Abstract
A regulated shift from the production of membrane to secretory forms of Immunoglobulin M (IgM) mRNA occurs during B cell differentiation due to the activation of an upstream secretory poly(A) site. U1A plays a key role in inhibiting the expression of the secretory poly(A) site by inhibiting both cleavage at the poly(A) site and subsequent poly(A) tail addition. However, how the inhibitory effect of U1A is alleviated in differentiated cells, which express the secretory poly(A) site, is not known. Using B cell lines representing different stages of B cell differentiation, we show that the amount of U1A available to inhibit the secretory poly(A) site is reduced in differentiated cells. Undifferentiated B cells have more total U1A than differentiated cells and a greater proportion of this is not associated with the U1snRNP. We show that this is available to inhibit poly(A) addition at the secretory poly(A) site using cold competitor RNA oligos to de-repress poly(A) addition in nuclear extracts from the respective cell lines. In addition, endogenous non-snRNP associated U1A—immunopurified from the different cell lines—inhibits poly(A) polymerase activity proportional to U1A recovered, suggesting that available U1A level alone is responsible for changes in its inhibitory effect at the secretory IgM poly (A) site.
Keywords: U1A protein, non snRNP-bound U1A, polyadenylation, IgM secretory poly(A) site
INTRODUCTION
B cell development begins in bone marrow after birth. Stem cells undergo a series of rearrangements of heavy chain and light chain to become immature B cells that express intact IgM receptor in surface membrane. The immature B cells that survive negative selection migrate to the periphery lymphoid tissue to become mature B cells. If they receive the correct set of signals from antigen and T cells, mature B cells will proliferate, undergo class switching and antibody affinity maturation, and then differentiate into antibody-secreting plasma cells (for review, see Goldsby and Osborne 2000).
The IgM precursor mRNA contains two alternatively used poly(A) sites: an upstream (secretory) poly(A) site and downstream (membrane) poly(A) site (Alt et al. 1980; Early et al. 1980). In immature B cells and mature B cells, the membrane mRNA form is produced, and the secretory poly(A) site is inactive (Lamson and Koshland 1984). Upon differentiation, the secretory poly(A) site is activated and the secretory form of mRNA is expressed (Galli et al. 1987, 1988; Peterson et al. 1991; Takagaki et al. 1996). In addition, the secretory mRNA’s stability is increased (Mason et al. 1988; Cox and Emtage 1989). U1A protein has been shown to inhibit activation of the secretory poly(A) site both on the level of cleavage and of poly(A) addition, the latter resulting in destabilization (Phillips et al. 2001, 2004). To inhibit cleavage it binds to two novel U1A binding motifs downstream of the secretory poly(A) site, thereby occluding the binding of Cleavage stimulatory Factor (CstF) to downstream GU-rich motifs and inhibiting formation of the cleavage/polyadenylation complex (Phillips et al. 2001, 2004). To inhibit poly(A) addition, it binds three novel U1A binding motifs upstream of the secretory poly(A) site and inhibits poly(A) polymerase activity (Phillips et al. 2001; Phillips and Gunderson 2003). Thus U1A is a key regulator of the expression of the secretory poly(A) site.
Evidence from in vivo studies with reporter constructs suggests that the inhibitory effect of U1A decreases upon B cell differentiation (Phillips et al. 2001). This would fit with a model in which the secretory poly(A) site is inhibited by U1A in undifferentiated cells that produce mRNA encoding the heavy chain of a transmembrane receptor and then activated or de-repressed in differentiated cells that produce secretory mRNA encoding the heavy chain of secreted antibody.
Not all U1A in the cell is available to inhibit poly(A) polymerase. The majority of U1A in the cell is associated with the U1snRNP and unable to bind mRNA (Gunderson et al. 1997). A decrease in the ratio of U1A to the snRNP associated protein, B′ has been observed during differentiation, suggesting that less U1A is available in differentiated cells (Milcarek et al. 2003). Furthermore, U1A has been reported to exist in a complex with other proteins, which may modulate its effect (O’Connor et al. 1997; Lutz et al. 1998). Therefore, a thorough investigation of the availability of U1A to inhibit poly(A) addition during B cell differentiation is necessary to understand the mechanism for the decrease in the inhibitory effect of U1A during differentiation.
We used B cell lines that represent different stages of B cell differentiation to examine the changing availability and capacity of U1A to inhibit poly(A) polymerase during B cell differentiation. We show here that not only the absolute level of U1A decreases relative to GAPDH, but also the proportion of U1A that is not snRNP-bound decreases as B cells differentiate. Using competition with cold RNA oligos that de-repress inhibition by U1A in nuclear extracts from the different cell types, we show that de-repression is proportional to the amount of non-snRNP-bound U1A levels. Furthermore, endogenous non-snRNP-bound U1A immunoprecipitated from the different cells lines with their individual U1A associated factors inhibits recombinant poly(A) polymerase activity in proportion to the amount of U1A recovered. Taken together, these result show that changing levels of the U1A protein itself regulate the inhibition of secretory poly(A) site expression during B cell differentiation.
RESULTS
B cell lines that represent different stages of B cell differentiation produce a graded ratio of secretory to membrane μ-mRNA
To investigate the changing availability and capacity of U1A to inhibit poly(A) addition during B cell differentiation, we used B cell lines that represent different stages of B cell differentiation. These are J558L, M12.4.1, and WEHI 231, which we used previously (Phillips et al. 2001). J558Ls are a plasmacytoma cell line that has lost endogenous IgM heavy chain (μ) and produce only the secretory form of μ chain mRNA from a transfected gene construct (Oi et al. 1983; Mason et al. 1988), M12.4.1 cells produce IgG2a and aproximately twofold secretory versus membrane μ-mRNA from a transfected μ chain construct (Kim et al. 1979) and WEHI 231 is an immature B cell line that produces more endogenous membrane than secretory μ mRNA (Mason et al. 1988).
We first confirmed that the cell lines used in this study produced a graded ratio of secreted to membrane μ-mRNA and thus adequately represent differing levels of inhibition of expression of the secretory poly(A) site. For this we used RNase protection assays (RPA) of poly(A)+ RNA extracted from the respective cell lines. J558L and M12.4.1 cells do not produce endogenous μ-heavy chain. We therefore transfected these with the pΔ3 plasmid containing the μ-heavy chain gene (Grosschedl and Baltimore 1985). Eight micrograms of plasmid were transfected into 106 cells, respectively, and poly(A)+ mRNA was extracted 24 h later. WEHI 231 produce endogenous μ-heavy chain and poly(A)+ RNA was extracted from these cells without trans-fection. RPA was performed using an in vitro transcribed uniformly 32P-labeled anti-sense probe spanning positions 2085–1730 of the μ-heavy chain and the secretory poly(A) site at 1997 and the 5′ splice site for splicing of the membrane exons at 1812, and includes 34 nt of polylinker sequence. Figure 1A ▶ shows the regions spanned by the probe and the positions of the protected fragments representing the membrane and secretory mRNA. After overnight hybridization of the poly(A)+ RNA with the probe and subsequent RNase digestion to remove single-stranded RNA, the resulting protected fragments were resolved on 8% denaturing PAGE and quantitated by PhosphorImager analysis (see Fig. 1B ▶). Ratios of secretory to membrane mRNA were calculated after adjustment for the length of the protected fragments and these results ± SD are presented below the lanes of each respective sample in Figure 1B ▶. J558L cells produce 38 ± 8 times more secretory than membrane mRNA from the transfected plasmid pμΔ3 containing the immunoglobulin M heavy chain gene, M12.4.1 produce 2.3 ± 0.3 times more secretory than membrane mRNA from pμΔ3 while WEHI 321 endogenously produce less than half the amount of secretory mRNA to membrane mRNA, with a ratio of 0.4 ± 0.1. This confirms that the cell lines used in this study produce a graded ratio of secretory to membrane μ-mRNA that correlates with the stage of differentiation, with the least differentiated producing the lowest ratio and the most differentiated producing a very high ratio.
FIGURE 1.
B cell lines that represent different stages of B cell differentiation produce a graded ratio of secretory to membrane μ-mRNA. RNase protection assays using an in vitro–transcribed, uniformly 32P-labeled anti-sense probe spanning positions 2085–1730 of the μ-heavy chain, and the secretory poly(A) site at 1997 and the 5′ splice site for splicing of the membrane exons at 1812. (A) Schematic diagram of the probe and the protected fragments representing the membrane (mem) and secretory (sec) mRNA. (B) RPA with poly(A)+ RNA extracted from the respective cell lines. Eight micrograms of pΔ3 plasmid containing the μ-heavy chain gene were transfected into 106 J558L and M12.4.1 cells 24 h previous to harvest. For WEHI 231 the results are for the endogenous μ-heavy chain. Protected fragments were resolved on 8% denaturing PAGE and quantitated by phosphorimager analysis. Ratios of secretory to membrane mRNA (sec:mem) were calculated after adjustment for the length of the protected fragments and results of triplicates ± SD are presented below each lane, respectively.
The total amount of U1A decreases with stage of differentiation
As a first approach, we measured the total U1A levels in J558L, M12.4.1, and WEHI 231 cell lines. Whole cells were lysed and sonicated in SDS-loading buffer at the concentration of 104/μL and run on 12% SDS-PAGE. The samples were eletrotransfered and immunostained by rabbit poly-clonal antibody specific for U1A and mouse polyclonal antibody specific for GAPDH. The bands were visualized using horseradish peroxidase conjugated anti-rabbit (for U1A) or anti-mouse (for GAPDH) secondary antibody and ECL reagents.
In order to accurately compare the U1A levels in the different cells lines we first titrated extract to determine the linear range of the assay. We found that 1 μL, 2 μL, and 3 μL for each cell extract was linear (see Fig. 2A ▶, lanes 1–3 [J558L], lanes 4–6 [M12.4.1], and lanes 7–9 [WEHI 231] both U1A and corresponding comparable GAPDH level). These bands were scanned and quantitated in Image-Quant. We calculated the ratio of U1A:GAPDH in triplicate ± SD for each cell line and normalized these to the value for J558L (i.e., J558L was set to 1). We found that the undifferentiated (M12.4.1, 1.39 ± 0.11 SD) and immature B cell (WEHI 231, 2.2 ± 0.31 SD) lines both had significantly more total U1A relative to GAPDH than differentiated cells (J558L = 1.0) and that WEHI 231 had significantly more U1A than M12.4.1 (Fig. 2A ▶, cf. M12.4.1 [lanes 4–6]/WEHI 231 [lanes 7–9] and J558L [lanes 1–3] and quantitation thereof in the bar graph). Thus, the amount of U1A is ranked according to differentiation stage WEHI 231 > M12.4.1 > J558L. This is in line with the inhibitory effect of U1A on poly(A) site expression found in previous in vivo studies that showed the same ranking (Phillips et al. 2001). This suggests that changing U1A levels between the cell lines may be responsible for the changing inhibitory effect.
FIGURE 2.
U1A levels decrease upon differentiation. (A) In whole cell extracts: Western blotting using U1A antibody 856 and anti-GAPDH antibody as control. (Lanes 1–3) J558L, (lanes 4–6) M12.4.1, (lanes 7–9) WEHI 231. Bands were quantitated by PhosphorImager analysis, and U1A values were calculated relative to GAPDH in each case as shown in the bar graph. The relative total U1A level in J558L was set as 1.0. Triplicates ± SD. Cell lines are indicated. (J) J558L, (M) M12.4, (W) WEHI231. (B) U1A protein compartmentalization during B cell differentiation. U1A protein in nuclear extracts (NE) and cytoplasmic extracts (CE) was detected by Western blotting using U1A antibody 856. (Lanes 1,2) CE and NE of J558L, (lanes 3,4) CE and NE of M12.4.1, (lanes 5,6) CE and NE of WEHI 231. For comparison, the loaded cytoplasmic extracts (CE) and nuclear extracts (NE) have the same amount of total protein. Bands were quantitated by PhosphorImager analysis and normalized to J558L NE (set as 1.0) in each case, and the ratio of CE:NE was calculated for each cell line as shown in the bar graph. Cell lines are indicated. (J) J558L, (M) M12.4, (W) WEHI231. Triplicates ± SD (C) Nuclear U1A protein decreases upon differentiation. Western blotting using anti-U1A antibody 856. (Lanes 1–3) J558L, (lanes 4–6) M12.4.1, (lanes 7–9) WEHI 231. Loaded were 1, 2, and 3 μL of each nuclear extract containing the same amount of total protein. The U1A bands were quantitated by phosphorimagery, and values were normalized to J558L (set as 1.0) as shown in the bar graph. Cell lines are indicated. (J) J558L, (M) M12.4, (W) WEHI231. Triplicates ± SD.
U1A retains the same distribution between the nucleus and cytoplasm at different stages of differentiation
As RNA processing occurs in the nucleus and U1A is known to shuttle between the cytoplasm and nucleus (Kambach and Mattaj 1992), we next determined the availability of U1A to the RNA processing apparatus by comparing U1A levels in nucleus and cytoplasm of the different cell lines. Cytoplasmic extracts (CE) and nuclear extracts (NE) were prepared from ~2 × 107 cells using a modified Dignam et al. (1983) method specific for a small number of lymphoid cells (Virtanen and Chen 1990). The CE and NE of each cell line with equal amounts of total protein were loaded in 12% SDS-PAGE for comparison. Western blotting shows that the nucleus has more U1A than the cytoplasm for all three B cell lines (Fig. 2B ▶, cf. lanes 1 and 2, 3 and 4, and 5 and 6). Bands were quantitated by phosphorimagery and normalized to J558L NE (set as 1.0) in each case and the ratio of CE:NE was calculated for each cell line (see bar graph in Fig. 2B ▶). Upon differentiation, the U1A protein level in both the nucleus and cytoplasm decreases (Fig. 2B ▶, cf. CE: WEHI 231, M12.4.1, and J558L [lanes 5,3,1] and NE: WEHI 231, M12.4.1, and J558L [lanes 6,4,2 and bar graph]). However, the proportion of U1A in CE to that in NE remains almost equivalent (Fig. 2B ▶, CE:NE). Thus there is no selective accumulation of U1A in either compartment during B differentiation and we can rule out selective accumulation in the cytoplasm as a means of restricting the availability of U1A to the RNA processing apparatus.
U1A in nuclear extracts from the different cells lines follows the same differentiation-specific decreasing pattern as seen for whole cells
In order to accurately calibrate the relative U1A protein level in nuclear extracts of the above lymphoid cell lines and HeLa cell lines, we once again loaded 1 μL, 2 μL, and 3 μL of each nuclear extract containing the same amounts of total protein (determined to be in the linear range of the assay) onto 12% SDS-PAGE and carried out Western blotting as before (see Fig. 2C ▶). The bands in Figure 2C ▶ were quantitated and, once again, normalized to J558L (set as 1.0). Our results showed that U1A levels decreased in nuclear extracts from cell lines from immature (WEHI 231, 2.12 ± 0.17), through mature (M12.4.1, 1.48 ± 0.10) to differentiated B cell (J558L, 1.0) following the B cell differentiation pathway (Fig. 2C ▶, cf. WEHI 231 [lanes 7–9], M12.4.1 [lanes 4–6], and J558L [lanes 1–3], and corresponding bar graph). In addition, we found that B cells at all stages of differentiation have more nuclear U1A than HeLa cells (0.55 ± 0.02) (data not shown).
The ratio of U1A to U170K and to U1 snRNA decreases as B cells differentiate
We wanted to investigate how much U1A is available to regulate polyadenylation of IgM secretory mRNA. U1snRNP associated U1A is not available to regulate poly-adenylation (Gunderson et al. 1997). In the fully assembled U1snRNP, U1A binds to loop 2 and U170K binds to loop 1 (Will et al. 1993), and the stoichiometry of U1A/70K is 1:1 (Will and Luhrmann 2001). U1A that is available to regulate mRNA poly(A) addition binds mRNA directly and will not be present in the U1snRNP. Thus an investigation of the ratio of U1A/U170K and U1A/U1snRNP in the various cells lines should provide clues as to whether the proportion of U1A that is not snRNP-associated changes during B cell differentiation.
To more accurately compare the differences in the ratio of U170K and U1snRNA to U1A between nuclear extracts from the cell lines representing different stages of B cell differentiation, we artificially adjusted the loading of the samples so that amounts of nuclear extracts contained equal amounts of U1A. In other words, we added 1.82, 3.64, and 5.46 μL of HeLa extracts; 1, 2, and 3 μL of J558L extracts; 0.67, 1.34, and 2.01 μL of M12.4.1; and 0.47, 0.94, and 1.4 μL of WEHI 231 extract. Bands were scanned and quantitated in Image-Quant as before. The ratios of nuclear U1A to U170k for the different cell extracts were normalized to J558L (set as 1.0). First we noticed that all B cells have a greater amount of U1A relative to 70K than HeLa cells (U1A:70K ratio = 0.71 ± 0.03 SD) (Fig. 3A ▶, cf. HeLa [lanes 1–3] and J558L [lanes 4–6], M12.4.1 [lanes 7–9], and WEHI 231 [lanes 10–12]). Furthermore, the ratio of U1A to 70K decreased with differentiation stage (Fig. 3A ▶, cf. lanes 10–12 and 7–9 and 4–6). WEHI 231 have a relative U1A to 70K ratio of 2.06 ± 0.11 SD; M12.4.1, 1.25 ± 0.06 SD; while the differentiated cells have a normalized ratio of 1.0 (Fig. 3B ▶). Thus the stoichiometry of U1A relative to 70K is greater in undifferentiated B cells. As the stoichiometry in U1snRNP is 1:1, these results suggest that there is more U1A that is not snRNP-bound in undifferentiated B cells and therefore more likely to be available to bind and regulate polyadenylation of mRNA.
FIGURE 3.
Undifferentiated cells have a greater ratio of nuclear U1A to U170K and to U1snRNA. (A) Western blotting using anti-U1A and anti-U170K. (Lanes 1–3) HeLa, (lanes 4–6) J558L, (lanes 7–9) M12.4.1, (lanes 10–12) WEHI 231. Triplicates ± SD (B) Quantitation of A. The U1A and 70K bands in A were quantitated in ImageQuant and values were normalized to J558L (set as 1.0). Cell lines are indicated. Triplicates ± SD (C) Western blotting against U1A and (D) Northern blotting against U1snRNA in nuclear extracts from the cell lines as indicated. (E) Quantitation of C and D. The bands in C and D were quantitated in ImageQuant and the ratios of nuclear U1A to U1snRNA were calculated and normalized to J558L (set as 1.0). Triplicates ± SD.
We next examined the stoichiometry of U1A to U1snRNA. When we compared extracts with equal protein amounts, we found U1snRNP levels to be equivalent, showing that it is the U1A levels that change, not the U1snRNA levels (data not shown). However, to more accurately determine the U1A to U1snRNA ratio, we once again artificially adjusted the loading of the amount of extract so that the samples contained equal amounts of U1A (Fig. 3C ▶, U1A content by Western blotting, lanes 1–4). We measured the U1snRNA level in the different B cell lines by Northern blotting. Titration was performed to check that the bands fell within the linear range before quantitation by phosphorimage and normalization to J558L (set to 1.0). As can be seen in Figure 3D ▶, the U1snRNA amount in the artificially adjusted volumes increases upon differentiation (Fig. 3D ▶, cf. lanes 4,3,2) showing that the relative ratio of U1A to U1snRNA decreases. The quantitation is shown in Figure 3E ▶. The relative U1A to U1snRNA ratio is 1.95 ± 0.12 for WEHI 231 and 1.18 ± 0.05 for M12.4.1, and J558L is 1.0. A value of 0.9 ± 0.01 was obtained for HeLa cells, once again showing that B cells in general have a higher ratio of U1A to U1snRNP than HeLa cells (Fig. 3E ▶, HeLa cells). This is consistent with results obtained above for the U1A to 70K ratio and further suggests that undifferentiated B cells have a greater proportion of U1A that is not snRNP bound and therefore available to regulate polyadenylation of mRNA.
Undifferentiated cells have a greater proportion of non-snRNP bound U1A than differentiated cells
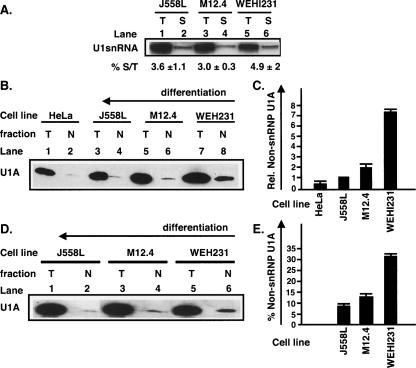
To directly measure the proportion of non-snRNP bound U1A in the different extracts, we used the fact that all snRNAs (except U6) have a unique nucleoside 2,2,7 tri-methylguanosine cap (TMG) (Will and Luhrmann 2001), which can be specifically recognized by anti-TMG antibodies. We employed an immunoprecipitation strategy to separate U1A that is snRNP bound from that which is not. We used anti-TMG-coupled agarose beads to immunoprecipitate U1snRNP-associated U1A from the nuclear extracts and leave the non-snRNP-bound U1A in supernatant. This way we could directly measure the proportion of non-snRNP U1A in the different cell extracts. We found that two rounds of immunoprecipitation with 20μL of TMG beads were sufficient to deplete >95% of U1snRNA as tested by Northern blotting of the supernatant (S) and comparing it with U1snRNA present in nondepleted extracts (T) (Fig. 4A ▶, % S/T). With this optimized protocol, we compared the proportion of U1A that was not bound in the U1snRNP (i.e., non-snRNP associated U1A (N) remaining in the supernatant) with total input U1A (T) for HeLa, J558L, M12.4.1, and WEHI 231 nuclear extracts of equal amounts of total protein. As can be seen in Figure 4 ▶, the nuclear U1A (T) decreases upon differentiation, confirming the results obtained above (cf. Fig. 4B ▶, lanes 7,5,3,1). The non-snRNP-associated U1A (N) also decreases upon differentiation but to a greater extent (Fig. 4B ▶, cf. lanes 8,6,4,2). The relative non-snRNP-bound U1A levels were quantitated and normalized to J558L (Fig. 4C ▶). We found 6.95 ± 0.14-fold more and 2.0 ± 0.2-fold more non-snRNP associated U1A in WEHI 231 (immature) and M12.4.1 (mature undifferentiated), respectively, than in J558L (differentiated) cells.
FIGURE 4.
Undifferentiated B cells have more non-snRNP-bound U1A and a greater proportion of all U1A is non-snRNP-associated. (A) Northern blotting against U1snRNA. (Lanes 1,3,5) T, inputted U1snRNA from J558L, M12.4.1, and WEHI 231 nuclear extracts. (Lanes 2,4,6) S, U1snRNA left in supernatant after immunoprecipitation. (B) Undifferentiated B cells have more non-snRNP-bound U1A. TMG immunoprecipitation starting with nuclear extracts of same amount of total proteins. (Lanes 1,3,5,7) T, inputted nuclear U1A from HeLa, J558L, M12.4.1, and WEHI 231. (Lanes 2,4,6,8) Non-snRNP-bound U1A left in supernatant after immunoprecipitation (N). U1A bands are detected by Western blotting. (C) Quantitation of B. The non-snRNP-associated U1A (N) bands were quantitated in ImageQuant and normalized to the non-snRNP-associated U1A from J558Ls. Triplicates ± SD (D) A greater proportion of all U1A is non-snRNP associated in undifferentiated cells. TMG immunoprecipitation starting with artificially adjusted volume of nuclear extract to contain equal amounts of nuclear U1A. (Lanes 1,3,5) T, inputted nuclear U1A from J558L, M12.4.1, and WEHI 231. (Lanes 2,4,6) N, non-snRNP-bound U1A left in supernatant after immunoprecipitation. (E) Quantitation of D. The percentage of non-snRNP-bound U1A relative to total U1A was calculated and normalized to the value for J558L. Triplicates ± SD.
To obtain an accurate comparison of the ratios of non-snRNP-bound U1A to total U1A for each extract, we again artificially adjusted the input amount of the three B cell nuclear extracts to equalize the starting amount of U1A. We subjected these to two rounds of TMG-beads immunoprecipitation followed by Western blotting as before (Fig. 4D ▶) and quantitated the proportion of non-snRNP-bound U1A (N/T) in each case (see Fig. 4E ▶). We found that the proportion of non-snRNP-bound U1A in the various cell lines decreased upon differentiation (31.6 ± 0.6% for WEHI 231, 13 ± 0.4% for M12.4.1, and 8.45 ± 0.2% for J558L) (Fig. 4D ▶, cf. lanes 5,3,1 and 6,4,2). Taken together, these results show that both the proportion and the total amount of non-snRNP U1A decreases upon differentiation. Thus the pool of U1A that may be available to regulate mRNA polyadenylation correlates with the level of expression of the μ-secretory poly(A) site. This fits with the model that the inhibitory effect of U1A is greatest in undifferentiated cells and is relieved upon differentiation allowing expression of the μ-secretory poly(A) site and production of the secreted antibody.
Endogenous U1A in undifferentiated B cell nuclear extracts has a greater inhibitory effect on polyadenylation than that in differentiated cells
We next investigated if endogenous U1A is available to inhibit poly(A) addition. For this we tested whether we could de-repress the inhibitory effect of U1A on polyadenylation in nuclear extracts from the various cell lines. We also wanted to know if the level of de-repression achieved in each cell extract correlated with differentiation stage and therefore the amount of non-snRNP-associated U1A. For this we performed in vitro specific poly(A) assays in nuclear extracts from the different cell types and used a cold competitor to de-repress poly(A) activity in each extract respectively. For polyadenylation we used an α-32P-labeled μ-secretory RNA substrate (1790–2030) representing pre-cleaved mRNA that was synthesized by in vitro transcription as previously described (Phillips et al. 2001). For the cold competitor we used a synthesized 30-mer RNA oligo (SL2) that spans stem-loop 2 of U1snRNP and includes the consensus U1A binding site (Klein Gunnewiek et al. 2000). To control for nonspecific effects of addition of stem-loop RNA oligos to the reaction we used the MS2 stem-loop RNA that does not bind U1A.
We first compared the poly (A) efficiency using HeLa cell and B cell nuclear extracts and 30,000 cpm (50 fmol) radio-labeled mRNA substrate. Nuclear extracts from HeLa cells polyadenylated the RNA substrate to a greater extent than nuclear extracts from B cells in general, and nuclear extracts from undifferentiated B cells (WEHI 231 and M12.4.1) polyadenylated a greater percentage of the RNA substrate than those from differentiated B cells (J558L) (data not shown). The poor performance of nuclear extracts from differentiated cells has been previously reported (Virtanen and Sharp 1988). We nevertheless obtained sufficient poly-adenylation to be able to measure de-repression in each case. In vitro specific polyadenylation assays were performed using equal amounts of total protein in the nuclear extracts (8 μg). We found that we could achieve specific de-repression with 5 and 10 ng of SL2 RNA, and that beyond 20 ng nonspecific effects came into operation as judged by an effect of MS2 RNA at this concentration (data not shown). Five (5) and 10 ng of SL2 RNA represents a 10-and 20-fold molar excess of cold competitor, respectively, over the radiolabeled RNA polyadenylation substrate. We found that we could specifically de-repress all three cell nuclear extracts significantly with 5 and 10 ng of SL2 RNA. However, the extent of de-repression was significantly lower for J558L (differentiated) than for either M12.4.1 (mature undifferentiated) or WEHI 231 (immature undifferentiated) nuclear extracts, and M12.4.1 significantly lower than WEHI231 (Fig. 5A ▶, cf. J558L [lanes 1–3], M12.4.1 [lanes 4–6], and WEHI 231 [lanes 7–9]). It can be seen that for this range, MS2 RNA has no effect (Fig. 5A ▶; WEHI 231 MS2 control [lanes 10–12]). Unreacted probe is in lane 13 (no extract). We quantitated the poly(A) tails by PhosphorImager analysis and normalized the set of values obtained to that from “zero cold competitor” for each nuclear extract. For 5 and 10 ng of SL2 cold competitor we found the results to be 1.25 ± 0.06 SD and 1.15 ± 0.04 for J558L, 1.40 ± 0.10 and 1.60 ± 0.04 for M12.4.1, and 2.03 ± 0.04 and 2.29 ± 0.10 SD for WEHI 231 (see Fig. 5 ▶). In other words, we could de-repress WEHI 231 approximately fourfold more than J558L and twofold more than M12.4.1. These results correlate with differentiation stage, with undifferentiated cells being the most repressed, and differentiated cells with almost no discernable repression. From this we conclude that endogenous U1A is available to inhibit poly(A) addition in context with other factors present in nuclear extracts. Furthermore, taken together with the findings in Figure 4 ▶, we conclude that the level of repression correlates with the differentiation stage and the amount of non-snRNP-associated U1A in each case.
FIGURE 5.
The extent of de-repression with SL2 RNA is larger in undifferentiated cells. In vitro specific poly(A) assays in nuclear extracts from the different cell types as indicated. For polyadenylation we used 50 fmol (30,000 cpm) of an α-32P-labeled μ-secretory RNA substrate (1790–2030) representing precleaved mRNA that was synthesized by in vitro transcription as previously described (Phillips et al. 2001). SL2 is a synthesized RNA 30-mer oligo that spans stem-loop 2 of U1snRNP and includes the consensus U1A binding site. This was used to de-repress poly(A) activity in each extract, respectively. To control for nonspecific effects of addition of stem-loop RNA oligos to the reaction the MS2 stem-loop RNA that does not bind U1A was used. (A) In vitro specific polyadenylation with nuclear extracts from the indicated cell lines. Zero (0), 5, and 10 ng SL2 representing a 0-, 10-, and 20-fold molar excess of cold competitor, respectively, over the radiolabeled RNA polyadenylation substrate, were added to consecutive samples. (Lanes 1–3) J558L, (lanes 4–6) M12.4.1, and (lanes 7–9) WEHI 231. Controls: WEHI 231 NE, MS2 RNA added (lanes 10–12) and probe alone, no extract (lane 13). (B) Quantitation of A. Comparison of the increased percentage of polyadenylation with added SL2 RNA. Poly(A) tails were quantitated by phosphorimagery. Values were normalized to the value from the poly(A) tail with no SL2 RNA added for each set of samples from the same extract. Triplicates ± SD.
Non-snRNP-associated U1A’s ability to inhibit poly(A) polymerase activity correlates with differentiation stage
To directly test whether endogenous non-snRNP-associated U1A complexes could inhibit poly(A) polymerase activity, we added the non-snRNP-associated U1A obtained from the nuclear extracts from the cells representing different stages of differentiation, as in Figure 4D ▶, into nonspecific poly(A) assays with recombinant poly(A) polymerase. For this assay we once again started with volumes of HeLa and the three B cells nuclear extracts that contained the same amount of nuclear U1A to test if a greater proportion of U1A is available to inhibit poly(A) addition in undifferentiated cells. The amount of inputted U1A was estimated to be ~300 ng in each case by comparison with a standard curve (data not shown). After the snRNP-associated U1A was removed by two rounds of TMG immunoprecipitation as before, the non-snRNP-bound U1A that remained in the supernatant was immunopurified with the anti-U1A 856 antibody bound to Sepharose beads (see Materials and Methods). This non-snRNP-bound U1A was then eluted from the beads and neutralized to give a final volume of 13.5 μL. The amount of eluted non-snRNP-bound U1A was measured by Western blotting and compared to a standard curve. Eight, 18, 27, and 66 ng were obtained for HeLa, J558L, M12.4.1, and WEHI 231, respectively, confirming the differentiation stage progression in the proportion of non-snRNP-bound U1A between the cell lines (see Fig. 6A ▶). We also did a silver stain to see what other proteins were coprecipitated in complex with U1A (see Fig. 6B ▶). We found a prominent band at 32 kDa (U1A), which was not present in the no-extract lane (lane 10) and increased from HeLa (lane 11), J558L (lane 12) to M12.4.1 (lane 13) to WEHI 231 (lane 14) (Fig. 6B ▶, cf. lanes 11–14). There were a number of prominent bands that were also present in the no-extract control. These did not inhibit poly(A) polymerase activity (see Fig. 6C ▶, lane 7) and therefore were deemed nonspecific antibody-associated bands. There were two bands in the 45 kDa region that were not present in the no-extract lane (lane 10) but were present to the same degree in the extract lanes (lanes 11–14). However, a 45 kDa was visible in the extract only, no antibody lanes (lanes 1–4), suggesting that this too is a nonspecific protein that binds beads alone. This also did not inhibit poly(A) polymerase activity (Fig. 6C ▶, lane 8). We therefore conclude that U1A is the major specific protein eluting under this protocol.
FIGURE 6.
The percentage of polyadenylated IgM RNA tail in nonspecific poly(A) assay correlates with the proportion of non-snRNP-bound U1A immunopurified from the nuclear extracts. Starting amounts of extract were artificially adjusted to contain equal amounts of input nuclear U1A. (A) Western blot and quantitation of non-snRNP-bound U1A from the different nuclear extracts (lanes 1–4) by comparison with a standard curve of recombinant U1A (rU1A) (lanes 5–8) after immunodepletion with anti-TMG antibodies, immunoprecipitation with anti-U1A 856 antibody, and elution. (B) Silver stain of proteins obtained after immunoprecipitation and elution. (C) Nonspecific poly(A) assay with immunopurified samples from nuclear extracts and recombinant poly(A) polymerase. (Lane 1) No PAP, (Lanes 2–8) 50 ng PAP. Lane 2, PAP alone; lanes 3–6, PAP plus 2μL of eluate from HeLa (lane 3), J558L (lane 4), M12.4.1 (lane 5), and WEHI 231 (lane 6); lane 7, PAP plus eluate from a mock immunoprecipitation with no extract; lane 8, PAP plus eluate from a mock immunoprecipitation with no antibody. (D) Quantitation of C. Poly(A) tails were quantitated by phosphorimagery and expressed as percentage of the poly(A) tail obtained with the mock immunoprecipitated and eluted sample (no antibody control, lane 8). Triplicates ± SD.
For the nonspecific polyadenylation assay, we found that 50 ng recombinant bovine poly(A) polymerase (PAP) produced a robust poly(A) tail on the 30,000 rpm radiolabeled IgM secretory RNA substrate (Fig. 6C ▶, lane 2). We then introduced 2 μL of the eluted complexes from the different cell lines into the assay to assess the extent of inhibition induced in each case. We found that HeLa eluted complexes were able to induce a very small difference in the length and abundance of the poly(A) tail compared with the “no extract” control and no antibody control (Fig. 6B ▶, cf. lanes 3 and 7,8). The effect of the eluted complexes from the B cell lines was in general greater than for HeLa cells, with the equal amounts of eluate inducing a progressively greater inhibition of poly(A) tails production from J558L to M12.4.1 to WEHI 231, respectively (Fig. 6C ▶, cf. lanes 3–6). The processed and unprocessed RNA substrates were quantitated by phosphorimagery and the amount of poly(A) tail was calculated and expressed as a percentage of the unreacted probe (see lane 1). An average of 83.6% ± 0.5% of IgM secretory RNA substrate is processed when mock eluted samples were added (lanes 7,8). Two microliters of HeLa eluted samples can reduce the percentage to 47.1% ± 2.5%, J558L to 25.9% ± 1.0%, M12.4.1 to 16.9% ± 0.8%, and WEHI 231 to 6.5% ± 0.4% (Fig. 6D ▶). From these results we conclude that eluted complexes from undifferentiated B cells inhibit poly(A) polymerase to a greater extent than differentiated cells and that the inhibitory effect correlates with the proportion of non-snRNP-bound U1A from the B cells in their respective stages of differentiation.
DISCUSSION
U1A plays a key role in the regulation of the expression of the secretory poly(A) site. Therefore, it is essential to discover whether U1A available to regulate polyadenylation at the secretory poly(A) site changes during B cell differentiation. We have shown that (1) total U1A levels decrease upon differentiation both in whole cells and in the nucleus, (2) the proportion of U1A that is not bound in U1snRNP and therefore available to inhibit the expression of the secretory poly(A) site is reduced upon differentiation, and (3) the U1A not bound to U1snRNP is able to inhibit poly(A) addition to the secretory poly(A) site in vitro.
Total U1A as well as the proportion that is non-snRNP associated decreases with B cell differentiation
The alternative processing of the IgM heavy chain pre-mRNA is finely balanced with a weak secretory poly(A) site in competition with a weak splicing reaction to produce the secretory or membrane form of mRNA, respectively. A twofold change in strength of either reaction can result in a complete change in mRNA product even in cells that normally produce the opposite mRNA (Peterson and Perry 1989; Peterson 1992). We show here that the total U1A levels decrease upon differentiation both in whole cells and in the nucleus. Mature undifferentiated (M12.4.1, 1.39 ± 0.11 SD) and immature B cell (WEHI 231, 2.2 ± 0.31 SD) lines both had significantly more total U1A relative to GAPDH than differentiated cells (J558L = 1.0). Thus we found a 55% decrease from immature to differentiated B cells and a 30% decrease from mature undifferentiated to differentiated B cells. The latter are in agreement with results from mature human B cells induced to differentiate with IL-6 where a 29% decrease in U1A relative to an snRNP protein was observed (Milcarek et al. 2003). However as a large proportion of the cell’s U1A is bound up in the U1snRNP and unavailable to bind the secretory poly(A) site, it is deceiving to examine changes in total U1A alone. We found that not only did the level of total U1A decrease but also the proportion of U1A that is not snRNP associated decreases upon differentiation. When the amount of non-snRNP-bound U1A was compared, the changes were much more dramatic with immature B cells (WEHI 231) having sevenfold and maturing undifferentiated (M12.4.1) having twofold more non-snRNP-associated U1A than differentiated cells (J558L). As the alternative processing is finely balanced (Peterson and Perry 1989; Peterson 1992), this amount of change in the abundance of a polyadenylation inhibitory factor alone could conceivably change the mRNA product from secreted to membrane mRNA. However, other changes in factors affecting the process have been documented that would presumably also contribute to the switch in mRNA product during B cell differentiation.
We examined a number of other parameters to document the change in non-snRNP-associated U1A. We found a change in stoichiometry of U1 snRNA and the snRNP-associated 70K protein relative to U1A without a change in U1snRNA levels in these cells. This is similar to the approach taken by Milcarek et al. (2003) in mature human B cells, and our results are in agreement with theirs. However, they did not measure non-snRNP-associated U1A as we have done in the Trimethylcap immunodepletion experiments.
The U1A not bound to U1snRNP is able to inhibit poly(A) addition to the secretory poly(A) site in vitro
Although U1A protein predominantly exists as a component of U1snRNP, it enters the nucleus by a separate Ran independent route (Kambach and Mattaj 1992). Once in the nucleus, it can presumably bind either U1snRNP or mRNA, either its own or heterologous mRNA, such as the IgM secretory mRNA. As the proportion of U1A theoretically available to bind to mRNA is not necessarily a measure of U1A activity on an mRNA, we also wanted to investigate if endogenous U1A from the various cell lines is able to inhibit poly(A) addition at the secretory poly(A) site and if this inhibition correlates with the proportion of non-snRNP-associated U1A. We demonstrated with the de-repression experiments in Figure 5 ▶ that there is a proportion of U1A that actively inhibits polyadenylation at the secretory poly (A) site in its native context in nuclear extracts. Furthermore, non-snRNP-associated U1A immunoprecipitated from each of the extracts is able to inhibit recombinant poly(A) polymerase activity on the secretory poly(A) site substrate. Once again, the inhibition correlates with the proportion of U1A recovered. Thus these results show that endogenous U1A actively suppresses polyadenylation at the secretory poly(A) site and that this suppression is alleviated upon differentiation.
The next question is whether U1A acts alone or in conjunction with other proteins. In the de-repression experiments, U1A is present in the context of other endogenous proteins in the various cell extracts, and the inhibitory effect could be mediated or modulated by accessory proteins. Others have reported that RNA-free U1A is present in a complex with other proteins (O’Connor et al. 1997; Lutz et al. 1998). We therefore considered the possibility that U1A that binds mRNA might also be in complex with other proteins that might modulate its activity on that mRNA. We anticipated that these accessory proteins would co-precipitate with U1A obtained from the various cell extracts in Figure 6 ▶ and would continue to affect U1A activity. However, our silver staining of the products of the immunoprecipitations show U1A as the major band that is not also present in the “no extract” and “no antibody” control, suggesting that U1A is the major player in this inhibitory effect. Furthermore, we find that the inhibitory effect correlates with the proportion of U1A obtained, diminishing the possibility that a developmentally induced factor excessively modulates U1A activity at a particular stage of differentiation. Thus, the results we obtained suggest a more direct mechanism: The level of U1A available to bind the secretory mRNA is reduced upon differentiation, thus alleviating its inhibitory effects on both cleavage and poly(A) addition. We cannot completely rule out the possibility that an accessory protein that facilitates the effect of U1A may not be coprecipitated with our protocol or visible by silver staining. Nevertheless, the inhibitory effect correlates with the abundance of the U1A protein, and its levels change during differentiation, thus offering a straightforward explanation of the data.
We have shown that both total U1A and the proportion of non-snRNP U1A decreases upon differentiation, offering a simple mechanism for how U1A inhibition of the secretory poly(A) site is alleviated upon B cell differentiation, leading to the activation of this poly(A) site and the production of secreted antibody.
MATERIALS AND METHODS
Cell culture and preparation of whole cell extracts
HeLa, J558L, and WEHI 231 cells were obtained from the European Collection of Animal Cell Cultures (ECACC). M12.4.1 cells were the gift from K.J. Kim (Kim et al. 1979). J558L, M12.4.1, and WEHI 231 were cultured in RPMI 1640 (Gibco) with 5% fetal calf serum (Gibco), 1× nonessential amino acids (Sigma), 1 mM sodium pyruvate (Sigma), 55 mM mecaptoethanol (Gibco), and penicillin/streptomycin (100 units/mL). Cells were harvested during exponential growth, washed by 1× PBS twice, and lysed and sonicated in SDS loading buffer, and then their proteins were subjected to 12% SDS-PAGE separation and Western blotting.
RNase protection assays
The plasmid containing the IgM heavy chain gene (pμΔ3; Grosschedl and Baltimore 1985) was transfected into M12.4.1 cells and J558L cells in triplicate. Poly(A) mRNA was extracted 17 h later from these and WEHI 231 cells using a Quickprep micro mRNA preparation kit from Pharmacia as previously described (Phillips et al. 1996). mRNA levels were measured by RNase protection analysis according to Melton et al. (1984). Poly(A)+ RNA was hybridized overnight at 45° C with 100,000 cpm uniformly 32P-labeled μ anti-sense RNA spanning positions 2085–1730, which includes the 5′ splice site (1810) and the secretory poly(A) site (1998), in the presence of 5 μg tRNA. Single-stranded RNA was digested using 50 units RNase T1 and 0.5 μg RNase A for 30 min at 37° C. Products were quantitated by phosphorimagery.
Preparation of cytoplasmic extract and nuclear extract
Cells were harvested during exponential growth and counted using a Fischer scientific haemocytometer. Cell nuclear extracts were prepared using the modified extraction procedure originally developed by Dignam et al. (1983) and modified for B lymphocytes by Virtanen and Chen (1990) with hypotonic buffer A containing 10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.2 mM PMSF, 0.2% Triton X-100, and buffer C containing 20 mM HEPES (pH 7.9), 25% glycerol, 0.35 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 0.4 u/μL RNase inhibitor (Promega). The protein concentration in cytoplasmic extract and nuclear extract was determined by the Bio-Rad protein assay.
Northern blot to detect U1 snRNA level
The anti-U1 snRNA probe used for Northern blotting was prepared by in vitro transcription from Rsa 1 digested p630 plasmid containing the human U1 snRNA gene cloned into pGEM 3Z+ (Gunderson et al. 1998). RNA probes were prepared by in vitro transcription as previously described (Phillips and Virtanen 1997). RNA samples were proteinase K treated, phenol-chloroform extracted, and ethanol precipitated, separated by 8% urea-acryl-amide denaturing PAGE, and electrotransfered to Amersham hybond-N+ membrane. The proteins were cross-linked to the membrane in a UV Stratalinker 2400 apparatus. The dried membrane was prehybridized for at least 1 h and hybridized against U1snRNA probe (final concentration 4,000 dpm/μL) overnight. The membrane then was washed with 2× SSC and exposed to PhosphorImager screen or Blue Lite autorad film (Bioexpress).
Trimethyl guanosine immunoprecipitation
Nuclear extracts were mixed with 10 μL or 20 μL anti-2,2,7-trimethylguanosine (TMG) agarose beads (Oncogene) in 50 μL or 200 μL NET buffer (50mM Tris at pH 7.6, 150 mM NaCl, 0.01% NP-40) and 0.4 U/μL RNasin (RNase inhibitor), incubated at 4° C for 1 h, and supernatants were recovered by centrifugation at 14,000 rpm for 5 min. The supernatants were mixed again with beads, and the steps above were repeated once more to deplete the extracts of >95% of U1snRNA.
In vitro specific polyadenylation assay
The polyadenylation assays in nuclear extracts were performed according to the protocol as previous described (Virtanen and Sharp 1988). For each reaction, 30,000 cpm of 32p-labeled μ-secretory RNA substrate IgM1790–2030 mutss (Virtanen and Sharp 1988) (spanning positions 1790–2030 of accession number V00818 and containing a mutated 5′ splice site, g/gtaaac to g/caaacc, shown to eliminate splicing to the membrane exons) (Peterson and Perry 1989) were used. As described by Klein Gunnewiek et al. (2000), 0–10 ng SL2 RNA and MS2 RNA were added as competitor and control, respectively.
Immunopurification of non-snRNP-bound U1A from nuclear extracts
Rabbit polyclonal antibody 856 specific for U1A (Kambach and Mattaj 1992) (kindly provided by Iain Mattaj, EMBL, Heidelberg) was coupled to CNBr-activated sepharose 4B bead according to the Amersham Bioscience’s recommended protocol. Seven micrograms protein per milliliter of swollen beads was used.
U1 snRNP-associated U1A was removed from nuclear extracts by TMG immunoprecipitation as above. The TMG-depleted supernatant after centrifugation was adjusted to a final volume of 400 μL with NET buffer and pretreated with 20 μL uncoupled Sepharose 4B bead for 20 min at room temperature with gentle rotation. The pretreated supernatant was incubated with 20 μL U1A antibody-coupled Sepharose 4B beads at room temperature for 2 h with gentle rotation. Beads were then subjected to extensive washing as follows: twice with high salt NET buffer (500mM NaCl) and once with NET buffer (150 mM NaCl).
Twenty microliters (v/v = 1:1) elution buffer (50 mM KCl, 150 mM glycine at pH 1.5, 0.1% Triton-X) was added to the washed bead and incubated for 5 min with gentle shaking. The eluted supernatant was neutralized by 2 μL (1/10 volume) of 1.0 M Tris (pH 8.0). Purified non-snRNP-bound U1A was separated on 12% SDS-PAGE, visualized by Western blotting or silver stain, scanned, and quantitated in ImageQuant.
Nonspecific polyadenylation assay with immunoprecipitated non-snRNP-bound U1A
Nonspecific poly(A) assays were performed as previously described (Phillips et al. 2001) with 50 ng of recombinant PAP and 30,000 cpm of 32P-labeled μ-secretory RNA substrate IgM1790–2030 mutss and incubated for 30 min at 37° C.
Acknowledgments
This work was supported by a National American Heart Association Scientist Development Grant (0430004N) to C.P. and NIH GM57286 to S.G.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2159506.
REFERENCES
- Alt, F.W., Bothwell, A.L.M., Knapp, M., Siden, E., Mather, E., Koshland, M., and Baltimore, D. 1980. Synthesis of secreted and membrane- bound immunoglobulin μ heavy chains is directed by mRNAs that differ at their 3′ ends. Cell 20: 293–301. [DOI] [PubMed] [Google Scholar]
- Cox, A. and Emtage, J.S. 1989. A 6-fold difference in the half-life of immunoglobulin μ heavy chain mRNA in cell lines representing two stages of B cell differentiation. Nucleic Acids Res. 17: 10439– 10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam, J.D., Lebowitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early, P., Rogers, J., Davis, M., Calame, K., Bond, M., Wall, R., and Hood, L. 1980. Two mRNAs can be produced from a single immunoglobulin μ gene by alternative RNA processing pathways. Cell 20: 313–319. [DOI] [PubMed] [Google Scholar]
- Galli, G., Guise, J.W., McDevitt, M.A., Tucker, P.W., and Nevins, J.R. 1987. Relative position and strengths of poly(A) sites as well as transcription termination are critical to membrane versus secreted μ-chain expression during B-cell development. Genes & Dev. 1: 471–481. [DOI] [PubMed] [Google Scholar]
- Galli, G., Guise, J., Tucker, P.W., and Nevins, J.R. 1988. Poly(A) site choice rather than splice site choice governs the regulated production of IgM heavy-chain RNAs. Proc. Natl. Acad. Sci. 85: 2439– 2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsby, R.A. and Osborne, BA. 2000. B-cell development, activation and differentiation. In Kuby immunology (eds. R.A. Goldsby et al.), pp. 268–300. W.H. Freeman & Co., New York.
- Grosschedl, R. and Baltimore, D. 1985. Cell-type specificity of immunoglobulin gene expression is regulation by at least three DNA sequence elements. Cell 41: 885–897. [DOI] [PubMed] [Google Scholar]
- Gunderson, S.I., Vagner, S., Polycarpou-Schwarz, M., and Mattaj, I.W. 1997. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes & Dev. 11: 761–773. [DOI] [PubMed] [Google Scholar]
- Gunderson, S.I., Polycarpou-Schwarz, M., and Mattaj, I.W. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct inter-action between U1 70K and poly(A) polymerase. Mol. Cell 1: 255– 264. [DOI] [PubMed] [Google Scholar]
- Kambach, C. and Mattaj, I.W. 1992. Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J. Cell Biol. 118: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.J., Kanellopoulos-Lanevin, C., Merwin, R.M., Sachs, D.H., and Asofsky, R. 1979. Establishment and characterisation of Balb/c lymphoma lines with B cell properties. J. Immunol. 122: 549– 554. [PubMed] [Google Scholar]
- Klein Gunnewiek, J.M., Hussein, R.I., van Aarssen, Y., Palacios, D., de Jong, R., van Venrooij, W.J., and Gunderson, S.I. 2000. Fourteen residues of the U1 snRNP-specific U1A protein are required for homodimerization, cooperative RNA binding, and inhibition of polyadenylation. Mol. Cell. Biol. 20: 2209–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson, G. and Koshland, M.E. 1984. Changes in J-chain and μ-chain RNA expression as a function of B-cell differentiation. J. Exp. Med. 160: 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, C.S., Cooke, C., O’Connor, J.P., Kobayashi, R., and Alwine, J.C. 1998. The snRNP-free U1A (SF-A) complex(es): Identification of the largest subunit as PSF, the polypyrimidine-tract binding protein- associated splicing factor. RNA 4: 1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, J.O., Williams, G.T., and Neuberger, M.S. 1988. The half-life of immunoglobulin mRNA increases during B-cell differentiation: A possible role for targeting to membrane-bound polysomes. Genes & Dev. 2: 1003–1010. [DOI] [PubMed] [Google Scholar]
- Melton, D.A., Krieg, P.A., Rebagliati, M.R., Maniatis, T., Zinn, K., and Green, M.R. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12: 7035–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milcarek, C., Martincic, K., Chung-Ganster, L.-H., and Lutz, C.S. 2003. The snRNP-associated U1A levels change following IL-6 stimulation of human B-cells. Mol. Immunol. 39: 809–814. [DOI] [PubMed] [Google Scholar]
- O’Connor, J.P., Alwine, J.C., and Lutz, C.S. 1997. Identification of a novel, non-snRNP protein complex containing U1A protein. RNA 3: 1444–1455. [PMC free article] [PubMed] [Google Scholar]
- Oi, V.T., Morrison, S.L., Herzenberg, L.A., and Berg, P. 1983. Immunoglobulin gene expression in transformed lymphoid cells. Proc. Natl. Acad. Sci. 80: 825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, M.L. 1992. Balanced efficiencies of splicing and cleavage-polyadenylation are required for μs and μm mRNA regulation. Gene Expr. 2: 319–327. [PMC free article] [PubMed] [Google Scholar]
- Peterson, M.L. and Perry, R.P. 1989. The regulated production of μm and μs mRNA is dependent on relative efficiencies of ms poly(A) site usage and to the Cμ4-to-M1 splice. Mol. Cell. Biol. 9: 726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, M.L., Gimmi, E.R., and Perry, R.P. 1991. The developmentally regulated shift from membrane to secreted μ mRNA production is accompanied by an increase in cleavage-polyadenylation efficiency but no measurable change in splicing efficiency. Mol. Cell. Biol. 11: 2324–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, C. and Gunderson, S.I. 2003. Sequences adjacent to the 5′ splice site control U1A binding upstream of the IgM heavy chain secretory poly(A) site. J. Biol. Chem. 278: 22102–22111. [DOI] [PubMed] [Google Scholar]
- Phillips, C. and Virtanen, A. 1997. The murine IgM secretory poly(A) site contains dual upstream and downstream elements which affect polyadenylation. Nucleic Acids Res. 25: 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, C., Schimpl, A., Dietrich-Goetz, W., Clements, J.B., and Virtanen, A. 1996. Inducible nuclear factors binding the IgM heavy chain pre-mRNA secretory poly(A) site. Eur. J. Immunol. 26: 3144–3152. [DOI] [PubMed] [Google Scholar]
- Phillips, C., Jung, S., and Gunderson, S.I. 2001. Regulation of nuclear poly(A) addition controls the expression of immunoglobulin M secretory mRNA. EMBO J. 20: 6443–6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, C., Pachikara, N., and Gunderson, S.I. 2004. U1A inhibits cleavage at the immunoglobulin M heavy-chain secretory poly(A) site by binding between the two downstream GU-rich regions. Mol. Cell. Biol. 24: 6162–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki, Y., Seipelt, R.L., Peterson, M.L., and Manley, J.L. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952. [DOI] [PubMed] [Google Scholar]
- Virtanen, A. and Chen, U. 1990. Cell-free system for polyadenylation using mouse B cell extracts. In Immunological methods vol. IV (eds. I. Lefkovits and B. Pernis), pp. 95–105. Academic Press, New York.
- Virtanen, A. and Sharp, P.A. 1988. Processing at immunoglobulin polyadenylation sites in lymphoid cell extracts. EMBO J. 7: 1421– 1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L. and Luhrmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Behrens, S.E., and Luhrmann, R. 1993. Protein compositionof mammalian spliceosomal snRNPs. Mol. Biol. Rep. 18: 121–126. [DOI] [PubMed] [Google Scholar]