Abstract
The U1 snRNP-A (U1A) protein has been known for many years as a component of the U1 snRNP. We have previously described a form of U1A present in human cells in significant amounts that is not associated with the U1 snRNP or U1 RNA but instead is part of a novel complex of non-snRNP proteins that we have termed snRNP-free U1A, or SF-A. Antibodies that specifically recognize this complex inhibit in vitro splicing and polyadenylation of pre-mRNA, suggesting that this complex may play an important functional role in these mRNA-processing activities. This finding was underscored by the determination that one of the components of this complex is the polypyrimidine-tract-binding protein-associated splicing factor, PSF. In order to further our studies on this complex and to determine the rest of the components of the SF-A complex, we prepared several stable HeLa cell lines that overexpress a tandem-affinity-purification-tagged version of U1A (TAP-tagged U1A). Nuclear extract was prepared from one of these cell lines, line 107, and affinity purification was performed along with RNase treatment. We have used mass spectrometry analysis to identify the candidate factors that associate with U1A. We have now identified and characterized PSF, p54nrb, and p68 as novel components of the SF-A complex. We have explored the function of this complex in RNA processing, specifically cleavage and polyadenylation, by performing immunodepletions followed by reconstitution experiments, and have found that p54nrb is critical.
Keywords: U1 snRNP, U1A, SF-A, polyadenylation
INTRODUCTION
The formation of mRNAs in the nuclei of eukaryotic cells involves several co- and post-transcriptional processing events, including 5′ end capping, RNA splicing, and 3′ end formation. 3′ end formation involves the tightly coupled steps of cleavage and polyadenylation. Polyadenylation stabilizes mRNA, enhances translation (Sachs et al. 1986; Ford et al. 1997; Wickens et al. 1997; Preiss and Hentze 1998), promotes transcription termination (Colgan and Manley 1997; Proudfoot 2004), and helps the transport of the mature mRNA to the cytosol (Huang and Carmichael 1996).
In mammalian cells, polyadenylation is dependent on a core upstream element with a consensus sequence of AAUAAA, or a close variant (Tian et al. 2005), located ~10–30 nt upstream of the cleavage site, and a core downstream element with a less conserved G/U- or U-rich sequence that serves as the binding site for cleavage stimulation factor (CstF). Cleavage and polyadenylation specificity factor (CPSF) specifically recognizes the core upstream element, and stimulates poly(A) polymerase (PAP). CstF binds to the core downstream element downstream of the cleavage site and is involved in the cleavage reaction. PAP adds adenosine residues to the 3′ ends of RNA. In addition to CPSF, CstF, and PAP, the protein factors involved in polyadenylation include cleavage factors I and II (CFI, CFII) and RNA polymerase II (Zhao et al. 1999; Edmonds 2002).
U small nuclear ribonucleoproteins (snRNPs; U1, U2, U4, U5, and U6) are RNA–protein complexes each of which contain small, U-rich, RNAs complexed with a set of seven Sm proteins and several particle-specific proteins (Will and Luhrmann 2001). Along with other less stably associated splicing factors, U snRNPs are assembled into the spliceosome to recognize and remove new introns emerging from the transcription machinery. Among the U snRNPs, the U1 snRNP plays a crucial role in 5′ splice site definition and choice. The U1 snRNP contains an RNA component (U1RNA), which interacts with the 5′-splice site via base-pairing. The U1 snRNP contains the Sm core proteins as do all the other U snRNPs, as well as U1-specific proteins: U1-70K, U1A, U1C (Lührmann et al. 1990).
The U1 snRNP specific polypeptide A (U1A) is a 32-kDa protein. It contains two RNA recognition motifs (RRM1 and RRM2), yet only RRM1 interacts specifically with stem–loop II of U1 RNA (Scherly et al. 1989, 1990; Lutz-Freyermuth et al. 1990; Allain et al. 1997). The C-terminal RRM2 of U1A does not bind to U1 RNA and no RNAs have been shown as yet to bind to it (Lu and Hall 1995; C.S. Lutz, unpubl.). U1A, as a specific protein in U1 snRNP, was first thought to be involved in an early step of splicing, but it is currently unknown what exact role U1A plays in splicing. Recent studies have revealed that U1A also influences polyadenylation. U1A autoregulation is one of the best-characterized examples of an “on/off switch” type of polyadenylation regulation (Gunderson et al. 1997). U1A also plays a positive role in supporting the interaction of the polyadenylation machinery with simian virus 40 late polyadenylation signal (SVL) (Lutz and Alwine 1994; Lutz et al. 1996). Anti-U1A antibodies inhibit SVL polyadenylation in vitro (Lutz and Alwine 1994; O’Connor et al. 1997). Interestingly, U1A does not need the involvement of U1 RNA or other components of the U1 snRNP to perform these functions. Taken together, these data suggest that U1A plays a more general role in pre-mRNA processing.
Although part of the U1 snRNP, a significant portion of the cellular U1A exists in a snRNP-free form in one or more novel complexes. The non-snRNP-associated form of U1A, called snRNP-free U1A (SF-A), was found to be complexed with a previously unrecognized set of non-snRNP proteins (O’Connor et al. 1997; Lutz et al. 1998). This SF-A complex migrated in a different series of fractions from the U1 snRNP in a 5%–30% sucrose gradient fractionation. A unique monoclonal antibody made against U1A, MAb 12E12, can recognize the SF-A complex, and this epitope is masked when U1A is bound to U1 RNA (O’Connor et al. 1997; Lutz et al. 2002). When MAb 12E12 was included in an in vitro processing reaction on SVL, both polyadenylation and splicing were inhibited (Lutz et al. 1998). One of the components of the SF-A complex was previously identified as PSF (polypyrimidine-tract-binding protein-associated splicing factor) (Lutz et al. 1998).
Recent progress in protein complex purification and mass spectrometry as well as the rapid growth of protein databases has allowed for the simple and efficient identification of the unknown components of complexes. Here we report the isolation and identification of the novel components of the SF-A complex using the tandem affinity purification (TAP) procedure (Rigaut et al. 1999) followed by MALDI-TOF/TOF. We have found that several auxiliary splicing factors—PSF, p54nrb, and p68 helicase—are also components of the SF-A complex. As a collection of auxiliary splicing factors yet also playing a functional role in polyadenylation, the SF-A complex may be a special adaptor between the processes of polyadenylation and splicing.
RESULTS
Identification of proteins interacting with U1A by tandem affinity purification
In order to identify the individual components of SF-A complex, we purified proteins associated with U1A using tandem affinity purification (TAP) (Rigaut et al. 1999). The TAP method is a double affinity chromatography purification technique that involves appending two epitopes to a protein of interest containing two immunoglobulin-binding domains of protein A from Staphylococcus aureus, a cleavage site for the tobacco etch virus (TEV) protease, and the calmodulin-binding peptide (CBP). The target protein, potentially in a complex with interacting factors, will be purified by two consecutive affinity chromatographic steps under mild conditions (Puig et al. 2001).
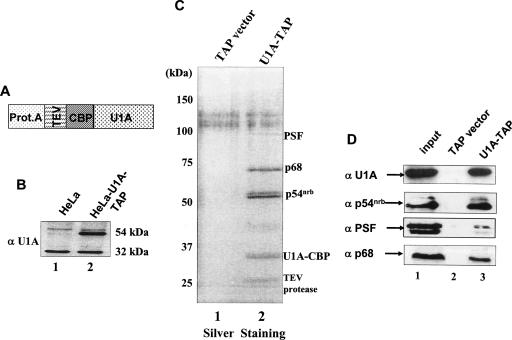
A plasmid expressing TAP-tagged U1A (Fig. 1A ▶) was transfected into HeLa cells. These U1A-TAP HeLa cell lines were clonally isolated as stable transfectants and maintained under puromycin selection continually. The TAP vector alone (empty vector) without a target protein was also transfected into HeLa cells at the same time to make a control stable cell line. The U1A-TAP stable cell line that was chosen for further analysis has an expression level of TAP-tagged U1A similar to that of endogenous U1A protein as visualized by Western blotting (Fig. 1B ▶). Nuclear extracts were prepared from the U1A-TAP stable cell line or the TAP vector cell line. Nuclear extracts prepared from both cell lines were active in in vitro polyadenylation reactions, demonstrating that the U1A-TAP does not alter the overall activity of this process (data not shown). For TAP purification, these nuclear extracts were next pretreated with RNase A, and then were applied to dual affinity chromatography according to the TAP protocol. The purified eluates contained a total of four main bands, including the target U1A-CBP protein, that were not present in TAP-vector-transfected cells (Fig. 1C ▶). These three non-U1A proteins were therefore candidates for U1A interacting proteins.
FIGURE 1.
Purification of U1A interacting proteins. (A) Schematic representation of the U1A-TAP construct. U1A cDNA was cloned into pZome-1N (Cellzome) so that an N-terminal TAP cassette was introduced in frame with the U1A cDNA. The TAP tag consisted of a calmodulin-binding peptide (CBP), a consensus TEV cleavage site (TEV), and the IgG-binding domain of protein A (Prot.A). (B) The U1A-TAP construct was transfected into HeLa cells; stable, clonally isolated cell lines were established; and aliquots of nuclear proteins from these stable cell lines were analyzed by Western blot analysis with the U1A-specific polyclonal antibody 310. Note that the tagged U1A migrated with ~20 kDa greater apparent mass than endogenous U1A. (C) Analysis of U1A interacting proteins. Proteins of TAP elutes from either TAPvector-transfected or U1A-TAP-transfected cells were resolved on a 12.5% SDS-PAGE gel and silver stained. The proteins in lane 2 were identified by mass spectrometry as indicated on the right. The nuclear extract used in this TAP purification is pretreated with RNase A (see Materials and Methods). The protein band at ~27 kDa is the recombinant TEV protease. (D) Verification of the identities of the proteins from TAP-U1A purification. Aliquots of TEV eluates were subjected to Western blot analysis with specific antibodies against p54nrb, PSF, and p68, respectively. Lane 1 in each panel represents 10% of input.
Bands representing putative U1A-binding proteins were excised and analyzed by MALDI-TOF/TOF. Comparison of the obtained peptide sequences with protein databases identified several proteins that previously had been linked to mRNA processing. Here we report the identification of three proteins—PSF, p54nrb, and p68 helicase—as interacting partners for U1A. The identification of these three proteins was based on peptide sequences obtained from each of the three samples individually showing complete identity to the human PSF, p54nrb, and p68 proteins. None of these proteins are defined components of the U1 snRNP, and the RNase A treatment prior to purification ensured that RNA did not bridge the interaction. The doublet that exists at ~54 kDa may correspond to two different forms of p54nrb; this is under investigation (data not shown). The isolation of this protein complex further suggested that U1A existed in one or more complexes other than U1 snRNP (O’Connor et al. 1997; Milcarek et al. 2003).
PSF had been previously identified as a component of the SF-A complex (Lutz et al. 1998). The presence of PSF in TAP eluates confirmed that PSF was a bona fide binding partner of U1A, and it also verified that TAP purification was an efficient way to purify associated proteins with the U1A target.
Both PSF and p54nrb were originally identified as splicing factors (Dong et al. 1993; Patton et al. 1993). PSF was first thought to be involved in defining the 3′-splice site, while p54nrb was initially considered as a splicing factor due to its high homology with the C-terminal region of PSF. We now know these two proteins exhibit multifunctional characteristics in a variety of nuclear process (Karhumaa et al. 2000; Zhang and Carmichael 2001; Shav-Tal and Zipori 2002; Kameoka et al. 2004; Song et al. 2005). The identification of p54nrb as an associated protein with U1A might suggest yet more functions to be ascribed to PSF and p54nrb.
p68 is a human DEAD-box helicase that plays an essential role in splicing (Liu 2002) and copurifies with pre-spliceosomes (Hartmuth et al. 2002).
To further validate the identities of these interacting proteins with U1A, aliquots of TEV eluates were used for Western blot analysis. Immunoblotting with U1A-, PSF-, p54nrb-, and p68-specific antibodies revealed the expected specific reactivity in the relevant eluate fractions (Fig. 1D ▶). We conclude that PSF, p54nrb, and p68 helicase were retained on the calmodulin beads by means of their direct interaction with U1A.
Analysis of the interactions among PSF, p54nrb, p68, and U1A by GST pull-down assay
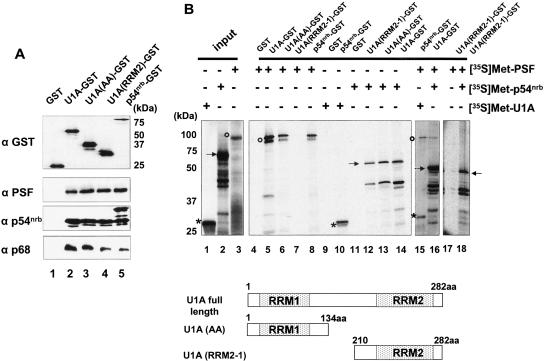
To investigate how proteins interact with each other and to begin to map interaction domains, we carried out GST pull-down assays with the aid of recombinant GST fusion proteins. The fusion proteins used in these experiments were the full-length GST-U1A; two U1A portions, GST-AA (U1A amino acids 1–134), and GST-RRM2-1 (U1A amino acids 210–282); and GST-p54nrb as can be seen in the Western blot using anti-GST antibody (Fig. 2A ▶, top). The fusion proteins were either incubated with nuclear extract, or with 35S-labeled candidate proteins prepared by in vitro transcription and translation in rabbit reticulocyte lysates (TNT; Promega). Under both conditions, RNase A was added to the reactions to avoid indirect interactions mediated by RNA. As shown in Figure 2A ▶ (bottom panels), PSF was selected from the nuclear extract by both GST-p54nrb and GST-U1A, including both RRM portions of U1A. p54nrb also coprecipitated with GST-U1A as well as with both portions of U1A. In addition, p54nrb also interacted with itself. In the GST pull-down experiments using in vitro transcribed and translated (TNT) products (Fig. 2B ▶), the in vitro translated, 35S-labeled p54nrb also coprecipitated with GST-U1A full-length as well as the two terminal parts of U1A, and vice versa (Fig. 2B ▶, lanes 12–14), which showed consistent results from the two different GST pull-down assays. However, no binding could be detected between 35S-labeled PSF and the C-terminal part of U1A (RRM2-1). The lack of binding using the 35S-PSF product in this experiment with U1A RRM2 may indicate that the interaction with U1A RRM2 is mediated through another protein, but this is only speculation at this point. The interaction of U1A RRM2 with PSF is not enhanced in the presence of p54nrb (Fig. 2B ▶, lanes 17,18). p54nrb is highly homologous to the C terminus of PSF (Dong et al. 1993; Yang et al. 1993). PSF and p54nrb were previously characterized as a heterodimer (Zhang et al. 1993), indicating that PSF and p54nrb are interacting proteins. This interaction was demonstrated again in our GST pull-down assays (Fig. 2B ▶, lane 8). In order to know whether PSF, p54nrb, and U1A were mutually influencing their binding to each other, we added all three proteins together in two independent GST pull-down assays. Under these conditions, these three proteins were retained together, suggesting that PSF, p54nrb, and U1A can interact simultaneously with each other (Fig. 2B ▶, lanes 15,16). These data are also consistent with the possibility that multiple dimeric complexes may coexist, and we cannot distinguish between these two possibilities at this time.
FIGURE 2.
Direct protein–protein interactions. (A) GST pull-down experiments with GST-tagged proteins and HeLa nuclear extract. Bacterially expressed GST, GST-U1A, GST-U1A (AA), GST-U1A (RRM2-1), or GST-p54nrb proteins (see Materials and Methods) individually were immobilized on glutathione-Sepharose beads and after purification were analyzed by 12.5% SDS-PAGE and Western blotting using an anti-GST antibody (top panel). These purified proteins were then incubated with HeLa nuclear extract. After extensive washing, proteins bound to the beads were eluted in protein sample buffer, resolved on 12.5% SDS-PAGE gels, and were analyzed by Western blotting. The antibodies used in the Western blot are indicated on the left. (B) GST pull-down experiments with 35S-labeled proteins. In vitro transcribed and translated [35S]Met-labeled U1A, p54nrb, and PSF (left) were tested for binding to GST, GST-U1A, GST-U1A (AA), GST-U1A (RRM2-1), and GST-p54nrb fusion proteins. PSF is indicated by open circles; p54nrb, by solid arrows; and U1A, by asterisks. A schematic representation of the structure of U1A and the two truncated mutants, U1A (AA) and U1A (RRM2-1), is shown below.
PSF, p54nrb, p68 helicase are components of the SF-A complex
From previous results, we have known that U1A exists in at least two distinct complexes (U1 snRNP bound and SF-A) (O’Connor et al. 1997; Lutz et al. 1998; Milcarek et al. 2003). The two forms of U1A migrated in different fractions on sucrose gradients. SF-A was found in fractions 7–11 of a 5%–30% sucrose gradient, while the U1 snRNP was found in fractions 17–23 (O’Connor et al. 1997). PSF was found in the same fractions where SF-A migrated, further indicative of its presence in the SF-A complex (Lutz et al. 1998).
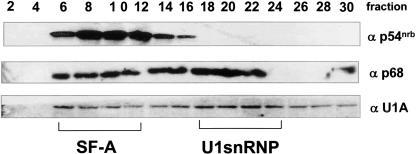
To detect whether p54nrb and p68 helicase co-sediment with U1A in the SF-A complex, we performed similar sucrose gradient fractionation using human 293T cell nucleoplasm, followed by Western blot analysis using p54nrb-, U1A-, and p68-specific antibodies (Fig. 3 ▶). The top of the gradient is fraction 1. p54nrb was enriched in fractions 6–12, where the SF-A complex migrated. Some of the p68 protein migrated along with the SF-A complex, while some p68 was found in fractions where the U1 snRNP migrated. Since recent research has revealed that p68 RNA helicase copurifies with pre-spliceosomes (Hartmuth et al. 2002), and that p68 functions in destabilizing the U1–5′-ss interactions, suggesting its role in the transition from pre-spliceosome to mature spliceosome (Liu 2002), we were not surprised to see p68 helicase also co-sedimenting with the U1 snRNP. We speculated that the presence of p68 helicase in the SF-A complex was dependent on protein–protein interactions, since it bound to U1A even in the presence of RNase A. Taken together, our results of sucrose gradient analysis further supported that p54nrb, p68, and PSF are components of the SF-A complex.
FIGURE 3.
Sucrose-gradient fractionation shows that p54nrb, p68, and U1A co-migrate in the SF-A-containing fractions. Sucrose gradients (5%–30%) were prepared using human 293T cell nucleoplasm. One-milliliter (1 mL) fractions were taken and 30 μL of every other fraction were separated by 12.5% SDS-PAGE and were blotted onto nitrocellulose. Three identical blots were probed with different antibodies: p54nrb, U1A 310, or p68 antibody. Smaller numbers represent fractions from the top of the gradient. The relative migration positions of the SF-A complex and the U1 snRNP are noted below.
Both PSF and p54nrb antibodies reduce in vitro polyadenylation activity
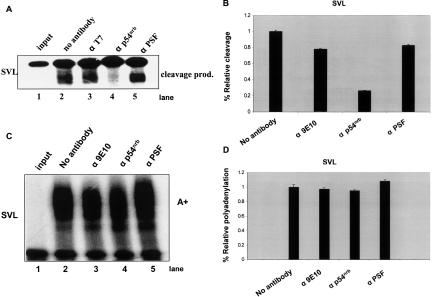
Since p54nrb and PSF are components of SF-A complex and antibodies to this complex inhibit polyadenylation (O’Connor et al. 1997), we next asked the possible role in polyadenylation that these two components individually might play. For this purpose, we included p54nrb monoclonal antibody and PSF antibody, as well as the c-myc tag 9E10 antibody as a control, into in vitro cleavage/polyadenylation reactions using the well-characterized SVL polyadenylation signal substrate RNA (dark bars in Fig. 4A,B ▶). We found that the p54nrb antibody strongly inhibited in vitro polyadenylation; the addition of p54nrb antibody to the reaction reduced the polyadenylation efficiency to only 15.7% as compared to the reaction without any antibody (Fig. 4B ▶). PSF antibody was less potent in this inhibition, but the PSF antibody caused a decrease of polyadenylation activity to about one-third of the control (Fig. 4B ▶), whereas the rabbit pre-bleed antibody had no effect on polyadenylation (Fig. 4C ▶). In contrast, the antibody 9E10 did not show any negative effect on polyadenylation, which confirmed that the inhibition was antibody-specific. To ensure that the inhibition was not only limited to SVL, the same in vitro cleavage/polyadenylation reactions were also performed using the well-characterized adenovirus L3 polyadenylation signal substrate RNA. The results were in agreement with reactions on the SVL substrate RNA (light gray bars in Fig. 4A,B ▶). It is also worth noting that the level of inhibition seen with the anti-p54nrb antibodies is comparable to that seen previously using a monoclonal U1A antibody that specifically recognizes the SF-A complex (O’Connor et al. 1997). Since the p54nrb and PSF antibodies caused a marked decrease in in vitro cleavage/polyadenylation reactions, we concluded that p54nrb and PSF affected polyadenylation in a significant way, but we wanted to further explore possible mechanisms for how this may take place.
FIGURE 4.
p54nrb and PSF are involved in in vitro polyadenylation. (A) Both p54nrb and PSF antibodies inhibited in vitro polyadenylation with SVL. Cleavage/polyadenylation reactions (see Materials and Methods) with SVL were performed in the presence or the absence of each of the antibodies as shown on the top of each lane. The reaction products were resolved on a 5% polyacrylamide gel containing 8 M urea and visualized by PhosphorImager analysis. Anti-c-myc-tag epitope antibody (9E10) was included as a control. (B) Quantitation of A. The same in vitro cleavage/polyadenylation reactions as panel B were repeated in three independent experiments on both SVL (dark bars) and L3 (gray bars). Polyadenylation efficiency was quantitated by PhosphorImager analysis and ImageQuant software. Percent relative polyadenylation was represented by the polyadenylation efficiency in the presence of antibodies vs. the total input RNA. Error bars represent SD. (C) Cleavage/polyadenylation reactions on SVL substrate RNAs were performed in the presence of the rabbit pre-bleed (pre) or anti-PSF antisera.
p54nrb antibody inhibits in vitro cleavage, but PSF antibody does not
Co- or post-transcriptional cleavage of mRNA precursor is an essential step in mRNA maturation, and is also the initial step of the 3′ end formation process. It involves the assembly of a functional cleavage/polyadenylation complex with cooperative interactions between the protein components and RNA cis-element sequences (Wilusz et al. 1990; Gilmartin and Nevins 1991; Murthy and Manley 1995; Wahle 1995; Ruegsegger et al. 1996). Based on the information that reconstitution of the poly(A) tail addition alone in vitro requires only purified CPSF and PAP (Bienroth et al. 1993), most regulation probably takes place during the cleavage step. Using cordycepin 5′-triphosphate, a nonhydrolyzable analog of ATP, we turned to cleavage assays performed in vitro to test whether PSF and p54nrb antibodies inhibited polyadenylation because of a reduction of the cleavage step. Again, the two antibodies were included separately in reactions, and cordycepin was included in the reaction. The cleavage product alone was seen when no antibody was added (Fig. 5A ▶, lane 2). Less than 30% of cleavage activity remained in the presence of p54nrb antibody in in vitro cleavage reactions on SVL (Fig. 5A ▶, lane 4), which suggested that this antibody dramatically reduced cleavage. To our surprise, the PSF antibody did not seem to have any effect on in vitro cleavage (Fig. 5A ▶, lane 5). Quantification of these results can be found in Figure 5B ▶. This may suggest that PSF does not play a role in the cleavage process.
FIGURE 5.
p54nrb antibody inhibits in vitro cleavage on SVL. (A) In vitro cleavage reactions (see Materials and Methods). The cleavage reactions were performed in the presence or the absence of antibodies to p54nrb or PSF, respectively. T7 antibody represents anti-T7 gene antibody (Novagen) (Lutz-Freyermuth et al. 1990). Cleavage products are indicated on the right. (B) Cleavage efficiency was quantitated by PhosphorImager analysis and ImageQuant software and represented the same way as relative polyadenylation. Standard deviation was calculated from three independent experiments. (C) p54nrb and PSF antibodies did not show significant inhibition on in vitro polyadenylation with precleaved SVL. 32P-labeled SVL RNA was in vitro transcribed from HpaI-linearized pSP65-SVL, and similar polyadenylation reactions were performed as shown in Figure 4B ▶. (D) The polyadenylation efficiency was quantitated by the polyadenylation efficiency in the presence of antibody vs. the absence of antibody. Error bars represent SD.
To separate the 3′ end processing into two independent reactions, cleavage and poly(A) addition, a precleaved SVL transcript was made, so that the cleavage was a dispensable step (see Materials and Methods). Using this precleaved SVL transcript, a similar in vitro polyadenylation reaction was performed (Fig. 5C,D ▶). The addition of either p54nrb antibody or PSF antibody had little effect on polyadenylation of a precleaved RNA, suggesting that p54nrb affected polyadenylation by affecting the cleavage step only.
Immunodepletions and purified protein reconstitutions can rescue polyadenylation activity
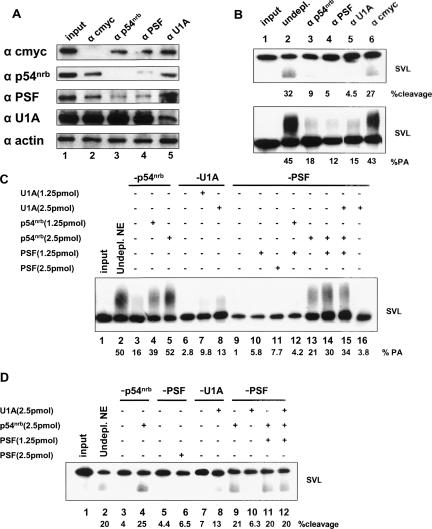
The antibody addition experiments were but a first step in understanding the role that these proteins may play in 3′ end formation of mRNAs. We next chose to use immunodepletions followed by reconstitutions with purified, recombinant proteins. Figure 6A ▶ shows the analysis of the immunodepleted nuclear extracts by Western blotting. The antibodies used in the individual immunodepletions are shown at the top of the panel, while the antibodies used for Western blotting are shown on the left. It is apparent that most antibodies specifically depleted the extract only for the protein for which each antibody is specific. Notably, an exception to this is found in Figure 6A ▶, lanes 3 and 4, where the antibody for p54nrb also depleted the extract of PSF and vice versa. Anti-actin antibody was used as a negative control for the Western blot (Fig. 6A ▶, bottom panel), and anti-c-myc was used as a negative control for immunodepletion (lane 2).
FIGURE 6.
Immunodepletion and reconstitution experiments reveal a critical role for p54nrb in polyadenylation. (A) Western blot of immunodepleted extracts. The specific proteins that were immunodepleted in each case are listed at the top of the figure; the antibodies used for Western blotting are shown to the left. Ten micrograms (10 μg) of nuclear extract was loaded in each lane. (B) Immunodepleted extracts were tested for their activity in cleavage alone (top) or cleavage/ polyadenylation (bottom). The percentages shown below are quantification of percent cleavage (top) or percent polyadenylation (bottom) of this particular experiment; upon three independent repeats of this experiment very similar results were obtained. (C) Reconstitution experiments of purified recombinant protein(s) to immunodepleted extracts, then used in in vitro polyadenylation reactions with SVL substrate RNA. (Lane 1) Input SVL RNA; (lane 2) regular, nondepleted nuclear extract; (lanes 3–5) p54nrb-depleted extract; (lanes 4,5) 1.25 and 2.5 pmol of purified GST-p54nrb additions, respectively; (lanes 6–8) U1A-depleted extract; (lanes 7,8) 1.25 and 2.5 pmol of purified GST-U1A additions, respectively; (lanes 9–16) PSF-depleted extracts; (lanes 10,11) 1.25 and 2.5 pmol of purified His-PSF additions, respectively; (lane 12) 1.25 pmol of His-PSF + 1.25 pmol of GST-p54nrb reconstitutions; (lane 13) 2.5 pmol of GST-p54nrb added back; (lane 14) 1.25 pmol of His-PSF + 2.5 pmol of GST-p54nrb reconstitutions; (lane 15) 1.25 pmol of His-PSF + 2.5 pmol of GST-p54nrb + 2.5 pmol of GST-U1A reconstitutions; (lane 16) 2.5 pmol of GST-U1A reconstitution. (D) Reconstitution experiments of purified recombinant protein(s) to immunodepleted extracts, then used in in vitro cleavage reactions with SVL substrate RNA. (Lane 1) Input SVL RNA; (lane 2) regular nuclear extract; (lanes 3,4) p54nrb-depleted nuclear extract (lane 4 also includes 2.5 pmol of GST-p54nrb reconstitutions); (lanes 5,6) PSF-depleted nuclear extract (lane 6 also includes 2.5 pmol of purified His-PSF); (lanes 7,8) U1A-depleted nuclear extract (lane 8 also includes 2.5 pmol of purified GST-U1A); (lanes 9–12) PSF-depleted nuclear extract (lane 10 also includes purified 2.5 pmol of GST-U1A, lane 11 also includes 1.25 pmol of His- PSF + 2.5 pmol of GST-p54nrb reconstitutions, lane 12 also includes 1.25 pmol of His-PSF + 2.5 pmol of p54nrb + 2.5 pmol of GST-U1A reconstitutions).
These immunodepleted extracts were next used in in vitro cleavage and polyadenylation reactions to test the functionality of the extracts on SVL substrate RNA (Fig. 6B ▶). Regular SVL polyadenylation signal substrate RNAs were used here, not precleaved. When extracts were immunodepleted by anti-p54nrb-, anti-PSF-, or anti-U1A-specific antibodies, in vitro cleavage reactions were reduced dramatically (Fig. 6B ▶, cf. top panel lanes 3–5 and lane 2 with no antibodies added). The c-myc immunodepleted extract had little effect on the cleavage reaction as expected. The same held true for the in vitro polyadenylation reactions (Fig. 6B ▶, bottom panel), where p54nrb, PSF, and U1A immunodepleted extracts had significantly reduced polyadenylation activity (lanes 3–5).
Next we added purified, recombinant proteins—individually and in combinations—to the immunodepleted extracts, and performed in vitro cleavage/polyadenylation assays (Fig. 6C ▶) and in vitro cleavage reactions alone (Fig. 6D ▶). We found that the addition of recombinant PSF rescued neither in vitro polyadenylation nor cleavage to any appreciable extent (Fig. 6C ▶, lanes 10,11; Fig. 6D ▶, lane 6). Recombinant U1A rescued in vitro polyadenylation and cleavage to some extent (Fig. 6C ▶, lanes 7,8 ; Fig. 6D ▶, lane 8), but the addition of recombinant p54nrb restored both polyadenylation and cleavage to pre-immunodepletion levels (Fig. 6C ▶, lanes 4,5; Fig. 6D ▶, lane 4). As shown in Figure 6A ▶, lane 4, immunodepletion of PSF also significantly reduced the levels of p54nrb. The addition of recombinant PSF alone to a PSF-depleted extract was thus not sufficient to restore cleavage and polyadenylation. We therefore added back recombinant PSF and p54nrb in combinations. The amount of recombinant PSF protein added relative to the amount of p54nrb was found to be very important; 1.25 pmol of p54nrb did not rescue polyadenylation, whereas 2.5 pmol did (Fig. 6C ▶ [cf. lanes 12 and 14]; Fig. 6D ▶, lane 11). This highlights the importance of p54nrb in our in vitro polyadenylation reactions. The combination of recombinant PSF, p54nrb, and U1A also restored in vitro polyadenylation and cleavage to nearly wild-type levels in a PSF-immunodepleted extract (Fig. 6C ▶, lane 15; Fig. 6D ▶, lane 12). These data suggest that p54nrb plays an important and not previously known role in 3′ end formation of mRNA.
DISCUSSION
Most of the fundamental functions of RNA metabolism in eukaryotic cells are carried out by multiprotein complexes such as those involved in transcription, splicing, turnover, and polyadenylation (for reviews, see Hirose and Manley 2000; Lemon and Tijan 2000). It has recently been established that these co- and post-transcriptional processing events are intimately linked (for review, see Neugebauer and Roth 1997; Lemon and Tijan 2000; Maniatis and Reed 2002; Proudfoot 2004). U snRNPs are RNA–protein complexes that are essential to mRNA splicing, and the U1A protein was first identified as a component of the U1 snRNP. We now know that at least 3%, and perhaps as much as 16%, of cellular U1A exists in the SF-A complex (O’Connor et al. 1997; Milcarek et al. 2003). The existence of the SF-A complex certainly suggests important roles that the complex may play.
We chose to use TAP purification to identify other components of the SF-A complex because of the high purity, low contaminants, and mild conditions that this procedure offered. Our U1A-TAP purification revealed three other candidates of the SF-A complex: p54nrb, PSF, and p68 helicase (Fig. 1 ▶). They are found in a complex not only because they copurified, but they also migrate in the same series of fractions in sucrose gradient analysis (Fig. 3 ▶), and they interact with each other via protein–protein interactions as revealed by GST pull-down assays (Fig. 2 ▶). Although all four components of the SF-A complex are known RNA-binding proteins and they contain RNA-binding motifs in their amino acid structures, the interactions between them are not due to RNA bridging. RNase A was added in all the purification and binding assays to rule out the possibility that RNA was involved. Without RNase A, other protein components of the U1 snRNP copurified with U1A in TAP purification, such as U1-70K (data not shown). The nuclear matrix protein matrin-3 was also isolated in the absence of RNase A with U1A-TAP (data not shown). We speculate that this copurification resulted from the association of inosine-containing RNA (I-RNA) with a complex of three proteins, p54nrb, PSF, and matrin-3 (Zhang and Carmichael 2001).
This work has also addressed several aspects of these proteins’ (U1A, p54nrb, and PSF) possible functions. PSF and p54nrb are multifunctional nuclear proteins (Shav-Tal and Zipori 2002). p54nrb was originally suspected to be involved in pre-mRNA splicing (Dong et al. 1993), and it was also the first RNA-binding protein described showing a strong preference for inosines that participated in I-RNA nuclear retention (Zhang and Carmichael 2001). The mouse homolog of p54nrb, NonO, was originally isolated as a transcription factor. We have shown here for the first time that p54nrb also plays a key role in cleavage/polyadenylation indicated by the antibody inhibition and immunodepletion/reconstitution experiments (Figs. 4 ▶–6 ▶ ▶). While the reconstitution experiments are compelling, it should be noted that this type of experiment does have its limitations. Although almost full recovery of both cleavage/polyadenylation activity and cleavage activity alone can be found with the addition of p54nrb to the immunodepleted extracts in our in vitro reactions, the regulation that occurs in vivo is likely much more complex and may require the presence of the other complex components.
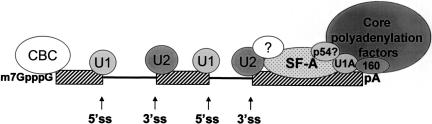
A role for PSF in splicing has been reported in several studies (Patton et al. 1993; Gozani et al. 1994; Lindsey et al. 1995). The actual functional roles that PSF and p54nrb actually play in mRNA splicing are unclear. The fact that p54nrb may also contribute to mRNA polyadenylation suggests further connections between splicing and polyadenylation through the SF-A complex and interactions with the polyadenylation machinery (see Fig. 7 ▶ for a proposed model). The model of exon definition (Berget 1995; Black 1995) is an attractive proposal for how the cell defines exons on a complex pre-mRNA. The last exon, with only a 3′-splice site and a polyadenylation signal, likely requires interactions between splicing and polyadenylation factors to ensure its proper definition.
FIGURE 7.
Model. The SF-A complex may be an adaptor between splicing and core polyadenylation factors. Not pictured for simplicity: SR proteins, U2AF, ASF/SF2. (5′ss) 5′-Splice site; (3′ss) 3′-splice site; (pA) polyadenylation signal; (U1 and U2) U1 and U2 snRNPs, respectively; lines indicate introns of a representative mammalian gene; striped boxes indicate exons of a representative mammalian gene.
In summary, these data provide evidence that p54nrb is involved in the regulation of polyadenylation, perhaps through affecting the assembly of polyadenylation machinery in the initial step, although a specific role is not known yet. p54nrb likely has a direct involvement in polyadenylation (perhaps by interacting with CPSF or CstF, either directly or indirectly), yet PSF may have no direct involvement. Instead, PSF may have an indirect involvement in polyadenylation, most likely through its interaction with p54nrb. Perhaps the role of PSF is to sequester p54nrb until it is required to augment polyadenylation. This is a matter for future investigations.
MATERIALS AND METHODS
Plasmid constructions
A plasmid encoding U1A-TAP was constructed by inserting U1A cDNA into EcoRI site of the mammalian expression vector pZome-1-N (Cellzome). Plasmids expressing His-tagged U1A, full-length U1A-GST fusion and both the N- and C-terminal halves, U1A (AA), U1A (RRM2-1)-GST, fusion proteins were previously described (Lutz et al. 1996). The p54nrb-GST construct was kindly provided by Gordon G. Carmichael (University of Connecticut Health Center). pSP65-SVL contains a 241-bp BamHI–BclI fragment that includes the SV-40 late polyadenylation signal inserted into pSP65 (Promega) at the BamHI site (Wilusz et al. 1988). Adenovirus L3 substrate was a gift of Jeffrey Wilusz and contains a 252-bp KpnI–DraI fragment containing the L3 polyadenylation signal of adenovirus type 5 inserted between the KpnI and HincII sites of pGEM 4 (Promega) (Wilusz et al. 1988). The pET9c-p54nrb used for making TNT products was kindly provided by Adrian Krainer (Cold Spring Harbor Laboratory).
Antibodies
The p68 antibody was a gift from Frances Fuller-Pace (University of Dundee, UK). The p54nrb antibody was purchased from BD Transduction Labs (Pharmingen). The PSF antibody was kindly provided by James Patton (Vanderbilt University). The anti-myc-tagged 9E10 antibody was purchased from Sigma. The rabbit anti-U1A polyclonal antibody 310 was previously described (Lutz and Alwine 1994).
Cell culture and nuclear extract
HeLa cells and 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 2 mM glutamine, and 1% penicillin at 37°C in a 5% CO2 atmosphere. HeLa cell nuclear extracts were prepared as described previously using cells grown in the laboratory or from the National Cell Culture Center (Moore 1990).
Sucrose-gradient fractionation
Sucrose gradients of 5%–30% (w/v) were prepared using 293T cell nucleoplasm as described previously (O’Connor et al. 1997).
TAP purification
For TAP purification experiments, HeLa cells were transfected using Cytofectene Transfection Reagent (BioRad), with 10 μg of TAP vector or U1A-TAP and were kept growing in the presence of 10 μg/mL of puromycin (Sigma). Stable transfectants were clonally isolated to establish the U1A-TAP HeLa cell lines used here. Nuclear extracts from vector alone or from U1A-TAP-transfected HeLa cells were prepared and subsequently adjusted to IgG-binding conditions as described (Rigaut et al. 1999). Two-and-a-half milliliters (2.5 mL) of nuclear extract was rotated overnight at 4°C with 100 μL of washed IgG beads (GE Healthcare/Amersham Biosciences Biotech) in the presence of 5 μL of RNase A (10 mg/ mL); the beads after binding were resuspended in TEV cleavage buffer, and 100 units of recombinant TEV enzyme (Invitrogen) was added to each of the mixtures. After rotating for 4 h at room temperature, the TEV eluates were adjusted to calmodulin-binding conditions and rotated for 2 h at 4°C with 100 μL of calmodulin affinity resin (Stratagene). After binding and washing, bound proteins were recovered by boiling the calmodulin beads in protein sample buffer and loaded onto a 12.5% SDS-PAGE gel. Proteins were detected using a silver staining kit (Invitrogen). After removal of the protein bands by excision, and subsequent trypsin digestion of the proteins, MALDI-TOF/TOF analysis was performed by the Center for Advanced Proteomics, UMDNJ–New Jersey Medical School.
GST pull-down assays
GST full-length U1A and subfragments of U1A, as well as GST-p54nrb fusion proteins, were expressed and purified as previously described (Gruda et al. 1993; Lutz et al. 1996). Pull-down assays with purified GST-U1A (full-length U1A), GST-N-terminal part of U1A (AA), GST-C-terminal part of U1A (RRM2-1), and GST-p54nrb were performed by incubating 2 μg of soluble recombinant fusion proteins with 10 μL of glutathione-Sepharose beads (GE Healthcare/Amersham Biosciences Biotech) in 0.5 mL of 1 × PBS for 30 min. In experiments in which nuclear extract was used, 50 μg of total protein of nuclear extract was added, or in experiments in which 35S-labeled products were used, 10 μL of in vitro transcribed and translated (TNT; Promega) product was used for the binding assays. Bound proteins were boiled in sample buffer, resolved by 12.5% SDS-PAGE, and either visualized with autoradiography (for TNT products) or immunoblotted with PFS, p68, or p54nrb antibodies (for nuclear extracts).
In vitro transcription of RNA substrates
All RNAs were prepared by in vitro transcription using SP6 or T7 RNA polymerase and [α-32P]UTP (PerkinElmer Life Sciences). The plasmids to make the transcripts were linearized by restriction digestion (SVL by DraI or HpaI for precleaved; L3 by HindIII). Briefly, the reaction mixture contained 1 μg of linearized DNA template, 20 units of RNase inhibitor (Pro-mega), 20 units of SP6 or T7 RNA polymerase (Promega) in the presence of 45 μCi of [α-32P]UTP, 50 μM of unlabeled UTP and GTP, and 500 μM each of unlabeled ATP, and CTP; as well as 500 μM cap analog (GE Healthcare/Amersham Biosciences Biotech) in transcription buffer. The reaction mixture was incubated at 37°C for 1 h. RNAs were then gel-purified from 5% polyacrylamide, 8 M urea gels by overnight crush elution in high salt buffer (0.4 M NaCl, 50 mM Tris at pH 8.0, 0.1% SDS) before use in reactions.
In vitro cleavage/polyadenylation and processing
A 12.5-μL in vitro polyadenylation reaction contained 10,000 cpm (~50 fmol) of gel-purified RNA, 3.25 μL of 10% polyvinyl alcohol, 1 mM ATP, 20 mM phosphocreatine, and 7.25 μL of nuclear extract. Reactions were incubated at 30°C for 1 h. Reaction products were then extracted with phenol/chloroform/isoamyl alcohol, precipitated with ethanol, and analyzed on 5% polyacrylamide gels containing 8 M urea. The results were visualized by autoradiography or using PhosphorImager analysis and ImageQuant software. Cleavage reactions were performed the same way as polyadenyla-tion reactions, except that 1 mM cordycepin 5′-triphosphate (Sigma) was supplemented to the reaction mixture, and ATP and phosphocreatine were added to a final concentration of 0.5 mM and 20 mM, respectively, which was different from polyadenylation reactions.
Immunodepletion of extracts
To deplete the U1A, p54nrb, PSF, and c-myc proteins from the HeLa nuclear extract, 5 μL of anti-U1A polyclonal antibody 310 (Lutz and Alwine 1994), anti-54nrb antibody (Affinity Bioreagents), anti-PSF antibody (Sigma), or c-myc antibodies (Santa Cruz) was added individually to 20 μL of HeLa nuclear extract and incubated on ice for 30 min. Ten microliters (10- μL) of prewashed GammaBind Plus Sepharose (GE Healthcare/ Amersham Biosciences) was added to the extract containing the antibody. The mixture was incubated on ice for 30 min, and the beads were removed by centrifugation. The supernatant representing the depleted nuclear extract was collected and analyzed by Western blotting and for polyadenylation activity.
Statistical analyses
Results are expressed as ± SD of the mean.
Acknowledgments
This work was supported by grants to C.S.L. from the American Cancer Society (RPG-00-265-01-GMC), Arthritis Investigator Award 2AI-LUT-A-5, NIH 5R03-HD042464-02, NIH 1R03-AR52038-01, and NSF MCB-0426195. We thank Gordon G. Carmichael, Frances V. Fuller-Pace, Magda M. Konarska, and James G. Patton for gifts of reagents. We also thank Charles C. Query, Vivian Bellofatto, Jeffrey Wilusz, and David Fritz for critical reading of the manuscript and helpful advice. Additionally, we thank all the members of the Lutz lab for technical assistance, helpful comments on the manuscript, and experimental suggestions.
This work is dedicated to the memory of Dr. Longwen Deng.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2213506.
REFERENCES
- Allain, F.H.T., Howe, P.W.A., Neuhaus, D., and Varani, G. 1997. Structural basis for the RNA-binding specificity of human U1A protein. EMBO J. 16: 5764–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget, S.M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270: 2411–2414. [DOI] [PubMed] [Google Scholar]
- Bienroth, S., Keller, W., and Wahle, E. 1993. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 12: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. 1995. Finding splice sites within a wilderness of RNA. RNA 1: 763–771. [PMC free article] [PubMed] [Google Scholar]
- Colgan, D. and Manley, J.L. 1997. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 11: 2755–2766. [DOI] [PubMed] [Google Scholar]
- Dong, B., Horowitz, D.S., Kobayashi, R., and Krainer, A.R. 1993. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 21: 4085–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds, M. 2002. A history of poly A sequences: From formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 71: 285–389. [DOI] [PubMed] [Google Scholar]
- Faig, O.Z. and Lutz, C.S. 2003. Novel specificity of anti-U1A auto-immune patient sera. Scand. J. Immunol. 57: 79–84. [DOI] [PubMed] [Google Scholar]
- Ford, L.P., Bagga, P.S., and Wilusz, J. 1997. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol. Cell. Biol. 17: 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin, G.M. and Nevins, J.R. 1991. Molecular analyses of two poly (A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol. Cell. Biol. 11: 2432–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani, O., Patton, J.G., and Reed, R. 1994. A novel set of spliceosome- associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 13: 3356–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruda, M.C., Zabolotny, J.M., Xiao, J.H., Davidson, I., and Alwine, J.C. 1993. Transcriptional activation by simian virus 40 large T antigen: Interactions with multiple components of the transcription complex. Mol. Cell. Biol. 13: 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson, S.I., Vagner, S., Polycarpou-Schwarz, M., and Mattaj, I.W. 1997. Involvement of the carboxyl terminus of vertebrate poly (A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes & Dev. 11: 761–773. [DOI] [PubMed] [Google Scholar]
- Hartmuth, K., Urlaub, H., Vornlocher, H.P., Will, C.L., Gentzel, M., Wilm, M., and Lührmann, R. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. 99: 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, Y. and Manley, J.L. 2000. RNA polymerase II and the integration of nuclear events. Genes & Dev. 14: 1415–1429. [PubMed] [Google Scholar]
- Huang, Y. and Carmichael, G.G. 1996. Role of polyadenylation in nucleo-cytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 6046–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka, S., Duque, P., and Konarska, M.M. 2004. p54nrb associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23: 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhumaa, P., Parkkila, S., Waheed, A., Parkkila, A.K., Kaunisto, K., Tucker, P.W., Huang, C.J., Sly, W.S., and Rajaniemi, H. 2000. Nuclear NonO/p54nrb protein is a nonclassical carbonic anhydrase. J. Biol. Chem. 275: 16044–16049. [DOI] [PubMed] [Google Scholar]
- Lemon, B. and Tijan, R. 2000. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 14: 2551–2569. [DOI] [PubMed] [Google Scholar]
- Lindsey, L.A., Crow, A.J., and Garcia-Blanco, M.A. 1995. A mammalian activity required for the second step of pre-messenger RNA splicing. J. Biol. Chem. 270: 13415–13421. [DOI] [PubMed] [Google Scholar]
- Liu, Z.R. 2002. p68 RNA is an essential human splicing factor that acts at the U1snRNA-5′ splice site duplex. Mol. Cell. Biol. 22: 5443–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. and Hall, K.B. 1995. An RBD that does not bind RNA: NMR secondary structure determination and biochemical properties of the C-terminal RNA binding domain from the human U1A protein. J. Mol. Biol. 247: 739–752. [DOI] [PubMed] [Google Scholar]
- Lührmann, R., Kastner, B., and Bach, M. 1990. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta 1087: 265–292. [DOI] [PubMed] [Google Scholar]
- Lutz, C.S. and Alwine, J.C. 1994. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes & Dev. 8: 576–586. [DOI] [PubMed] [Google Scholar]
- Lutz, C.S., Murthy, K.G.K., Schek, N., O’Connor, J.P., Manley, J.L., and Alwine, J.C. 1996. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes & Dev. 10: 325–337. [DOI] [PubMed] [Google Scholar]
- Lutz, C.S., Cooke, C., O’Connor, J.P., Kobayashi, R., and Alwine, J.C. 1998. The snRNP-free U1A (SF-A) complex(es): Identification of the largest subunit as PSF, the polypyrimidine-tract binding protein- associated splicing factor. RNA 4: 1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, C.S., McClain, M.T., Harley, J.B., and James, J.A. 2002. Anti- U1A monoclonal antibodies recognize unique epitope targets of U1A which are involved in the binding of U1 RNA. J. Mol. Recognit. 15: 163–170. [DOI] [PubMed] [Google Scholar]
- Lutz-Freyermuth, C., Query, C.C., and Keene, J.D. 1990. Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem–loop II of U1 RNA. Proc. Natl. Acad. Sci. 87: 6393–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Milcarek, C., Martincic, K., Chung-Ganster, L.H., and Lutz, C.S. 2003. The snRNP-associated U1A levels change following IL-6 stimulation of human B-cells. Mol. Immunol. 39: 809–814. [DOI] [PubMed] [Google Scholar]
- Moore, C.L. 1990. Preparation of mammalian extracts active in polyadenylation. Methods Enzymol. 181: 49–74. [DOI] [PubMed] [Google Scholar]
- Murthy, K.G.K. and Manley, J.L. 1995. The 160 kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′ end formation. Genes & Dev. 9: 2672–2683. [DOI] [PubMed] [Google Scholar]
- Neugebauer, K.M. and Roth, M.B. 1997. Transcription units as RNA processing units. Genes & Dev. 11: 3279–3285. [DOI] [PubMed] [Google Scholar]
- O’Connor, J.P., Alwine, J.C., and Lutz, C.S. 1997. Identification of a novel, non-snRNP protein complex containing U1A protein. RNA 3: 1444–1455. [PMC free article] [PubMed] [Google Scholar]
- Patton, J.G., Porro, E.B., Galceran, J., Tempst, P., and Nadal-Ginard, B. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes & Dev. 7: 393–406. [DOI] [PubMed] [Google Scholar]
- Preiss, T. and Hentze, M.W. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392: 516–520. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. 2004. New perspectives on connecting messenger mRNA processing with transcription. Curr. Opin. Cell Biol. 16: 272–278. [DOI] [PubMed] [Google Scholar]
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado- Nilsson, E., Wilm, M., and Seraphin, B. 2001. The Tandem Affinity Purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Ruegsegger, U., Beyer, K., and Keller, W. 1996. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 271: 6107–6113. [DOI] [PubMed] [Google Scholar]
- Sachs, A., Bond, M., and Kornberg, R. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: Domain structure and expression. Cell 45: 827–835. [DOI] [PubMed] [Google Scholar]
- Scherly, D., Boelens, W., van Venrooij, W.J., Dathan, N.A., Hamm, J., and Mattaj, I.W. 1989. Identification of the RNA binding segment of human U1A protein and definition of its binding site on U1 snRNA. EMBO J. 8: 4163–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherly, D., Boelens, W., Dathan, N.A., van Venrooij, W.J., and Mattaj, I.W. 1990. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B′. Nature 345: 502–506. [DOI] [PubMed] [Google Scholar]
- Shav-Tal, Y. and Zipori, D. 2002. PSF and p54nrb/NonO-multi-functional nuclear proteins. FEBS Lett. 531: 109–114. [DOI] [PubMed] [Google Scholar]
- Song, X., Sun, Y., and Garen, A. 2005. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc. Natl. Acad. Sci. 102: 12189–12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, B., Hu, J., Zhang, H., and Lutz, C.S. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle, E. 1995. Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 270: 2800–2808. [DOI] [PubMed] [Google Scholar]
- Wickens, M., Anderson, P., and Jackson, R.J. 1997. Life and death in the cytoplasm: Messages from the 3′ end. Curr. Opin. Genet. Dev. 7: 220–232. [DOI] [PubMed] [Google Scholar]
- Will, C.L. and Lührmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Wilusz, J., Feig, D.I., and Shenk, T. 1988. The C proteins of heterogeneous nuclear ribonucleoprotein complexes interact with RNA sequences downstream of polyadenylation cleavage sites. Mol. Cell. Biol. 10: 4477–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, J., Shenk, T., Takagaki, Y., and Manley, J.L. 1990. A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol. Cell. Biol. 10: 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.S., Hanke, J.H., Carayannopoulos, L., Craft, C.M., Capra, J.D., and Tucker, P.W. 1993. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol. Cell. Biol. 13: 5593–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. and Carmichael, G.G. 2001. The fate of ds RNA in the nucleus: A p54nrb-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465–475. [DOI] [PubMed] [Google Scholar]
- Zhang, W.W., Zhang, L.X., Busch, R.K., Farres, J., and Busch, H. 1993. Purification and characterization of a DNA-binding heterodimer of 52 and 100 kDa from HeLa cells. Biochem. J. 290: 267– 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]