the genome. Linkage disequilibrium provides a theoretical basis for performing candidate gene or whole-genome association studies to analyze complex disease. However, to constructively interrogate SNPs for these studies, technologies with sufficient throughput and sensitivity are required. A plethora of suitable and reliable methods have been developed, each of which has its own unique advantage. The characteristics of the most promising genotyping and polymorphism scanning technologies are presented. These technologies are examined both in the context of complex disease investigation and in their capacity to face the unique physical and molecular challenges (allele amplification, loss of heterozygosity and stromal contamination) of solid tumor research.
INTRODUCTION
Molecular biology has undergone a dramatic transformation as it has evolved into the field of genomics. Combined public and private efforts to sequence the human genome have resulted in a first draft representing ∼75% of the genome and an estimated 30 000–40 000 genes (1,2). One of the more promising applications of this massive genetic data set lies in developing a better understanding of complex diseases. Current efforts towards this end attempt to analyze polymorphism data across the entire human genome. To accelerate this process, improvements in polymorphism detection (genotyping) and our understanding of the genetic diversity among humans are required.
Polymorphism and linkage disequilibrium
The haploid human genome consists of approximately 3 billion bp of deoxyribonucleic acids. On average, aligned stretches of human DNA differ at <0.1% of orthologous nucleotide positions (3–6). In addition to single base substitution polymorphism, sequence variation within (and among) species or populations may be revealed as small insertion/deletions (indels) or variation in copy number of repeated motifs. From an evolutionary standpoint, polymorphism represents the transient state between the origin of genetic variation by mutation and the loss of variation by fixation of either the ancestral or derived state. This transient state can, in certain circumstances, persist for quite some time (e.g. when there is balancing selection that maintains polymorphism). Today, the expression ‘single nucleotide polymorphism’ (SNP) is often applied to variable sites for which the rarer base is present within the population at >1% frequency, whereas germline polymorphisms with a frequency <1% are typically referred to as ‘mutations’. We will use these definition for this review.
It has been well documented that Mendelian diseases (e.g. cystic fibrosis) result from defects in one gene. However, it is suspected that most common diseases (e.g. hypertension) are ‘polygenic’, meaning that variation in the presence/absence of the disease is attributable, at least in part, to polymorphism of multiple interacting genes. This phenotypic variation may also reflect environmental variation, as well as gene-by-environment interactions that may be complex and unpredictable.
An allele of one of these ‘susceptibility genes’ may contribute to disease in certain genetic and environmental contexts, but not in others. This complexity creates a monumental task in identifying and characterizing the combinations of genetic variants relevant to disease. Population genetics provides a theoretical basis for addressing part of this puzzle by constructing a mathematical framework to help describe the creation, maintenance and distribution of genotypes. Data analysis in the context of population genetic models should help identify susceptibility genes (7).
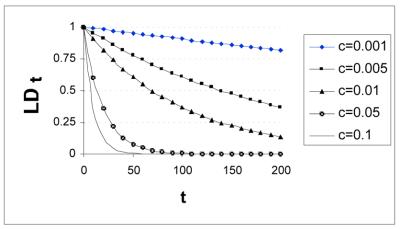
A fundamental concept intrinsic to these genetic models is ‘linkage disequilibrium’ (LD), the correlation of character states among polymorphic sites (8). The simplest explanation for LD is linkage coupled with insufficient passage of time to randomize the character states by meiotic recombination. Consider the case where a polymorphism is already present at Locus A. When a mutation occurs nearby (Locus B), the derived character state (B1) will be associated with one or the other character state of Locus A (i.e. A0 or A1). In this case, assume that B1 is initially linked to A1. If there is never recombination between the A and B loci, then as B1 increases in frequency, it will always be associated with A1. That is, there is a 100% chance that an individual carrying B1 also carries A1. LD can decay between A and B, however, if recombination occurs between the two loci: the rate of decay will be a function of recombination rate and the number of generations. [Assuming selective neutrality and no recurrent mutation, LDt = (1 – c)t LD0 (where c is recombination frequency, LD0 is initial LD and LDt is LD after t generations) (Fig. 1), see Lynch and Walsh (8) for a more detailed discussion]. Thus, the predictive value of the B allele with respect to the Locus A allele depends on the age of the polymorphism, the relative frequencies of the two A alleles when the B1 allele first arises (LD will be stronger if the linked A1 allele is rarer) and the recombination rate between A and B.
Figure 1.
Effects of recombination rate on LD. Plot depicting how LD decays more quickly with increased recombination rate with LD (dependent variable) as a function of time in generations (the independent variable) and recombination rate (c).
LD can also be maintained by natural selection, even for polymorphisms that are not syntenic. That is, particular combinations of character states may be selectively favored over others. Population structure can also produce LD, as can recent admixture of formerly isolated populations. For a fuller review of LD in the context of population genetic theory, see Nordborg and Tavare (9).
These principles form the conceptual foundation for association studies. Here, geneticists attempt to correlate SNP patterns with phenotypes (e.g. normal versus disease) to directly associate SNPs that act either as markers and/or are causative for disease. It is useful, here, to distinguish between co-segregation and LD. If a pair of polymorphic loci are separated by <50 cM, linked alleles of those loci will tend to co-segregate during meiosis. However, association studies depend on population data; implicit is the passing of numerous generations. Thus, co-segregation during meiosis is necessary, but insufficient, for the validity of LD association studies used for the purpose of mapping. Given this caveat, a genomic region characterized by high LD may have many polymorphisms segregating non-independently in the population. In such regions, a single polymorphic site can be used as a marker for the entire set of loci. Therefore, when attempting to identify a susceptibility gene across the entire genome, regions of relatively higher LD could require fewer loci to be genotyped, since a single locus can represent a larger set. Though most LD-based methodologies pinpoint only a single genomic region, one novel approach exists that reflects the polygenic nature of complex traits by assessing the effects of linkage and/or LD of markers in multiple genomic regions (10).
However, questions persist regarding the utility of LD in association studies. For example, since the degree of LD can vary throughout the genome, some regions may be refractory to association studies. If the marker and allelic variation are in a ‘recombination hotspot’, then the degree of LD may have degraded to such an extent over many generations that it is no longer informative. Fortunately, it appears that LD follows a block-like structure in the human genome, with large regions of low allelic diversity (high LD) flanked by small regions of recombination hotspots (11–13). If true across the entire genome, this would minimize the potential problem of genomic variability, thus arguing for the utility of LD in association studies.
Other potential logistical problems arise from variation in recombination rate. Several population genetics models predict a decrease in polymorphism levels in regions of reduced recombination (14,15). This prediction has empirical support (16–18). Thus, while large areas of reduced recombination could, in principal, maintain LD among more sites, fewer of those sites will tend to be polymorphic. Further, because reduced recombination also decreases the effectiveness of natural selection on newly arising deleterious mutations (14,19,20), compensatory insertions that increase the distance between genes might be selectively favored (21).
Even amongst its proponents, considerable discussion remains as to the most effective way to utilize LD and SNPs in association studies (10). Some issues include: the use of SNPs versus haplotypes (11,22,23), the utility of familial related individuals (24,25) and population selection, (i.e. large outbred population versus small isolated population) (26–30). Due to advances in technology, experimental evidence is beginning to influence these debates. For instance, part of the debate on population selection centers on the degree of LD in various populations. Experimental evidence suggests that the degree of LD in a small isolated population that has not been recently formed may be similar to that of a large outbred population (31–33). This result suggests that, depending on the population’s history, certain small isolated populations may not provide the advantage of a higher degree of LD; however, such populations may still be valuable due to their reduced genetic variation (27).
Despite these lingering questions, association studies using LD to identify susceptibility genes have achieved some recent success. Proof of principle experiments using a high-density SNP map in genomic regions already known to contain susceptibility genes for complex diseases (including Parkinson’s, Alzheimer’s, psoriasis, migraine, type II diabetes and Crohn’s disease) have confirmed known genes or identified new ones (13,34–40).
These successful association studies are ‘candidate gene’ approaches, where the location of the susceptibility gene is either already known or suspected. A more global approach, whole genome analysis, attempts to identify the location of unknown susceptibility genes across the whole genome, without bias to initial candidate locations. This experimental design is based on a two-step LD methodology (41). In the first step, a low-density SNP map (i.e. ‘picket fence’) of the whole genome is used to identify a region(s) containing the potential susceptibility gene(s). Once the region is identified, a high-density SNP map is used to home in on the susceptibility gene within the region (i.e. analagous to a candidate gene approach). The success of this approach greatly depends upon the degree of LD across the human genome, since this will determine the number and location of SNPs required to make meaningful predictions (estimates range from 100 000– 200 000 to as many as 1 000 000 SNPs) (31–33,42,43).
In order to generate a useful SNP map of the whole genome as described above, a subset of representative SNPs must be culled from a well-characterized map of millions of SNPs. At present, this endeavor requires tremendous resources, so several private and public organizations recently created the SNP Consortium to pool their data. As of February 2001, only a small fraction of the estimated 11 million SNPs (44) were listed as identified and positioned (45). This discovery process remains a significant challenge, demanding high-throughput platforms and novel techniques to identify known and new SNPs.
Genetic analysis of solid tumors
High-throughput polymorphism detection technologies hold great promise for the characterization of complex genetic diseases. In order to be effective, a given technology needs to be compatible with the molecular and physical characteristics of the disease itself. Solid tumor based cancer research illustrates both the opportunities and barriers to SNP discovery and identification.
Cancers arise from the accumulation of inherited polymorphisms (i.e. SNPs and mutations) and/or sporadic somatic polymorphisms (i.e. non-germline polymorphisms) in cell cycle, DNA repair, and growth signaling genes. Knowledge of these molecular changes can influence patient management. For instance, members of certain ethnic groups have a higher risk of carrying SNPs in cancer genes such as BRCA1, BRCA2 or APC. These SNPs confer an increased risk of developing breast, ovarian, prostate or colon cancers (46–53) and patient management would benefit from increased vigilance in testing. Somatic polymorphisms, such as those in the p53 gene, influence both clinical outcome and response to therapy (54–60). The precise nature of the p53 mutation, therefore, may alter treatment protocols and other clinical considerations (61–65).
In addition to the inherited and sporadic polymorphisms, many tumors exhibit aneuploidy and chromosomal instability in which the diploid structure of the genome is corrupted. Chromosomal rearrangement is common, and the genome may exhibit amplification of alleles, or conversely the loss of an allele—‘loss of heterozygosity’ (LOH). These changes further serve to characterize the unique molecular signature of the tumor, and consequently influence patient management as well. It is imperative, therefore, that detection techniques have the ability to accurately identify and describe these changes.
The physical characteristics of a solid tumor also makes mutation detection more difficult than SNP analysis on germline samples. Because solid tumors contain a mixture of both tumor cells and stromal (i.e. non-tumor) cells, a polymorphism present in the tumor sample may represent only a minor fraction (as little as 15%) of the total DNA. In contrast, for a germline sample with a SNP present, at least half of the sample being tested will contain that variant. As a result, a solid tumor sample requires detection strategies with higher sensitivity.
Criteria for evaluating polymorphism detection technologies
There is a smorgasbord of polymorphism detection technologies available, and their utility depends upon the experimental objectives (and tastes) of the researcher. The most useful criteria for evaluating a given technology include throughput, sensitivity/specificity, quantitative ability, sample requirements and cost.
Many parameters can increase throughput, such as multiplexing and sample pooling. Multiplexing means that multiple reactions can be performed simultaneously in the exact same reaction environment. Assays with very high multiplexing capabilities can process thousands of reactions in parallel (e.g. DNA microarrays). Conversely, assays with low multiplexing capabilities compensate by processing a large number of separate reactions simultaneously (e.g. Taqman assays). SNPs that are of low frequency in a population are less likely to be found when querying germline samples individually. In this case, pooling of samples can increase effective throughput and also increase the chances that a low-frequency SNP is identified per reaction.
In clinical applications, sensitivity (positivity in the presence of disease) is defined as the ability of a test to give a positive finding when the patient truly has the disease:
sensitivity = 100 × (true positives)/(true positives + false negatives) (66,67).
Technologies used to detect germline polymorphisms in a sample from an individual subject require much lower sensitivity than technologies used to detect sporadic mutations in a solid tumor sample. Pooled germline samples may require greater sensitivity than either of the above. Therefore, different applications can have different sensitivity requirements.
Specificity (negativity in the absence of disease) is defined as the ability of a test to give a negative finding when the patient is free of the disease:
specificity = 100 × (true negatives)/(true negatives + false positives) (66,67).
As a result, assays with low specificity are more prone to false positive results. Although false negative and false positive results are both generally undesirable, a false positive tends to be more deleterious. This is because a false negative only removes the sample from its appropriate group, whereas a false positive not only removes the sample from its appropriate group, but also places it in the wrong group: a double error. As a result, it is sometimes prudent to sacrifice sensitivity for greater specificity (68).
The demand for quantitative genotyping is rapidly growing. Quantitative genotyping assays on pooled samples of defined populations are being used to generate comprehensive allele frequencies on thousands of SNPs throughout the genome (69,70). In addition, the abundance of an allele that undergoes amplification or LOH can be important in characterizing tumors (71,72).
The total number of genotypes that can be performed is limited by the amount of sample available. When sample DNA is in short supply, PCR amplification of each SNP region is commonly employed. However, the PCR step can generate cross contamination or allele dropout artifacts, affecting the overall specificity and sensitivity. Alternatively, PCR-based whole genome amplification techniques may increase the amount of total DNA (73–75), however they exhibit amplification bias as high as 4–6 orders of magnitude (76). This bias can lead to variability in multiplexing, dropout of some target sequences and greatly diminished capacity of the assay to measure relative allele abundance. Therefore, there is a need for an amplification method that is inclusive of all genes and can more adequately maintain the relative abundance of targets during amplification. A promising approach pioneered by Paul Lizardi uses the strand displacement properties of polymerases to generate hyperbranched structures (J.M.Lage, J.Leamon, T.Pejovic, S.Hamann, M.Lacey, D.Dillo, R.Segraves, B.Vossbrink, A.González, D.Pinkel, D.G.Albertson, J.Costa and P.M.Lizardi, manuscript submitted) (76). This mechanism is similar to rolling circle amplification (RCA) (see below), except that a linear strand of DNA is used as a target instead of a circularized fragment. This technique is reported to generate 20–30 µg of product from as little as 100 fg of human genomic DNA, results in an amplification bias of <3-fold across entire chromosomes and is compatible with use in detecting gene losses (76).
Finally, the cost of genotyping can be significant, especially on a genome wide scale. Assuming that 200 000 polymorphic loci need to be interrogated for a complete genome scan, at current prices it would cost ∼$20 000 per individual for such an analysis (43). This cost makes whole genome analysis only accessible to a select research market—mainly pharmaceutical companies. However, analogous to Moore’s Law in computer processing power, the price per polymorphism should decrease as technology advances and utility increases. Until then costs can be decreased by less dense coverage of the genome and utilizing other approaches such as positional cloning to refine searches to smaller regions of the genome (40). Depending on the genetic component in the disease under study and the number of family members or affected sibling pairs, this hybrid approach may or may not be successful.
Technologies to detect known polymorphisms
Most techniques for detecting known polymorphisms are variations on a few standard methodologies (Table 1). Since the reaction products of many of these techniques can be displayed in several formats, it is expedient to focus discussion on the basic techniques first, then to review detection and display methods.
Table 1. Comparison of high-throughput techniques to identify known polymorphisms and mutations.
| Technique | Advantages | Current limitations |
|---|---|---|
| Microarrays | (i) Scan for polymorphisms in thousands of loci | (i) Specificity determined by hybridization: |
| –difficult to distinguish all polymorphisms | ||
| –difficult to detect low level polymorphisms | ||
| (ii) Each new DNA target requires a new array fabrication | ||
| (iii) Currently requires PCR amplification | ||
| Real-time PCR, | (i) Closed tube: | (i) Limited multiplexing capabilities: |
| Taqman assay, Molecular beacons | –minimizes cross contamination–readily automated | –can be extended though by use of molecular beacons instead of linear probes |
| (ii) Target amplification | (ii) Specificity determined by hybridization: | |
| (iii) Probe amplification and detection in single step | –difficult to distinguish all polymorphisms | |
| (iii) Intrinsic requirement for PCR amplification | ||
| Primer extension, Minisequencing | (i) Straightforward primer design (ii) Adaptable to MALDI–TOF detection, avoids need for labeling | (i) Cannot detect mononucleotide repeat insertions and deletions. (ii) Currently requires PCR amplification |
| Invader assay | (i) No PCR amplification required; isothermal reaction | (i) Limited multiplexing capabilities |
| (ii) Closed tube: | (ii) Cannot detect small insertions and deletions | |
| –minimizes cross contamination | (iii) Requires very pure probes to avoid false positives | |
| –readily automated | ||
| (iii) Probe amplificaiton and detection in single step | ||
| Ligase detection reaction, Oligonucleotide ligation assay | (i) High multiplexing capabilities (ii) High level of sensitivity and specificity | (i) Requires PCR amplification either before (PCR/LDR) or after (LDR/PCR) ligation step |
| (iii) Detects all varieties of point mutations and small insertions and deletions | ||
| Padlock probes, | (i) No PCR amplification required | (i) Requires very long probes |
| Rolling circle amplification | (ii) Compatible with in situ applications | (ii) Limited multiplexing capabilities |
Most techniques can be broadly classified as either hybridization-based or enzyme-based. Thorough and useful reviews of these techniques are available (77,78). In general, enzymatic approaches contain an additional level of specificity in discriminating polymorphisms, and usually have fewer sequence limitations than hybridization-based approaches.
Hybidization technologies
Microarrays. DNA microarrays designed to distinguish single nucleotide differences are generally based on the principle of ‘sequencing by hybridization’ and utilize a set of ‘tiling’ oligonucleotides (79–82). The general methodology is somewhat complex and involves the pooling and processing of PCR amplicons that are subsequently hybridized to a DNA microarray and visualized. Due to the massively parallel nature of DNA microarrays, this methodology is theoretically capable of genotyping thousands of polymorphisms simultaneously. Despite the high levels of scanning capacity and success in unknown polymorphism analysis (see ‘Microarray’ in ‘Techniques to identify unknown polymorphisms’ below), this application suffers significant limitations in detecting known polymorphisms. Recent experiments using microarrays demonstrated a 97% accuracy on only 65% of the SNPs surveyed (68). Unfortunately, even 97% accuracy may be inadequate for association studies. Furthermore, high false positive rates of 11–21% have been observed with this technology, limiting its utility in both SNP and tumor analysis (3). In addition, design and fabrication of microarrays are expensive, making it financially prohibitive to alter polymorphism sets. As a result, users are confined to the set of genotypes established by the manufacturer. In order to overcome this significant shortcoming, variations on this technique are being developed. One promising general methodology attempts to compare the annealing of matched versus mismatched probes to targets (‘probe’ typically refers to the DNA immobilized on the surface, whereas ‘target’ generally refers to DNA in solution) over a range of hybridization conditions. In DASH (dynamic allele-specific hybridization), this is achieved by monitoring hybridization over a range of temperatures (83), while in microelectric chip arrays, each probe is coupled to an electrical contact which sends a current to denature target–probe hybrids (84,85). Due to the massively parallel potential of microarrays, there is a strong motivation to further develop this technology for known polymorphism detection.
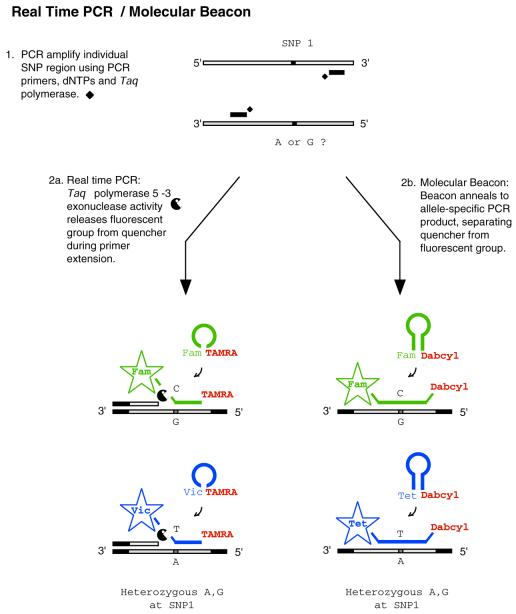
Real-time PCR. This dramatically different hybridization technique utilizes Taqman™ DNA probes to detect PCR products in real-time (86) (Fig. 2). Typically, a Taqman™ probe contains a fluorescent reporter at the 5′ end and a fluorescence resonance energy transfer (FRET) moiety at the 3′ end, which quenches the fluorescent signal of the reporter. The probe sequence is complementary to the PCR amplicon and is designed to anneal at the extension temperature. During extension, the 5′→3′ exonuclease activity of Taq DNA polymerase I cleaves the probe, emitting signal due to the separation of the reporter from the quencher. Since Taq DNA polymerase I can presumably cleave a matched as well as mismatched probe, discrimination of a polymorphism is determined solely by hybridization and not by the ability of the enzyme to discriminate. Because the enzyme does not confer specificity in detection, this technique is classified as hybridization-based. Depending on the optical thermocycler platform, up to 384 reactions can be monitored for each cycle without removing any sample, and its simplicity makes it readily amenable to robotic automation.
Figure 2.
Real-time PCR/molecular beacons assays. Generalized scheme for genotyping using real-time PCR. PCR product is detected by using linear DNA probes (2a) or Molecular Beacons (2b). Abbreviations: Fam [5-(&6)-carboxyfluorescein]; Tet (tetrachloro-6-carboxyfluorescein); Vic (a proprietary fluorescent dye produced by Applied Biosystems); Dabcyl [4-(4′dimethylaminophenylazo) benzoic acid]; and TAMRA [5-(&6-)-carboxytetramethylrhodamine].
However, due to the limited number of compatible reporter–quencher sets available, only two sequences can be confidently probed per reaction, which significantly lowers multiplexing capabilities with the Taqman™ probe. A variation of the common format that addresses this problem replaces the Taqman™ probe with a Molecular Beacon™ (87,88) (Fig. 2). A Molecular Beacon™ is similar to a Taqman™ probe except that the 3′ and 5′ ends are complementary. When the Molecular Beacon™ is free in solution it forms a stem–loop structure which brings the reporter and quencher into immediate proximity of one another. Once hybridized to the amplicon, the stem–loop structure opens, resulting in fluorescent signal. Molecular Beacons™ are more compatible with multiplexing, since the fluorescent group is quenched via direct energy transfer. This provides a larger set of compatible reporter–quencher combinations (88). Despite this inherent advantage, Molecular Beacons™ appear to be used less frequently, most likely due to the higher cost of synthesis and additional design parameters, though neither appears to be a dramatic shortcoming.
The sensitivity and specificity of real-time PCR is dependent on the ability of the probe to discriminate single base differences. Molecular Beacons™ are inherently more sensitive and have higher specificity than linear probes, since the stem–loop formation thermodynamically competes with the amplicon for hybridization (89). Although addition of a minor groove binder (MGB™) moiety to the 3′ end of the Taqman™ probe increases the specificity of the probe to such an extent that analysis can be run in end-point mode rather than real-time (E.Winn-Deen, Celera Genomics, Rockville, MD, personal communication). For end-point mode, samples are amplified in a thermocycler and then transferred to an optical thermocycler for detection. This results in a significant increase in throughput for the optical device. Sensitivity can be enhanced further by using peptide nucleotide analogs to preferentially suppress amplification of the abundant allele (90). This suppression has significant utility when attempting to detect spontaneous mutations in solid tumors with high stromal contamination. Another example of the application of real-time PCR in tumor analysis is known as ‘digital PCR’. In this elegant approach, samples are serially diluted to a single molecule allowing for an accurate quantification of LOH, albeit at the cost of dramatically reducing throughput (91,92).
Enzymatic technologies
Nucleotide extension. The primer extension assay represents one of the simplest techniques for known polymorphism detection (93). Existing in numerous variations (also known as minisequencing, SNuPE, GBA, APEX, AS-PE capture, FNC, TDI or PROBE) this assay typically involves the single base extension of an oligonucleotide by a polymerase (94) (Fig. 3). In the common format, an oligonucleotide is designed to anneal immediately upstream of the polymorphism locus and differentially labeled fluorescent dideoxynucleotides are utilized as substrates for polymerase extension. The fluorescent signal emitted corresponds to the nucleotide incorporated and thus the sequence of the polymorphism. The main advantages of primer extension include its simplicity and accuracy in distinguishing between heterozygous and homozygous genotypes. Since targets need to be PCR amplified, this technique suffers from the same limitations as most other enzymatic-based methodologies, but also encounters an additional disadvantage in that PCR reagents must be removed. False negatives due to mis-priming can occur, but are typically rare since the PCR step provides an orthogonal component that greatly minimizes this artifact. Numerous variations of primer extension have been developed to circumvent these limitations (77), including a mass spectrometry based approach, MassEXTEND™, which is discussed in further detail in the ‘Reaction product detection and display’ section below. A significant variation, pyrosequencing, also utilizes polymerase extension but monitors the generation of pyrophosphate for detection (95).
Figure 3.
Single nucleotide extension assay. Generalized scheme for genotyping using single nucleotide extension on a PCR amplified target. In this example products are detected using either fluorescent labeling (3a and 4a) or mass spectrometry (3b and 4b).
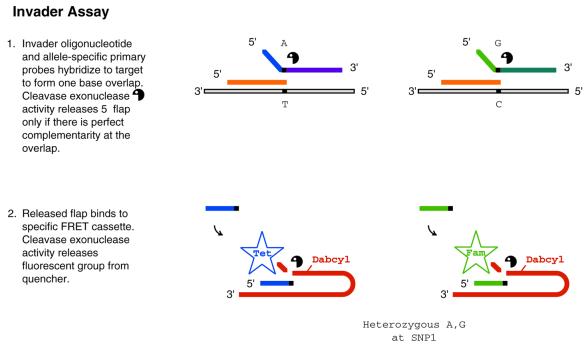
Cleavage. The Invader™ assay represents a nuclease-based approach for known polymorphism detection (96,97) (Fig. 4). It utilizes the exonuclease activity of Cleavase VIII (98) on overlapping oligonucleotide strands. Two oligonucleotides, an ‘invader’ probe and either a wild-type or mutant primary probe, overlap each other at a single nucleotide position on the template only if they are complementary to the polymorphism being queried. Cleavage occurs when the specific overlapping conformation is present, freeing an oligonucleotide referred to as a ‘flap’. This flap can be detected in a multiplex manner by size, mass or sequence (99,100). In the most common format the flap participates in a second cleavage assay with another complementary target, causing release of a fluorescent signal. The major advantage of this assay is that the same flap may bind to many targets, generating a cascading signal amplification and thereby obviating the need for PCR amplification. As with real-time PCR, the Invader™ assay is a single-tube one-step reaction. However, in the common format using fluorescent signal, it is uniplex, as only one genotype can be performed per reaction. Converting the assay to a solid support may expand the throughput capabilities (101), potentially enhancing the utility of this approach.
Figure 4.
Invader assay. Generalized scheme for genotyping using the Invader assay. In this example, target is not amplified, instead the probe product (i.e. flap) is used to amplify signal associated with detection of queried genotype by use of a second Invader cleavage (2). Abbreviation: Dabcyl [4-(4′dimethylaminophenylazo) benzoic acid].
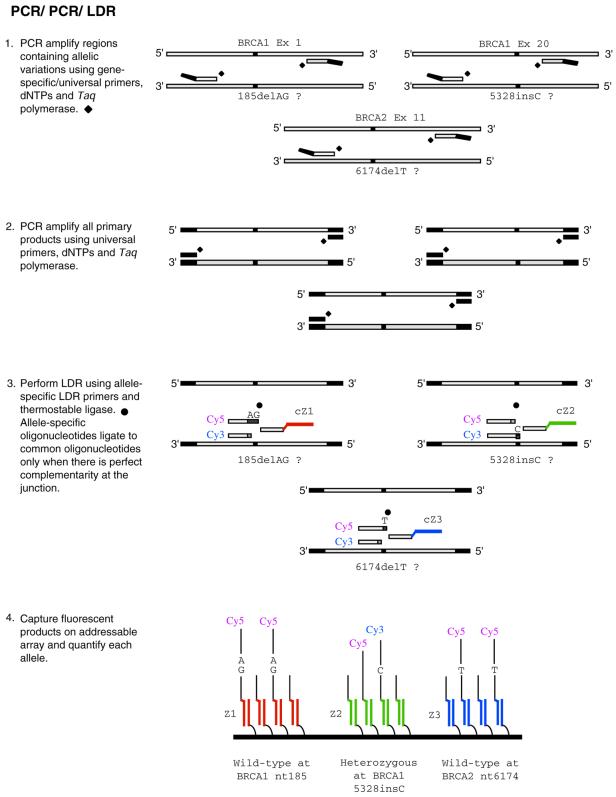
Ligation. Ligation-based technologies represent some of the most specific assays due to the high specificity of T4 ligase (oligo ligation assay) and even higher specificity of thermostable ligases (ligation detection reaction, LDR) (102–109). In this approach, two primers are designed to anneal adjacent to one another on the target of interest (Fig. 5). Generally, the upstream primer (discriminating primer) contains a fluorescent label at the 5′ end, with the 3′ nucleotide overlapping the polymorphic base. The fluorescent signal corresponds to the allele being queried at the 3′ position of the discriminating primer. When the discriminating primer forms a perfect complement with the target at the junction, the ligase covalently attaches the adjacent downstream primer (common primer). The resulting product is approximately twice as long as each of the individual primers and can be easily monitored for detection by means of capillary electrophoresis or by display on a microarray. The strength of this assay lies in its unsurpassed sensitivity and specificity. The thermostable Tth ligase can discriminate a G·T mismatch (the most common and difficult to detect) 1:200 fold against the correct complement (109). Furthermore, it offers some of the highest multiplexing capabilities to date (110) (D.R.Walt and N.Shen, unpublished results, Chips to Hits Conference, 2001). These assays are typically coupled to a PCR amplification step either before the ligation (target amplification, Fig. 5) or after the ligation (probe amplification, Fig. 6), and hence suffer the same disadvantages encountered by other techniques that utilize PCR. Since target amplification is not required for ligation, the development of higher sensitivity LDR detection formats may eliminate this disadvantage.
Figure 5.
PCR/PCR/LDR. Generalized scheme for genotyping using a ligation-based assay. In this example target is first amplified in a two-step PCR process (i.e. probe amplification) followed by ligation detection. Products are displayed on a ‘universal’ DNA microarray. Abbreviations: Cy3 (cyanine3) and Cy5 (cyanine5).
Figure 6.
LDR/PCR. Generalized scheme for genotyping using a ligation-based assay. In this example, the probe product is amplified (i.e. probe amplification) and products are displayed on a ‘universal’ DNA microarray. Abbreviation: Cy3 (cyanine3).
In a clever variation of LDR known as RCA, PCR amplification is replaced by a novel amplification of DNA by a polymerase on a circular template (111). In RCA, the discriminating and common primer sequences are located at either end of the same oligonucleotide. Ligation of these two sequences results in a circle locked around the template strand (‘padlock probe’) (112,113). Since it is circular, this oligonucleotide can then be copied repeatedly by a strand displacing DNA polymerase without dissociating from the template. Addition of another primer allows for formation of branched structures resulting in rapid amplification of the probe product (114). The main advantage of this technique is that PCR amplification is avoided. Recently, a modest number of padlock probes were multiplexed with no observed cross reactivity (J.Baner and U.Landegren, University of Uppsala, Uppsala, Sweden, personal communication). Further development in this direction will undoubtedly enhance the utility of this technique.
Reaction product detection and display. A variety of methodologies exist for detecting and displaying the products of a given assay. Since these display methods affect throughput and sensitivity, the merits of any given technique need to be evaluated in the context of the detection scheme. One commonly used methodology includes synthetically labeling products with fluorescent molecules and analyzing their differential physical properties (size, charge and mass). Gel and capillary electrophoresis typically use this method, but in most cases capillary electrophoresis is a more desirable platform due to its higher throughput. Conversely, mass spectrometry based approaches can identify DNA products without any synthetic chemical modifications, although mass tags are often employed to aid in allele discrimination (115). The general technique requires preparation of extremely clean samples followed by ionization and then detection based on the charge and mass properties of the product (116–119). The technique is sensitive enough to detect a single base extension product, although it is sometimes difficult to distinguish two different alleles simultaneously in the same reaction. The MassEXTEND™ reaction solves this problem by modifying the primer extension assay. In MassEXTEND™ the primer is designed 2–3 nt upstream of the polymorphic site, and the reaction contains a single ddNTP, which queries for a polymorphic single nucleotide position, plus the three remaining dNTPs (69) (Fig. 3). The shift in primer position relative to the common format provides an internal control against false positives due to false priming. Addition of dNTPs allows for the allele containing the common variant to be extended by multiple bases past the polymorphic locus. This results in a greater separation of peaks during detection and hence significantly better resolution of the two alleles. A disadvantage of both capillary eletrophoresis and mass spectrometry is that samples are processed serially, potentially limiting throughput capabilities. Nevertheless, mass spectrometry is capable of genotyping at approximately one polymorphism per second, and there have been successful attempts of coupling mass spectrometry to an array format (116,120). Further, with the advent of 384-capillary electrophoresis array machines, throughput has been significantly increased for this display method.
Advantages of fluorescence include cost and ease of detection, but it has limited multiplex capabilities in solution due to spectral overlap of the fluorescent labels. Consequently, assays with high multiplexing capabilities require an additional separation step when fluorescent labels are used for detection. Microarrays represent an ideal format for fluorescence detection due to their massively parallel throughput capabilities. In general, there are two different types of microarray formats for polymorphism detection: ‘position-based’ microarrays and ‘disordered’ microarrays.
With position-based microarrays, each unique oligonucleotide is covalently attached to the solid array surface at a fixed and defined location. Therefore, a genotype is determined based on the wavelength of the fluorescence and its position on the array. For example, in ligation-based assays, a unique oligonucleotide sequence tag (which we termed ‘zip-code’) can be added to the common primer and then hybridized to its complementary zip-code at a known position on a ‘universal’ microarray (121) (Figs 4 and 5). Using this platform and a ligation-based assay, pooled samples have been used to facilitate a high-throughput analysis of known low frequency germline mutations (122). Alternatively, instead of forming products in solution and then hybridizing to the array, the products can be generated on the solid support itself. Such a methodology has been coupled to nucleotide extension assays and the Invader assay in an attempt to increase multiplexing capabilities (101,123–125).
For disordered microarrays, reaction products are coupled to a medium, such as an oligonucleotide bead with a specific fluorescent signal, and then displayed on a solid surface. Since the bead is free to localize anywhere on the surface, presence of the product is identified by reading the unique fluorescent signature emitted, as opposed to reading a defined and fixed position on the array. Display on the solid surface is used to separate the beads so they can be individually interrogated for fluorescence. For example, a ligation product with a sequence tag can be captured in solution by beads that contain the complementary sequence tag and a unique fluorescent signature (126–128). Next, beads with captured product are isolated and displayed on a surface. The surface is then scanned and each unique fluorescent signature corresponds to the presence of a particular product. This final identification step can be difficult. However, once optimized, this approach can yield massively parallel assays that can be coupled to more sensitive enzymatic-based detection schemes (129).
Techniques to identify unknown polymorphisms
SNPs are often of low frequency within a population, so the vast majority of individually queried germline samples will be negative. To address this challenge, unknown samples can be pooled, provided that a highly sensitive technology is used (3,4,6) (Table 2). Pooling increases the level of throughput, resulting in a higher positive rate per reaction, and increasing the chance that an individual reaction will be informative (130).
Table 2. Comparison of techniques to identify unknown polymorphisms and mutations.
| Technique | Advantages | Current limitations |
|---|---|---|
| Sanger dideoxysequencing | (i) Detects any mutation up to 600 bp/reaction | (i) Difficult to detect low level mutations |
| (ii) As rapid as SSCP and DGGE but more accurate | (ii) Multiple reactions for large genes | |
| Variation detection array (VDA) | (i) High-throughput screen; uses direct hybridization (ii) Can screen large sequence blocks | (i) False positive rate of 11–21% in large scale screens |
| (ii) Difficulty detecting polymorphisms in mononucleotide repeats | ||
| (iii) Does not detect frameshift mutations | ||
| SSCP | (i) Detects low level mutations | (i) Misses 30% of possible mutations |
| (ii) Rapid, does not require extra enzymatic steps | (ii) Cannot distinguish missense from silent polymorphisms | |
| (iii) Does not locate position of polymorphism | ||
| (iv) Can miss mutation near common polymorphism | ||
| CSGE, DGGE, CDGE, DHPLC | (i) Detects low level polymorphisms | (i) Large scale screen missed 13% of polymorphisms |
| (ii) Rapid, does not require extra enzymatic steps | (ii) Cannot distinguish missense from silent polymorphisms | |
| (iii) Technically challenging to reproduce results | ||
| (iv) Requires GC clamp; limited to small fragments | ||
| (v) Does not locate position of polymorphism | ||
| ddF, REF | (i) Detects virtually all possible mutations | (i) Difficult to detect low level mutations |
| (ii) Cannot distinguish missense from silent polymorphisms | ||
| Cleavase | (i) Heteroduplex not required | (i) High background |
| (ii) Does not locate position of polymorphism | ||
| (iii) Requires optimization for each mutation | ||
| Chemical cleavage method (CCM) | (i) Identifies approximate position of mutation | (i) Labor intensive |
| (ii) High sensitivity | (ii) Chemical hazard | |
| T4 Endo VII, MutY | (i) Identifies approximate position of most mutations | (i) Difficult to detect low level mutations |
| (ii) Identifies missense, frameshift and nonsense mutations | (ii) High background observed depending on sequence | |
| Thermostable Endonuclease V–DNA ligase | (i) Identifies approximate position of mutation, identifies 98% of polymorphisms | (i) Does not detect transition mutations in GGCG or RCGC sequences |
| (ii) Identifies missense, frameshift and nonsense mutations, up to 1750 bp/reaction | (ii) New technique | |
| (iii) Detects low level mutations; 1 in 20 | ||
| (iv) In combination with sequencing, most rapid screen to directly identify mutation |
Direct sequencing. Sanger dideoxysequencing can detect any type of unknown polymorphism and its position, when the majority of DNA contains that polymorphism. However, direct sequencing has missed polymorphisms and mutations when the DNA is heterozygous (131,132). Further, direct sequencing has only limited utility for analysis of solid tumors or pooled samples of DNA due to low sensitivity (133). Once a sample is known to contain a polymorphism in a specific region, direct sequencing is particularly useful for identifying a polymorphism and its specific position. Even if the identity of the polymorphism cannot be discerned in the first pass, multiple sequencing attempts have proven quite successful in elucidating sequence and position information. Furthermore, since many techniques that are capable of identifying the position of a polymorphism are incapable of providing sequence information, Sanger sequencing has utility as a second step to locate and identify the exact base altered in a gene region previously identified as polymorphic.
Electrophoretic mobility assays. Classic methods detect unknown polymorphisms by observing the different electrophoretic migration behaviors of homoduplex versus heteroduplex DNA. These methods include confirmation sensative gel electrophoresis (CSGE) (134,135), denaturing-gradient gel electrophoresis (DGGE) (136–139), constant denaturing capillary electrophoresis (CDCE) (140,141), dideoxy fingerprinting (ddF) (142) and restriction endonuclease fingerprinting (REF) (143) (Table 2). A similar approach, denaturing high-performance liquid chromatography (DHPLC) also differentiates homoduplex from heteroduplex DNA, but is based on separation by ion-pair reverse-phase liquid chromatography on alkylated non-porous (styrene divinylbenzene) particles (144). Finally, single-stranded conformational polymorphism (SSCP) (145–148) is similar to the above but is based on the conformational properties of a single-stranded DNA. Although these techniques contain some very desirable characteristics and have been extensively used for scanning small gene regions, all fall short in either desired throughput or sensitivity for both whole genome association studies and mutational analysis of solid tumors. Coupling these techniques to 384-well DHPLC and capillary electrophoresis has dramatically improved throughput of these techniques for many applications (149–152). To characterize the polymorphism, the techniques that can identify the position of a polymorphism (ddF and REF) are not sensitive enough to reliably detect low level polymorphisms in pooled or solid tumor samples. The other techniques are more rapid and can detect low level polymorphisms but do not identify the approximate position of the polymorphism. As a result multiple rounds of dideoxysequencing may be required to identify the sequence of the polymorphism.
Microarray. The recent development of DNA microarray technology has established unprecedented levels of throughput. Variation detection arrays (VDA) apply this new technology to scan large sequence blocks and identify regions containing unknown polymorphisms (3,80,153). This methodology suffers from the same limitations in fabrication and design as observed in known polymorphism analysis, but has demonstrated much greater success in the context of unknown polymorphism detection for both SNP and tumor analysis. For example, in a proof of principle experiment, a GeneChip was used to interrogate lung tumor samples for mutations in p53, a gene mutated in ∼50% of all cancers. The experiment was performed in a simulated unknown discovery mode and was able to identify 88% of the known missense mutations and 80% of all known polymorphisms (153). These results compare with the more traditional method of dideoxysequencing, which detected 76% of the known mutations present. With respect to SNP analysis, a recent study of chromosome 21 successfully identified approximately half of the estimated number of common SNPs (frequency of 10–50%) across the entire chromosome (68). The experimental design required a sacrifice in sensitivity in order to minimize false positives. This explains the decrease in successful identification from 80 to 50% for the chromosome 21 SNP analysis when compared with the lung tumor study previously mentioned. In addition, the utility of this approach needs to be evaluated in the context of rare SNPs (frequency ∼1%). Since ∼50% of the common SNPs in the human genome are refractory to detection by this approach, alternative techniques will most likely be required for a more complete identification of SNPs. Improvements in variant methodologies, such as DASH and microelectric chip arrays, may enhance its utility.
Cleavage. Unknown polymorphisms can also be identified by the cleavage of mismatches in DNA–DNA heteroduplexes. This can be achieved either chemically [chemical cleavage method (CCM); 154–156], or enzymatically (T4 Endo nuclease VII, MutY cleavage or Cleavase; 157–159). Typically, at least two samples are PCR amplified (one sample can be sufficient for solid tumor samples with high levels of stromal contamination), denatured and then hybridized to create DNA–DNA heteroduplexes of the variant strands. Enzymes cleave adjacent to the mismatch and products are resolved via gel or capillary electrophoresis. Unfortunately, the cleavage enzymes often nick complementary regions of DNA as well. This increases background noise, lowers specificity, and reduces the pooling capacity of the assay.
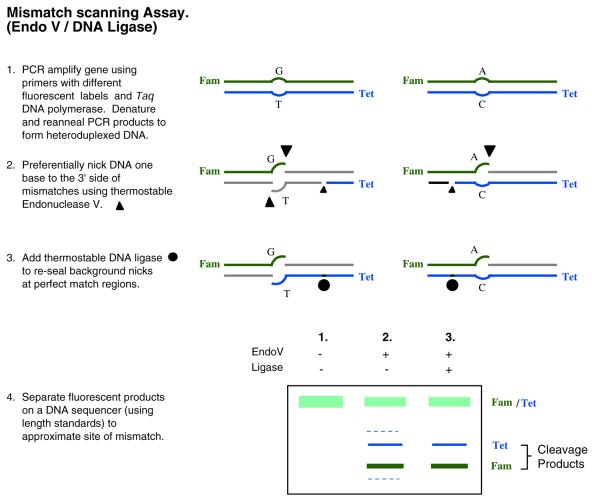
Cleavage/ligation. One way to improve signal-to-noise in the cleavage assay is to follow the cleavage with a ligation step to seal spurious nicks (Fig. 7). Unfortunately, many enzymes that are commonly used to detect mismatches are incompatible with this solution. Enzymes such as MutY do not generate re-ligatable ends (158), while enzymes such as T4 Endonuclease VII or a combination of MutH, MutS and MutL cleave far from the mismatch (157,159,160), so ligase would reseal all of the latter nicks. One technique addresses this issue by combining the ability of thermostable Endonuclease V (Endo V) enzyme to recognize and nick mismatched DNA, with the high fidelity of thermostable DNA ligase to suppress nicks at matched DNA (161). Endo V can nick either or both strands of the mismatch. Unlike the previous cleavage enzymes mentioned, Endo V nicks DNA close to the mismatched base (162). This allows the thermostable ligase to effectively discriminate between perfectly matched and mismatched regions of the DNA (108) and to ligate only perfectly matched nicks. This results in greatly reduced background noise. This method has very high sensitivity, and can distinguish one mutant sequence in a 20-fold excess of unaltered DNA. Further, since it can locate the approximate position of the polymorphism, it is readily compatible with follow-up dideoxysequencing to identify the exact polymorphism sequence. In addition, multiple polymorphisms from the same fragment can be detected simultaneously. This can be used to infer the position of a novel mutation relative to a known SNP, and potentially discriminate a missense from a silent mutation. To date, a few refractory sequences (GGCG and RCGC) have been identified. Nevertheless, evaluation of the SNP database suggests that the combined Endo V/ligase assay is capable of identifying 98% of the polymorphisms typically observed in the human genome (161). Since products are detected by means of capillary electrophoresis, samples are currently processed sequentially. However, due to its ability to minimize background noise, this technique is more amenable to pooled samples, effectively increasing its throughput capabilities. Overall, since it is a relatively new technique, its reliability and utility need to be established as it is more broadly applied.
Figure 7.
Mismatch scanning assay. Generalized scheme for assaying unknown polymorphisms using a Endo V/ligation-based approach. In this example products are displayed using gel electrophoresis. In lane 2, dashed bands represent products due to spurious nicking which are sealed in the presence of thermostable ligase, lane 3. Abbreviations: Fam [5-(&6)-carboxyfluorescein]; Tet (tetrachloro-6-carboxyfluorescein).
CONCLUSION
Numerous platforms exist for both known and unknown polymorphism detection, each with application-specific advantages. Since each technique has limitations unique to its underlying technology, different combinations of complementary technologies will most likely be necessary for robust data acquisition. Technology selection will depend on the particular experimental design and sample being queried, and will most aptly reflect considerations in throughput, sensitivity and specificity.
The promise of large-scale polymorphism analysis of the human genome is approaching critical mass. Validation of association-based genomic analysis is an ongoing, heuristic process that hinges on continued advances in SNP discovery techniques. Indeed, increasingly robust polymorphism detection assays are striving to meet the high-throughput and high-sensitivity demands of both whole genome association and tumor-based studies. Once validated, it is hoped that mined data will help to elucidate presently unidentified biological pathways associated with disease, ultimately yielding practical applications in drug discovery and diagnostics. If successful, this paradigm shift in the study of the genetic basis of disease will accelerate our understanding of complex diseases, and profoundly impact the human condition.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Jianmin Huang and Yu-wei Cheng of the Barany laboratory and Charles Cantor, Aravinda Chakravarti, Robert Cotton, Radoje Drmanac, Bill Efcavitch, Derek Gordon, Ken Kinzler, Fred Kramer, Pui-Yan Kwok, Ulf Landegren, Eric Lander, Ken Livak, Paul Lizardi, Mohamed Noor, Jurg Ott, Alex Pearlman, Bruce Neri, Alan Roses, Steve Sommer, Lloyd Smith, Thierry Soussi, Eric Spitzer, Mary Steed, Ann-Christine Syvänen, Bill Thilly, Bert Vogelstein, Bruce Wallace, David Ward, Emily Winn-Deen and Bob Waterston for their assistance in the preparation and critical evaluation of the manuscript. Work in the Barany laboratory is sponsored in part by a sponsored research grant from Applied Biosystems Inc., for which F.B. also serves as a consultant.
REFERENCES
- 1.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A., Gocayne,J.D., Amanatides,P., Ballew,R.M., Huson,D.H., Wortman,J.R., Zhang,Q., Kodira,C.D., Zheng,X.H., Chen,L., Skupski,M., Subramanian,G., Thomas,P.D., Zhang,J., Gabor Miklos,G.L., Nelson,C., Broder,S., Clark,A.G., Nadeau,J., McKusick,V.A., Zinder,N., Levine,A.J., Roberts,R.J., Simon,M., Slayman,C., Hunkapiller,M., Bolanos,R., Delcher,A., Dew,I., Fasulo,D., Flanigan,M., Florea,L., Halpern,A., Hannenhalli,S., Kravitz,S., Levy,S., Mobarry,C., Reinert,K., Remington,K., Abu-Threideh,J., Beasley,E., Biddick,K., Bonazzi,V., Brandon,R., Cargill,M., Chandramouliswaran,I., Charlab,R., Chaturvedi,K., Deng,Z., Di Francesco,V., Dunn,P., Eilbeck,K., Evangelista,C., Gabrielian,A.E., Gan,W., Ge,W., Gong,F., Gu,Z., Guan,P., Heiman,T.J., Higgins,M.E., Ji,R.R., Ke,Z., Ketchum,K.A., Lai,Z., Lei,Y., Li,Z., Li,J., Liang,Y., Lin,X., Lu,F., Merkulov,G.V., Milshina,N., Moore,H.M., Naik,A.K., Narayan,V.A., Neelam,B., Nusskern,D., Rusch,D.B., Salzberg,S., Shao,W., Shue,B., Sun,J., Wang,Z., Wang,A., Wang,X., Wang,J., Wei,M., Wides,R., Xiao,C., Yan,C. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 2.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W., Funke,R., Gage,D., Harris,K., Heaford,A., Howland,J., Kann,L., Lehoczky,J., LeVine,R., McEwan,P., McKernan,K., Meldrim,J., Mesirov,J.P., Miranda,C., Morris,W., Naylor,J., Raymond,C., Rosetti,M., Santos,R., Sheridan,A., Sougnez,C., Stange-Thomann,N., Stojanovic,N., Subramanian,A., Wyman,D., Rogers,J., Sulston,J., Ainscough,R., Beck,S., Bentley,D., Burton,J., Clee,C., Carter,N., Coulson,A., Deadman,R., Deloukas,P., Dunham,A., Dunham,I., Durbin,R., French,L., Grafham,D., Gregory,S., Hubbard,T., Humphray,S., Hunt,A., Jones,M., Lloyd,C., McMurray,A., Matthews,L., Mercer,S., Milne,S., Mullikin,J.C., Mungall,A., Plumb,R., Ross,M., Shownkeen,R., Sims,S., Waterston,R.H., Wilson,R.K., Hillier,L.W., McPherson,J.D., Marra,M.A., Mardis,E.R., Fulton,L.A., Chinwalla,A.T., Pepin,K.H., Gish,W.R., Chissoe,S.L., Wendl,M.C., Delehaunty,K.D., Miner,T.L., Delehaunty,A., Kramer,J.B., Cook,L.L., Fulton,R.S., Johnson,D.L., Minx,P.J., Clifton,S.W., Hawkins,T., Branscomb,E., Predki,P., Richardson,P., Wenning,S., Slezak,T., Doggett,N., Cheng,J.F., Olsen,A., Lucas,S., Elkin,C., Uberbacher,E., Frazier,M. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 3.Halushka M.K., Fan,J.B., Bentley,K., Hsie,L., Shen,N., Weder,A., Cooper,R., Lipshutz,R. and Chakravarti,A. (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nature Genet., 22, 239–247. [DOI] [PubMed] [Google Scholar]
- 4.Li W.H. and Sadler,L.A. (1991) Low nucleotide diversity in man. Genetics, 129, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephens J.C., Schneider,J.A., Tanguay,D.A., Choi,J., Acharya,T., Stanley,S.E., Jiang,R., Messer,C.J., Chew,A., Han,J.H., Duan,J., Carr,J.L., Lee,M.S., Koshy,B., Kumar,A.M., Zhang,G., Newell,W.R., Windemuth,A., Xu,C., Kalbfleisch,T.S., Shaner,S.L., Arnold,K., Schulz,V., Drysdale,C.M., Nandabalan,K., Judson,R.S., Ruano,G. and Vovis,G.F. (2001) Haplotype variation and linkage disequilibrium in 313 human genes. Science, 293, 489–493. [DOI] [PubMed] [Google Scholar]
- 6.Wang D.G., Fan,J.B., Siao,C.J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J., Kruglyak,L., Stein,L., Hsie,L., Topaloglou,T., Hubbell,E., Robinson,E., Mittmann,M., Morris,M.S., Shen,N., Kilburn,D., Rioux,J., Nusbaum,C., Rozen,S., Hudson,T.J., Lander,E.S. et al. (1998) Large-scale identification, mapping and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarti A. (1999) Population genetics—making sense out of sequence. Nature Genet. (Suppl. 1), 21, 56–60. [DOI] [PubMed] [Google Scholar]
- 8.Lynch M. and Walsh,B. (1997) Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- 9.Nordborg M. and Tavare,S. (2002) Linkage disequilibrium: what history has to tell us. Trends Genet., 18, 83–90. [DOI] [PubMed] [Google Scholar]
- 10.Hoh J., Wille,A. and Ott,J. (2001) Trimming, weighting and grouping SNPs in human case-control association studies. Genome Res., 11, 2115–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daly M.J., Rioux,J.D., Schaffner,S.F., Hudson,T.J. and Lander,E.S. (2001) High-resolution haplotype structure in the human genome. Nature Genet., 29, 229–232. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein D.B. (2001) Islands of linkage disequilibrium. Nature Genet., 29, 109–111. [DOI] [PubMed] [Google Scholar]
- 13.Rioux J.D., Daly,M.J., Silverberg,M.S., Lindblad,K., Steinhart,H., Cohen,Z., Delmonte,T., Kocher,K., Miller,K., Guschwan,S., Kulbokas,E.J., O’Leary,S., Winchester,E., Dewar,K., Green,T., Stone,V., Chow,C., Cohen,A., Langelier,D., Lapointe,G., Gaudet,D., Faith,J., Branco,N., Bull,S.B., McLeod,R.S., Griffiths,A.M., Bitton,A., Greenberg,G.R., Lander,E.S., Siminovitch,K.A. and Hudson,T.J. (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nature Genet., 29, 223–228. [DOI] [PubMed] [Google Scholar]
- 14.Charlesworth B., Morgan,M.T. and Charlesworth,D. (1993) The effect of deleterious mutations on neutral molecular variation. Genetics, 134, 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maynard Smith J. and Haigh,J. (1974) The hitchhiking effect of a favorable gene. Genet. Res., 23, 23–35. [PubMed] [Google Scholar]
- 16.Begun D.J. and Aquadro,C.F. (1991) Molecular population genetics of the distal portion of the X chromosome in Drosophila: evidence for genetic hitchhiking of the yellow-achaete region. Genetics, 129, 1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berry A.J., Ajioka,J.W. and Kreitman,M. (1991) Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics, 129, 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charlesworth B. (1996) Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res., 68, 131–149. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. (1974) The evolutionary advantage of recombination. Genetics, 78, 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill W.G. and Robertson,A. (1966) The effect of linkage on limits to artificial selection. Genet. Res., 8, 269–294. [PubMed] [Google Scholar]
- 21.Comeron J.M. and Kreitman,M. (2000) The correlation between intron length and recombination in Drosophila. Dynamic equilibrium between mutational and selective forces. Genetics, 156, 1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader J.S. (2001) The relative power of SNPs and haplotype as genetic markers for association tests. Pharmacogenomics, 2, 11–24. [DOI] [PubMed] [Google Scholar]
- 23.Judson R. and Stephens,J.C. (2001) Notes from the SNP vs. haplotype front. Pharmacogenomics, 2, 7–10. [DOI] [PubMed] [Google Scholar]
- 24.Bacanu S.A., Devlin,B. and Roeder,K. (2000) The power of genomic control. Am. J. Hum. Genet., 66, 1933–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risch N.J. (2000) Searching for genetic determinants in the new millennium. Nature, 405, 847–856. [DOI] [PubMed] [Google Scholar]
- 26.Reich D.E., Cargill,M., Bolk,S., Ireland,J., Sabeti,P.C., Richter,D.J., Lavery,T., Kouyoumjian,R., Farhadian,S.F., Ward,R. and Lander,E.S. (2001) Linkage disequilibrium in the human genome. Nature, 411, 199–204. [DOI] [PubMed] [Google Scholar]
- 27.Shifman S. and Darvasi,A. (2001) The value of isolated populations. Nature Genet., 28, 309–310. [DOI] [PubMed] [Google Scholar]
- 28.Pritchard J.K. and Rosenberg,N.A. (1999) Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet., 65, 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terwilliger J.D. and Goring,H.H. (2000) Gene mapping in the 20th and 21st centuries: statistical methods, data analysis and experimental design. Hum. Biol., 72, 63–132. [PubMed] [Google Scholar]
- 30.Abbott A. (2000) Manhattan versus Reykjavik. Nature, 406, 340–342. [DOI] [PubMed] [Google Scholar]
- 31.Kruglyak L. (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet., 22, 139–144. [DOI] [PubMed] [Google Scholar]
- 32.Eaves I.A., Merriman,T.R., Barber,R.A., Nutland,S., Tuomilehto-Wolf,E., Tuomilehto,J., Cucca,F. and Todd,J.A. (2000) The genetically isolated populations of Finland and sardinia may not be a panacea for linkage disequilibrium mapping of common disease genes. Nature Genet., 25, 320–323. [DOI] [PubMed] [Google Scholar]
- 33.Taillon-Miller P., Bauer-Sardina,I., Saccone,N.L., Putzel,J., Laitinen,T., Cao,A., Kere,J., Pilia,G., Rice,J.P. and Kwok,P.Y. (2000) Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nature Genet., 25, 324–328. [DOI] [PubMed] [Google Scholar]
- 34.Hewett D., Samuelsson,L., Polding,J., Enlund,F., Cantone,K., See,C.G., Smart,D., Chadha,S., Inerot,A., Enerback,C., Montgomery,D., Chavda,S., Christodolou,C., Robinson,P., Matthews,P., Plumpton,M., Dykes,C., Wahlstrom,J., Swanbeck,G., Martinsson,T., Roses,A., Riley,J. and Purvis,I. (2002) Identification of a psoriasis susceptibility candidate gene by linkage disequilibrium mapping with a localized single nucleotide polymorphism map. Genomics, 79, 305–314. [DOI] [PubMed] [Google Scholar]
- 35.Martin E.R., Lai,E.H., Gilbert,J.R., Rogala,A.R., Afshari,A.J., Riley,J., Finch,K.L., Stevens,J.F., Livak,K.J., Slotterbeck,B.D., Slifer,S.H., Warren,L.L., Conneally,P.M., Schmechel,D.E., Purvis,I., Pericak-Vance,M.A., Roses,A.D. and Vance,J.M. (2000) SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am. J. Hum. Genet., 67, 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin E.R., Scott,W.K., Nance,M.A., Watts,R.L., Hubble,J.P., Koller,W.C., Lyons,K., Pahwa,R., Stern,M.B., Colcher,A., Hiner,B.C., Jankovic,J., Ondo,W.G., Allen,F.H.,Jr, Goetz,C.G., Small,G.W., Masterman,D., Mastaglia,F., Laing,N.G., Stajich,J.M., Ribble,R.C., Booze,M.W., Rogala,A., Hauser,M.A., Zhang,F., Gibson,R.A., Middleton,L.T., Roses,A.D., Haines,J.L., Scott,B.L., Pericak-Vance,M.A. and Vance,J.M. (2001) Association of single-nucleotide polymorphisms of the tau gene with late-onset Parkinson disease. JAMA, 286, 2245–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy L.C., Hosford,D.A., Riley,J.H., Bird,M.I., White,N.J., Hewett,D.R., Peroutka,S.J., Griffiths,L.R., Boyd,P.R., Lea,R.A., Bhatti,S.M., Hosking,L.K., Hood,C.M., Jones,K.W., Handley,A.R., Rallan,R., Lewis,K.F., Yeo,A.J., Williams,P.M., Priest,R.C., Khan,P., Donnelly,C., Lumsden,S.M., O’Sullivan,J., See,C.G., Smart,D.H., Shaw-Hawkins,S., Patel,J., Langrish,T.C., Feniuk,W., Knowles,R.G., Thomas,M., Libri,V., Montgomery,D.S., Manasco,P.K., Xu,C.F., Dykes,C., Humphrey,P.P., Roses,A.D. and Purvis,I.J. (2001) Single-nucleotide polymorphism alleles in the insulin receptor gene are associated with typical migraine. Genomics, 78, 135–149. [DOI] [PubMed] [Google Scholar]
- 38.Scott W.K., Nance,M.A., Watts,R.L., Hubble,J.P., Koller,W.C., Lyons,K., Pahwa,R., Stern,M.B., Colcher,A., Hiner,B.C., Jankovic,J., Ondo,W.G., Allen,F.H.,Jr, Goetz,C.G., Small,G.W., Masterman,D., Mastaglia,F., Laing,N.G., Stajich,J.M., Slotterbeck,B., Booze,M.W., Ribble,R.C., Rampersaud,E., West,S.G., Gibson,R.A., Middleton,L.T., Roses,A.D., Haines,J.L., Scott,B.L., Vance,J.M. and Pericak-Vance,M.A. (2001) Complete genomic screen in Parkinson disease: evidence for multiple genes. JAMA, 286, 2239–2244. [DOI] [PubMed] [Google Scholar]
- 39.Altshuler D., Daly,M. and Kruglyak,L. (2000) Guilt by association. Nature Genet., 26, 135–137. [DOI] [PubMed] [Google Scholar]
- 40.Horikawa Y., Oda,N., Cox,N.J., Li,X., Orho-Melander,M., Hara,M., Hinokio,Y., Lindner,T.H., Mashima,H., Schwarz,P.E., del Bosque-Plata,L., Oda,Y., Yoshiuchi,I., Colilla,S., Polonsky,K.S., Wei,S., Concannon,P., Iwasaki,N., Schulze,J., Baier,L.J., Bogardus,C., Groop,L., Boerwinkle,E., Hanis,C.L. and Bell,G.I. (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nature Genet., 26, 163–175. [DOI] [PubMed] [Google Scholar]
- 41.Roses A.D. (2000) Pharmacogenetics and the practice of medicine. Nature, 405, 857–865. [DOI] [PubMed] [Google Scholar]
- 42.Collins A., Lonjou,C. and Morton,N.E. (1999) Genetic epidemiology of single-nucleotide polymorphisms. Proc. Natl Acad. Sci. USA, 96, 15173–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roses A. (2002) Pharmacogenetics place in modern medical science and practice. Life Sci., 70, 1471–1480. [DOI] [PubMed] [Google Scholar]
- 44.Kruglyak L. and Nickerson,D.A. (2001) Variation is the spice of life. Nature Genet., 27, 234–236. [DOI] [PubMed] [Google Scholar]
- 45.Sachidanandam R., Weissman,D., Schmidt,S.C., Kakol,J.M., Stein,L.D., Marth,G., Sherry,S., Mullikin,J.C., Mortimore,B.J., Willey,D.L., Hunt,S.E., Cole,C.G., Coggill,P.C., Rice,C.M., Ning,Z., Rogers,J., Bentley,D.R., Kwok,P.Y., Mardis,E.R., Yeh,R.T., Schultz,B., Cook,L., Davenport,R., Dante,M., Fulton,L., Hillier,L., Waterston,R.H., McPherson,J.D., Gilman,B., Schaffner,S., Van Etten,W.J., Reich,D., Higgins,J., Daly,M.J., Blumenstiel,B., Baldwin,J., Stange-Thomann,N., Zody,M.C., Linton,L., Lander,E.S. and Atshuler,D. (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 46.Abeliovich D., Kaduri,L., Lerer,I., Weinberg,N., Amir,G., Sagi,M., Zlotogora,J., Heching,N. and Peretz,T. (1997) The founder mutations 185delAG and 5382insC in BRCA1 and 6174delT in BRCA2 appear in 60% of ovarian cancer and 30% of early-onset breast cancer patients among Ashkenazi women. Am. J. Hum. Genet., 60, 505–514. [PMC free article] [PubMed] [Google Scholar]
- 47.Beller U., Halle,D., Catane,R., Kaufman,B., Hornreich,G. and Levy-Lahad,E. (1997) High frequency of BRCA1 and BRCA2 germline mutations in Ashkenazi Jewish ovarian cancer patients, regardless of family history. Gynecol. Oncol., 67, 123–126. [DOI] [PubMed] [Google Scholar]
- 48.Berman D.B., Costalas,J., Schultz,D.C., Grana,G., Daly,M. and Godwin,A.K. (1996) A common mutation in BRCA2 that predisposes to a variety of cancers is found in both Jewish Ashkenazi and non-Jewish individuals. Cancer Res., 56, 3409–3414. [PubMed] [Google Scholar]
- 49.Laken S.J., Petersen,G.M., Gruber,S.B., Oddoux,C., Ostrer,H., Giardiello,F.M., Hamilton,S.R., Hampel,H., Markowitz,A., Klimstra,D., Jhanwar,S., Winawer,S., Offit,K., Luce,M.C., Kinzler,K.W. and Vogelstein,B. (1997) Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nature Genet., 17, 79–83. [DOI] [PubMed] [Google Scholar]
- 50.Oddoux C., Struewing,J.P., Clayton,C.M., Neuhausen,S., Brody,L.C., Kaback,M., Haas,B., Norton,L., Borgen,P., Jhanwar,S., Goldgar,D., Ostrer,H. and Offit,K. (1996) The carrier frequency of the BRCA2 6174delT mutation among Ashkenazi Jewish individuals is approximately 1%. Nature Genet., 14, 188–190. [DOI] [PubMed] [Google Scholar]
- 51.Roa B.B., Boyd,A.A., Volcik,K. and Richards,C.S. (1996) Ashkenazi Jewish population frequencies for common mutations in BRCA1 and BRCA2. Nature Genet., 14, 185–187. [DOI] [PubMed] [Google Scholar]
- 52.Struewing J.P., Abeliovich,D., Peretz,T., Avishai,N., Kaback,M.M., Collins,F.S. and Brody,L.C. (1995) The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nature Genet., 11, 198–200. [DOI] [PubMed] [Google Scholar]
- 53.Struewing J.P., Hartge,P., Wacholder,S., Baker,S.M., Berlin,M., McAdams,M., Timmerman,M.M., Brody,L.C. and Tucker,M.A. (1997) The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N. Engl. J. Med., 336, 1401–1408. [DOI] [PubMed] [Google Scholar]
- 54.Broll R., Stark,A., Windhovel,U., Best,R., Strik,M.W., Schimmelpenning,H., Schwandner,O., Kujath,P., Bruch,H.P. and Duchrow,M. (1999) Expression of p53 and mdm2 mRNA and protein in colorectal carcinomas. Eur. J. Cancer., 35, 1083–1088. [DOI] [PubMed] [Google Scholar]
- 55.Bunz F., Hwang,P.M., Torrance,C., Waldman,T., Zhang,Y., Dillehay,L., Williams,J., Lengauer,C., Kinzler,K.W. and Vogelstein,B. (1999) Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest., 104, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dameron K.M., Volpert,O.V., Tainsky,M.A. and Bouck,N. (1994) Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science, 265, 1582–1584. [DOI] [PubMed] [Google Scholar]
- 57.Heide I., Thiede,C., Sonntag,T., de Kant,E., Neubauer,A., Jonas,S., Peter,F.J., Neuhaus,P., Herrmann,R., Huhn,D. and Rochlitz,C.F. (1997) The status of p53 in the metastatic progression of colorectal cancer. Eur. J. Cancer., 33, 1314–1322. [DOI] [PubMed] [Google Scholar]
- 58.Prives C. and Hall,P.A. (1999) The p53 pathway. J. Pathol., 187, 112–126. [DOI] [PubMed] [Google Scholar]
- 59.Tortola S., Marcuello,E., Gonzalez,I., Reyes,G., Arribas,R., Aiza,G., Sancho,F.J., Peinado,M.A. and Capella,G. (1999) p53 and K-ras gene mutations correlate with tumor aggressiveness but are not of routine prognostic value in colorectal cancer. J. Clin. Oncol., 17, 1375–1381. [DOI] [PubMed] [Google Scholar]
- 60.Zou Z., Gao,C., Nagaich,A.K., Connell,T., Saito,S., Moul,J.W., Seth,P., Appella,E. and Srivastava,S. (2000) p53 regulates the expression of the tumor suppressor gene maspin. J. Biol. Chem., 275, 6051–6054. [DOI] [PubMed] [Google Scholar]
- 61.Aurelio O.N., Cajot,J.F., Hua,M.L., Khwaja,Z. and Stanbridge,E.J. (1998) Germ-line-derived hinge domain p53 mutants have lost apoptotic but not cell cycle arrest functions. Cancer Res., 58, 2190–2195. [PubMed] [Google Scholar]
- 62.Aurelio O.N., Kong,X.T., Gupta,S. and Stanbridge,E.J. (2000) p53 mutants have selective dominant-negative effects on apoptosis but not growth arrest in human cancer cell lines. Mol. Cell. Biol., 20, 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foster B.A., Coffey,H.A., Morin,M.J. and Rastinejad,F. (1999) Pharmacological rescue of mutant p53 conformation and function. Science, 286, 2507–2510. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y., Blandino,G., Oren,M. and Givol,D. (1998) Induced p53 expression in lung cancer cell line promotes cell senescence and differentially modifies the cytotoxicity of anti-cancer drugs. Oncogene, 17, 1923–1930. [DOI] [PubMed] [Google Scholar]
- 65.Webley K.M., Shorthouse,A.J. and Royds,J.A. (2000) Effect of mutation and conformation on the function of p53 in colorectal cancer. J. Pathol., 191, 361–367. [DOI] [PubMed] [Google Scholar]
- 66.Tietz N.W. (1986) Textbook of Clinical Chemistry. W.B. Saunders Company, Philadelphia, PA.
- 67.Wallach J. (2000) Interpretation of Diagnostic Tests. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 68.Patil N., Berno,A.J., Hinds,D.A., Barrett,W.A., Doshi,J.M., Hacker,C.R., Kautzer,C.R., Lee,D.H., Marjoribanks,C., McDonough,D.P., Nguyen,B.T., Norris,M.C., Sheehan,J.B., Shen,N., Stern,D., Stokowski,R.P., Thomas,D.J., Trulson,M.O., Vyas,K.R., Frazer,K.A., Fodor,S.P. and Cox,D.R. (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science, 294, 1719–1723. [DOI] [PubMed] [Google Scholar]
- 69.Buetow K.H., Edmonson,M., MacDonald,R., Clifford,R., Yip,P., Kelley,J., Little,D.P., Strausberg,R., Koester,H., Cantor,C.R. and Braun,A. (2001) High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl Acad. Sci. USA, 98, 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwok P.Y. (2000) Approaches to allele frequency determination. Pharmacogenomics, 1, 231–235. [DOI] [PubMed] [Google Scholar]
- 71.Shih I.M., Zhou,W., Goodman,S.N., Lengauer,C., Kinzler,K.W. and Vogelstein,B. (2001) Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res., 61, 818–822. [PubMed] [Google Scholar]
- 72.Thiagalingam S., Laken,S., Willson,J.K., Markowitz,S.D., Kinzler,K.W., Vogelstein,B. and Lengauer,C. (2001) Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc. Natl Acad. Sci. USA, 98, 2698–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Telenius H., Carter,N.P., Bebb,C.E., Nordenskjold,M., Ponder,B.A. and Tunnacliffe,A. (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics, 13, 718–725. [DOI] [PubMed] [Google Scholar]
- 74.Cheung V.G. and Nelson,S.F. (1996) Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc. Natl Acad. Sci. USA, 93, 14676–14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang L., Cui,X., Schmitt,K., Hubert,R., Navidi,W. and Arnheim,N. (1992) Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl Acad. Sci. USA, 89, 5847–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dean F.B., Hosono,S., Fang,L., Wu,X., Faruqi,A.F., Bray-Ward,P., Sun,Z., Zong,Q., Du,Y., Du,J., Driscoll,M., Song,W., Kingsmore,S.F., Egholm,M. and Lasken,R.S. (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA, 99, 5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwok P.Y. (2001) Methods for genotyping single nucleotide polymorphisms. Annu. Rev. Genomics Hum. Genet., 2, 235–258. [DOI] [PubMed] [Google Scholar]
- 78.Syvanen A.C. (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nature Rev. Genet., 2, 930–942. [DOI] [PubMed] [Google Scholar]
- 79.Drmanac R., Drmanac,S. and Deane,L. (2000) Sequencing and fingerprinting DNA by hybridization with oligonucleotide probes. In Meyers,R.A. (ed.), Encyclopedia of Analytical Chemistry. John Wiley & Sons Ltd, Chichester, UK, pp. 5232–5257.
- 80.Hacia J.G., Brody,L.C., Chee,M.S., Fodor,S.P. and Collins,F.S. (1996) Detection of heterozygous mutations in BRCA1 using high density oligonucleotide arrays and two-colour fluorescence analysis. Nature Genet., 14, 441–447. [DOI] [PubMed] [Google Scholar]
- 81.Lipshutz R.J., Morris,D., Chee,M., Hubbell,E., Kozal,M.J., Shah,N., Shen,N., Yang,R. and Fodor,S.P. (1995) Using oligonucleotide probe arrays to access genetic diversity. Biotechniques, 19, 442–447. [PubMed] [Google Scholar]
- 82.Guo Z., Guilfoyle,R.A., Thiel,A.J., Wang,R. and Smith,L.M. (1994) Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res., 22, 5456–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prince J.A., Feuk,L., Howell,W.M., Jobs,M., Emahazion,T., Blennow,K. and Brookes,A.J. (2001) Robust and accurate single nucleotide polymorphism genotyping by dynamic allele-specific hybridization (DASH): design criteria and assay validation. Genome Res., 11, 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Radtkey R., Feng,L., Muralhidar,M., Duhon,M., Canter,D., DiPierro,D., Fallon,S., Tu,E., McElfresh,K., Nerenberg,M. and Sosnowski,R. (2000) Rapid, high fidelity analysis of simple sequence repeats on an electronically active DNA microchip. Nucleic Acids Res., 28, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sosnowski R.G., Tu,E., Butler,W.F., O’Connell,J.P. and Heller,M.J. (1997) Rapid determination of single base mismatch mutations in DNA hybrids by direct electric field control. Proc. Natl Acad. Sci. USA, 94, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 87.Marras S.A., Kramer,F.R. and Tyagi,S. (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal., 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 88.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 89.Bonnet G., Tyagi,S., Libchaber,A. and Kramer,F.R. (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA, 96, 6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Orum H., Nielsen,P.E., Egholm,M., Berg,R.H., Buchardt,O. and Stanley,C. (1993) Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res., 21, 5332–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou W., Galizia,G., Lieto,E., Goodman,S.N., Romans,K.E., Kinzler,K.W., Vogelstein,B., Choti,M.A. and Montgomery,E.A. (2001) Counting alleles reveals a connection between chromosome 18q loss and vascular invasion. Nat. Biotechnol., 19, 78–81. [DOI] [PubMed] [Google Scholar]
- 92.Vogelstein B. and Kinzler,K.W. (1999) Digital PCR. Proc. Natl Acad. Sci. USA, 96, 9236–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Syvanen A.C. (1999) From gels to chips: ‘minisequencing’ primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum. Mutat., 13, 1–10. [DOI] [PubMed] [Google Scholar]
- 94.Syvanen A.C., Aalto-Setala,K., Harju,L., Kontula,K. and Soderlund,H. (1990) A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics, 8, 684–692. [DOI] [PubMed] [Google Scholar]
- 95.Nyren P., Pettersson,B. and Uhlen,M. (1993) Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal. Biochem., 208, 171–175. [DOI] [PubMed] [Google Scholar]
- 96.Lyamichev V.I., Kaiser,M.W., Lyamicheva,N.E., Vologodskii,A.V., Hall,J.G., Ma,W.P., Allawi,H.T. and Neri,B.P. (2000) Experimental and theoretical analysis of the invasive signal amplification reaction. Biochemistry, 39, 9523–9532. [DOI] [PubMed] [Google Scholar]
- 97.Kwiatkowski R.W., Lyamichev,V., de Arruda,M. and Neri,B. (1999) Clinical, genetic and pharmacogenetic applications of the Invader assay. Mol. Diagn., 4, 353–364. [DOI] [PubMed] [Google Scholar]
- 98.Hsu T.M., Law,S.M., Duan,S., Neri,B.P. and Kwok,P.Y. (2001) Genotyping single-nucleotide polymorphisms by the invader assay with dual-color fluorescence polarization detection. Clin. Chem., 47, 1373–1377. [PubMed] [Google Scholar]
- 99.Cooksey R.C., Holloway,B.P., Oldenburg,M.C., Listenbee,S. and Miller,C.W. (2000) Evaluation of the invader assay, a linear signal amplification method, for identification of mutations associated with resistance to rifampin and isoniazid in Mycobacterium tuberculosis. Antimicrob. Agents Chemother., 44, 1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Agarwal P., Oldenburg,M.C., Czarneski,J.E., Morse,R.M., Hameed,M.R., Cohen,S. and Fernandes,H. (2000) Comparison study for identifying promoter allelic polymorphism in interleukin 10 and tumor necrosis factor alpha genes. Diagn. Mol. Pathol., 9, 158–164. [DOI] [PubMed] [Google Scholar]
- 101.Wilkins Stevens P., Hall,J.G., Lyamichev,V., Neri,B.P., Lu,M., Wang,L., Smith,L.M. and Kelso,D.M. (2001) Analysis of single nucleotide polymorphisms with solid phase invasive cleavage reactions. Nucleic Acids Res., 29, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barany F. (1991) Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl Acad. Sci. USA, 88, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Landegren U., Kaiser,R., Sanders,J. and Hood,L. (1988) A ligase-mediated gene detection technique. Science, 241, 1077–1080. [DOI] [PubMed] [Google Scholar]
- 104.Housby J.N. and Southern,E.M. (1998) Fidelity of DNA ligation: a novel experimental approach based on the polymerisation of libraries of oligonucleotides. Nucleic Acids Res., 26, 4259–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sriskanda V. and Shuman,S. (1998) Specificity and fidelity of strand joining by Chlorella virus DNA ligase. Nucleic Acids Res., 26, 3536–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shuman S. (1995) Vaccinia virus DNA ligase: specificity, fidelity and inhibition. Biochemistry, 34, 16138–16147. [DOI] [PubMed] [Google Scholar]
- 107.Tong J., Barany,F. and Cao,W. (2000) Ligation reaction specificities of an NAD(+)-dependent DNA ligase from the hyperthermophile Aquifex aeolicus. Nucleic Acids Res., 28, 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tong J., Cao,W. and Barany,F. (1999) Biochemical properties of a high fidelity DNA ligase from Thermus species AK16D. Nucleic Acids Res., 27, 788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Luo J., Bergstrom,D.E. and Barany,F. (1996) Improving the fidelity of Thermus thermophilus DNA ligase. Nucleic Acids Res., 24, 3071–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barany F., Lubin,M. and Belgrader,P. (2000) US Patent 6,268,148.
- 111.Lizardi P.M., Huang,X., Zhu,Z., Bray-Ward,P., Thomas,D.C. and Ward,D.C. (1998) Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet., 19, 225–232. [DOI] [PubMed] [Google Scholar]
- 112.Baner J., Nilsson,M., Mendel-Hartvig,M. and Landegren,U. (1998) Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res., 26, 5073–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nilsson M., Malmgren,H., Samiotaki,M., Kwiatkowski,M., Chowdhary,B.P. and Landegren,U. (1994) Padlock probes: circularizing oligonucleotides for localized DNA detection. Science, 265, 2085–2088. [DOI] [PubMed] [Google Scholar]
- 114.Zhang D.Y., Brandwein,M., Hsuih,T.C. and Li,H. (1998) Amplification of target-specific, ligation-dependent circular probe. Gene, 211, 277–285. [DOI] [PubMed] [Google Scholar]
- 115.Fei Z., Ono,T. and Smith,L.M. (1998) MALDI–TOF mass spectrometric typing of single nucleotide polymorphisms with mass-tagged ddNTPs. Nucleic Acids Res., 26, 2827–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tang K., Opalsky,D., Abel,K., van den Boom,D., Yip,P., Del Mistro,G., Braun,A. and Cantor,C.R. (2002) Single nucleotide polymorphism analyses by MALDI–TOF MS. Intl J. Mass Spectrom., in press. [Google Scholar]
- 117.Haff L.A. and Smirnov,I.P. (1997) Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI–TOF mass spectrometry. Genome Res., 7, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fei Z. and Smith,L.M. (2000) Analysis of single nucleotide polymorphisms by primer extension and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom., 14, 950–959. [DOI] [PubMed] [Google Scholar]
- 119.Laken S.J., Jackson,P.E., Kinzler,K.W., Vogelstein,B., Strickland,P.T., Groopman,J.D. and Friesen,M.D. (1998) Genotyping by mass spectrometric analysis of short DNA fragments. Nat. Biotechnol., 16, 1352–1356. [DOI] [PubMed] [Google Scholar]
- 120.Tang K., Fu,D.J., Julien,D., Braun,A., Cantor,C.R. and Koster,H. (1999) Chip-based genotyping by mass spectrometry. Proc. Natl Acad. Sci. USA, 96, 10016–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gerry N.P., Witowski,N.E., Day,J., Hammer,R.P., Barany,G. and Barany,F. (1999) Universal DNA microarray method for multiplex detection of low abundance point mutations. J. Mol. Biol., 292, 251–262. [DOI] [PubMed] [Google Scholar]
- 122.Favis R., Day,J.P., Gerry,N.P., Phelan,C., Narod,S. and Barany,F. (2000) Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat. Biotechnol., 18, 561–564. [DOI] [PubMed] [Google Scholar]
- 123.Pastinen T., Raitio,M., Lindroos,K., Tainola,P., Peltonen,L. and Syvanen,A.C. (2000) A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res., 10, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Iannone M.A., Taylor,J.D., Chen,J., Li,M.S., Rivers,P., Slentz-Kesler,K.A. and Weiner,M.P. (2000) Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry, 39, 131–140. [PubMed] [Google Scholar]
- 125.Chen J., Iannone,M.A., Li,M.S., Taylor,J.D., Rivers,P., Nelsen,A.J., Slentz-Kesler,K.A., Roses,A. and Weiner,M.P. (2000) A microsphere-based assay for multiplexed single nucleotide polymorphism analysis using single base chain extension. Genome Res., 10, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang L., Tran,D.K. and Wang,X. (2001) BADGE, BeadsArray for the Detection of Gene Expression, a high-throughput diagnostic bioassay. Genome Res., 11, 1888–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iannone M.A., Consler,T.G., Pearce,K.H., Stimmel,J.B., Parks,D.J. and Gray,J.G. (2001) Multiplexed molecular interactions of nuclear receptors using fluorescent microspheres. Cytometry, 44, 326–337. [DOI] [PubMed] [Google Scholar]
- 128.Han M., Gao,X., Su,J.Z. and Nie,S. (2001) Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol., 19, 631–635. [DOI] [PubMed] [Google Scholar]
- 129.Steemers F.J., Ferguson,J.A. and Walt,D.R. (2000) Screening unlabeled DNA targets with randomly ordered fiber-optic gene arrays. Nat. Biotechnol., 18, 91–94. [DOI] [PubMed] [Google Scholar]
- 130.Shaw S.H., Carrasquillo,M.M., Kashuk,C., Puffenberger,E.G. and Chakravarti,A. (1998) Allele frequency distributions in pooled DNA samples: applications to mapping complex disease genes. Genome Res., 8, 111–123. [DOI] [PubMed] [Google Scholar]
- 131.Chadwick R.B., Conrad,M.P., McGinnis,M.D., Johnston-Dow,L., Spurgeon,S.L. and Kronick,M.N. (1996) Heterozygote and mutation detection by direct automated fluorescent DNA sequencing using a mutant Taq DNA polymerase. Biotechniques, 20, 676–683. [DOI] [PubMed] [Google Scholar]
- 132.Andreutti-Zaugg C., Scott,R.J. and Iggo,R. (1997) Inhibition of nonsense-mediated messenger RNA decay in clinical samples facilitates detection of human MSH2 mutations with an in vivo fusion protein assay and conventional techniques. Cancer Res., 57, 3288–3293. [PubMed] [Google Scholar]
- 133.Yan H., Kinzler,K.W. and Vogelstein,B. (2000) Tech.sight. Genetic testing—present and future. Science, 289, 1890–1892. [DOI] [PubMed] [Google Scholar]
- 134.Ganguly A. (2002) An update on conformation sensitive gel electrophoresis. Hum. Mutat., 19, 334–342. [DOI] [PubMed] [Google Scholar]
- 135.Ganguly A., Rock,M.J. and Prockop,D.J. (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc. Natl Acad. Sci. USA, 90, 10325–10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fodde R. and Losekoot,M. (1994) Mutation detection by denaturing gradient gel electrophoresis (DGGE). Hum. Mutat., 3, 83–94. [DOI] [PubMed] [Google Scholar]
- 137.Guldberg P. and Guttler,F. (1994) ‘Broad-range’ DGGE for single-step mutation scanning of entire genes: application to human phenylalanine hydroxylase gene. Nucleic Acids Res., 22, 880–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ridanpaa M. and Husgafvel-Pursiainen,K. (1993) Denaturing gradient gel electrophoresis (DGGE) assay for K-ras and N-ras genes: detection of K-ras point mutations in human lung tumour DNA. Hum. Mol. Genet., 2, 639–644. [DOI] [PubMed] [Google Scholar]
- 139.Ridanpaa M., Burvall,K., Zhang,L.H., Husgafvel-Pursiainen,K. and Onfelt,A. (1995) Comparison of DGGE and CDGE in detection of single base changes in the hamster hprt and human N-ras genes. Mutat. Res., 334, 357–364. [DOI] [PubMed] [Google Scholar]
- 140.Chen J. and Thilly,W.G. (1994) Use of denaturing-gradient gel electrophoresis to study chromium-induced point mutations in human cells. Environ. Health Perspect., 3, 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Khrapko K., Hanekamp,J.S., Thilly,W.G., Belenkii,A., Foret,F. and Karger,B.L. (1994) Constant denaturant capillary electrophoresis (CDCE): a high resolution approach to mutational analysis. Nucleic Acids Res., 22, 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sarkar G., Yoon,H.-S. and Sommer,S.S. (1992) Dideoxy fingerprinting (ddF): a rapid and efficient screen for the presence of mutations. Genomics, 13, 441–443. [DOI] [PubMed] [Google Scholar]
- 143.Liu B., Farrington,S.M., Petersen,G.M., Hamilton,S.R., Parsons,R., Papadopoulos,N., Fujiwara,T., Jen,J., Kinzler,K.W., Wyllie,A.H. et al. (1995) Genetic instability occurs in the majority of young patients with colorectal cancer. Nature Med., 1, 348–352. [DOI] [PubMed] [Google Scholar]
- 144.Underhill P.A., Jin,L., Lin,A.A., Mehdi,S.Q., Jenkins,T., Vollrath,D., Davis,R.W., Cavalli-Sforza,L.L. and Oefner,P.J. (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res., 7, 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hayashi K. (1991) PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl., 1, 34–38. [DOI] [PubMed] [Google Scholar]
- 146.Korn S.H., Moerkerk,P.T. and de Goeij,A.F. (1993) K-ras point mutations in routinely processed tissues: non-radioactive screening by single strand conformational polymorphism analysis. J. Clin. Pathol., 46, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Makino R., Yazyu,H., Kishimoto,Y., Sekiya,T. and Hayashi,K. (1992) F-SSCP: fluorescence-based polymerase chain reaction-single-strand conformation polymorphism (PCR–SSCP) analysis. PCR Methods Appl., 2, 10–13. [DOI] [PubMed] [Google Scholar]
- 148.Suzuki Y., Orita,M., Shiraishi,M., Hayashi,K. and Sekiya,T. (1990) Detection of ras gene mutations in human lung cancers by single-strand conformation polymorphism analysis of polymerase chain reaction products. Oncogene., 5, 1037–1043. [PubMed] [Google Scholar]
- 149.Larsen L.A., Johnson,M., Brown,C., Christiansen,M., Frank-Hansen,R., Vuust,J. and Andersen,P.S. (2001) Automated mutation screening using dideoxy fingerprinting and capillary array electrophoresis. Hum. Mutat., 18, 451–457. [DOI] [PubMed] [Google Scholar]
- 150.Kozlowski P. and Krzyzosiak,W.J. (2001) Combined SSCP/duplex analysis by capillary electrophoresis for more efficient mutation detection. Nucleic Acids Res., 29, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mitchelson K.R. (2001) The application of capillary electrophoresis for DNA polymorphism analysis. Methods Mol. Biol., 162, 3–26. [DOI] [PubMed] [Google Scholar]
- 152.Rozycka M., Collins,N., Stratton,M.R. and Wooster,R. (2000) Rapid detection of DNA sequence variants by conformation-sensitive capillary electrophoresis. Genomics, 70, 34–40. [DOI] [PubMed] [Google Scholar]
- 153.Ahrendt S., Halachmi,S., Chow,J., Wu,L., Halachmi,N., Yang,S., Wehage,S., Jen,J. and Sidransky,D. (1999) Rapid p53 sequence analysis in primary lung cancer using an oligonucleotide probe array. Proc. Natl Acad. Sci. USA, 96, 7382–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cotton R.G.H., Rodrigues,N.R. and Campbell,R.D. (1988) Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc. Natl Acad. Sci. USA, 85, 4397–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hansen L.L., Justesen,J. and Kruse,T.A. (1995) Detection of sequence variation by PCR. In Diefenbach,C.W. and Dveksler,G.S. (eds), PCR Primer: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, NY, pp. 275–286.
- 156.Haris I.I., Green,P.M., Bentley,D.R. and Giannelli,F. (1994) Mutation detection by fluorescent chemical cleavage: application to hemophilia B. PCR Methods Appl., 3, 268–271. [DOI] [PubMed] [Google Scholar]
- 157.Giunta C., Youil,R., Venter,D., Chow,C.W., Somers,G., Lafferty,A., Kemper,B. and Cotton,R.G. (1996) Rapid diagnosis of germline p53 mutation using the enzyme mismatch cleavage method. Diagn. Mol. Pathol., 5, 265–270. [DOI] [PubMed] [Google Scholar]
- 158.Xu J.F., Yang,Q.P., Chen,J.Y., van Baalen,M.R. and Hsu,I.C. (1996) Determining the site and nature of DNA mutations with the cloned MutY mismatch repair enzyme. Carcinogenesis, 17, 321–326. [DOI] [PubMed] [Google Scholar]
- 159.Youil R., Kemper,B.W. and Cotton,R.G. (1995) Screening for mutations by enzyme mismatch cleavage with T4 endonuclease VII. Proc. Natl Acad. Sci. USA, 92, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Smith J. and Modrich,P. (1996) Mutation detection with MutH, MutL and MutS mismatch repair proteins. Proc. Natl Acad. Sci. USA, 93, 4374–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huang J., Kirk,B., Favis,R., Soussi,T., Paty,P., Cao,W. and Barany,F. (2002) An endonuclease/ligase based mutation scanning method especially suited for analysis of neoplastic tissue. Oncogene, 21, 1909–1921. [DOI] [PubMed] [Google Scholar]
- 162.Yao M. and Kow,Y.W. (1994) Strand-specific cleavage of mismatch-containing DNA by deoxyinosine 3′-endonuclease from Escherichia coli. J. Biol. Chem., 269, 31390–31396. [PubMed] [Google Scholar]