Abstract
Replication of the segmented double-stranded (ds)RNA genome of rotavirus requires the viral RNA-dependent RNA polymerase (RdRP) to use 11 different (+)RNAs as templates for (−) strand synthesis. Complementary sequences proximal to the 5′ and 3′ termini are predicted to direct cyclization of the (+)RNAs by forming panhandle structures from which short highly conserved terminal sequences protrude as single-stranded tails. Cell-free replication assays indicate that such structural organization of the 5′- and 3′-ends is required for efficient dsRNA synthesis. Multiple specifically recognized elements exist at the 3′-end that promote dsRNA synthesis including RdRP-recruitment signals and a (−) strand initiation sequence. In contrast to the 3′-end, the role of the 5′-end has been less well defined. In this study, we determined that the 5′-end contains a base-specific recognition signal that plays an important role in the assembly of the RdRP and cofactors into a stable initiation complex for (−) strand synthesis. The 5′ recognition signal is associated with the G2 residue of the 5′-consensus sequence, a residue that shows absolute conservation among all rotavirus groups (A, B, and C) examined to date. From our results, we suggest that rotavirus (+)RNA cyclization, although likely initiated by 5′- 3′ nucleotide complementarity, may be stabilized by RdRP-dependent bridging. Given that synthesis of the (−) strand on the (+)RNA template will disrupt 5′–3′ nucleotide interactions, RdRP-dependent bridging may be the sole mechanism by which the dsRNA product can be held in the necessary cyclized conformation required for efficient multiple rounds of transcription.
Keywords: rotavirus, RNA replication, initiation, RNA secondary structure
INTRODUCTION
The molecular mechanisms that orchestrate packaging and replication of the segmented double-stranded (ds)RNA genome of viruses belonging to the Reoviridae are poorly understood. The sequence of events includes the gene-specific interaction of multiple viral (+)RNAs with newly formed previrion cores (i.e., assortment) followed by RNA replication, a process in which the (+)RNAs are used as templates for (−) strand synthesis to produce the dsRNA genome segments (for review, see Patton and Spencer 2000). The precision of the assortment process is such that the newly formed cores will contain a complete constellation of genome segments. This precision suggests that each species of viral (+)RNA contains its own unique packaging signal. However, since only a single type of viral RNA-dependent RNA polymerase (RdRP) directs the synthesis of all the dsRNA genome segments, the various (+)RNA templates can be anticipated to share common cis-acting signals that promote (−) strand synthesis.
Of the 12 distinct genera within the Reoviridae, rota-viruses have received most attention owing to their responsibility as primary agents causing acute dehydrating gastroenteritis in infants and young children (Parashar et al. 2003). Studies on the molecular biology of rotaviruses have been particularly advanced by the availability of virion-derived and recombinant rotavirus RdRPs that specifically recognize viral (+)RNAs in vitro and that efficiently synthesize dsRNAs from viral (+)RNAs in fully definable cell-free replication systems (Chen et al. 1994; Patton et al. 1997; Tortorici et al. 2003). The rotavirion is an icosahedron consisting of three layers of protein that surround 11 segments of dsRNA (Prasad et al. 1988). The innermost layer is formed by 60 dimers of the core lattice protein, VP2, arranged with T = 1 symmetry (Lawton et al. 1997b). One copy each of the viral RdRP, VP1, and the mRNA-capping enzyme, VP3, are positioned at each of the 12 pentamers of the VP2 lattice. The dsRNA genome segments are thought to be organized in the core such that each interacts with one particular pentamer and its associated RdRP-capping enzyme complex (Prasad et al. 1996). In the virion, the intermediate layer protein, VP6, surrounds the VP2 lattice, and the outer layer proteins, VP4 and VP7, surround the VP6 lattice (Prasad et al. 1988).
Rotavirus entry is accompanied by loss of the VP4–VP7 protein layer, producing transcriptionally active double-layered particles that direct synthesis of the 11 capped (+)RNAs (Imai et al. 1983; Lawton et al. 1997a). Only at their termini do the (+)RNAs share sequence homology (Desselberger and McCrae 1994). Notably, for the medically significant group A rotaviruses, the (+)RNAs begin with 5′-GGC(A/U)6–8-3′ (5′-consensus sequence [5′CS]) and in most cases end with 5′-UGUGACC-3′ (3′CS) (Kearney et al. 2004). Cell-free replication assays have been useful in identifying cis-acting signals in rotavirus (+)RNAs that promote (−) strand synthesis (Patton et al. 1996; Wentz et al. 1996). Such analyses have shown that the 3′CS is essential for dsRNA synthesis, with the 3′-terminal CC of this sequence crucial for forming the (−) strand initiation complex (Chen and Patton 2000; Chen et al. 2001). Other cis-acting signals have been identified in the (+)RNAs that enhance (−) strand synthesis, albeit to an extent less than the 3′CS. Such enhancement signals have been mapped to the 5′-end of (+)RNAs and to regions of the RNA immediately upstream of the 3′CS (Patton et al. 1996, 1999; Barro et al. 2001; Chen et al. 2001).
Computer modeling suggests that base-pairing in cis between the 5′ and the 3′ enhancement signals of rotavirus (+)RNAs leads to cyclization and the formation of panhandle structures from which the 3′CS extends as an un-base-paired tail (Chen and Patton 1998; Patton and Spencer 2001). The ability of the (+)RNAs to function as templates for dsRNA synthesis is inhibited if mutations are introduced into the RNA, which converts the 3′CS from a single-stranded tail to one that is predicted to base-pair to the 5′ terminus (Chen and Patton 1998). These and related observations suggest that an important role of the 5′–3′ panhandle may be to stabilize the (+)RNAs in a conformation that allows the ready interaction of the RdRP with the 3′-terminal CC and, thus, the efficient formation of the (−) strand initiation complex.
Cell-free replication assays with purified recombinant proteins have shown that the RdRP requires the core lattice protein, VP2, to achieve the replicase activity that directs (−) strand synthesis (Patton et al. 1997). Interestingly, the ratio of VP1:VP2 required to stimulate maximum replicase activity is 1:10, the same ratio of VP1:VP2 that forms each of the vertices of the core. Recent studies have pointed to a direct role for VP2 in forming the initiation complex for (−) strand synthesis (Tortorici et al. 2003). The essential role that VP2 plays in (−) strand initiation suggests a mechanism whereby genome replication can be linked to core assembly and therefore to the packaging of the newly made dsRNA products. Although the RdRP lacks enzymatic activity in the absence of VP2, the polymerase alone can bind specifically to recognition signals in viral (+)RNAs (Patton 1996). In the case of the gene 8 (g8) (+)RNA, polymerase binding sites appear to be located within the 3′-enhancement signal (Tortorici et al. 2003). Given that neither the sequence nor predicted secondary structures of the 3′-enhancement signal are conserved among the viral (+)RNAs, RdRP recognition signals may be to some extent gene specific and therefore have a function in the assortment process.
In this study, we have used cell-free replication systems to understand the importance of the 5′ region, particularly its 5′CS, on the template activity of the rotavirus g11 (+)RNA. The contribution of the 5′ region was assessed by analyzing the template activity of g11 (+)RNAs with mutated 5′-terminal sequences that, in some cases, had predicted secondary structures different than that of the wild-type g11 (+)RNA. The analysis revealed a sequence-dependent contribution of residues within the 5′CS on the formation of the (−) strand initiation complex. These data provide the first experimental evidence that specific recognition of both the 5′ and 3′ ends of rotavirus (+)RNAs is involved in dsRNA synthesis. We propose that the rotavirus replicase complex interacts with both ends as a mechanism favoring the use of full-length (+)RNAs for replication and to produce cyclized dsRNA products appropriate for use as templates for transcription within the core.
RESULTS
Cis-acting replication signals in the g11 (+)RNA
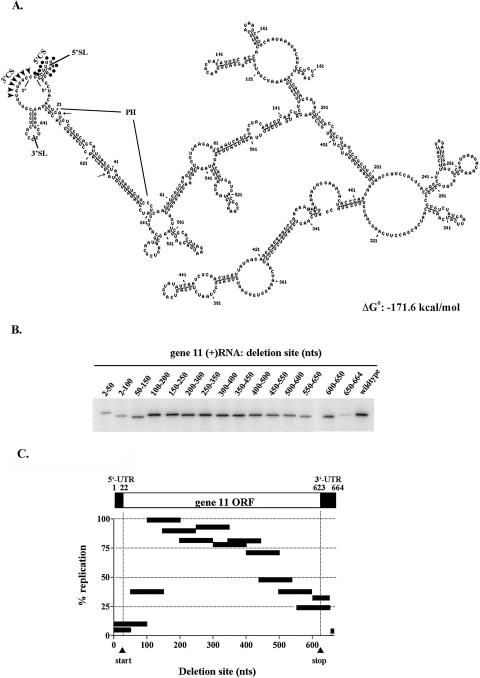
Computer modeling has indicated that portions of the 5′ and 3′-ends of at least some rotavirus (+)RNAs, such as those transcribed from g8 and g9, interact in cis to form modified panhandle structures (Patton and Spencer 2001). Such structures have been positively correlated with the efficient use of the (+)RNAs as templates for dsRNA synthesis (Chen and Patton 1998). As a first step in identifying the structural elements in the 664-nucleotide (nt) g11 (+)RNA of the rotavirus porcine strain CN86 (group A) that are important for (−) strand synthesis, the secondary structure of the RNA was predicted using the Massively Parallel Genetic Algorithm (MPGAfold) (Shapiro and Navetta 1994; Shapiro and Wu 1996; Shapiro et al. 2001a,b). In brief, MPGAfold provides a stochastic optimization procedure that generates a population of all possible stems for an RNA sequence, then mixes, combines, and recombines these stems in multiple iterations, ultimately developing lowest energy (best fit) and consensus structures. The algorithm operates in parallel upon a population of thousands to hundreds of thousands of RNA secondary structures in developing solutions for an RNA sequence. MPGAfold runs are independently repeated numerous times to identify a best fit or consensus structure and are executed on a high-performance parallel computer. The predicted secondary structure that corresponded to the best free-energy structure for the g11 (+)RNA at a 16K population (ΔG° = −171.6 kcal/mol) showed the presence of the following three features at the ends of the molecule: (1) a strong interaction near the 5′- and 3′-ends of the RNA, forming a relatively long stem (defined as the panhandle [PH]); (2) an open 5′–3′ terminus in which the 3′CS is un-base-paired; and (3) two hairpin loops, one located at the 5′ side (5′SL) and the other at the 3′ side (3′SL) of the open 5′–3′ terminus (Fig. 1A ▶). This 5′–3′ structural organization was predicted in 18 out of 20 MPGAfold runs. Notably, the initiating AUG codon (residues 22–24) for the NSP5 open reading frame (ORF) in the g11 (+)RNA is positioned near the 5′-end of the stretch of residues making up the 5′ side of the PH (5′PH). The terminating UAA codon (residues 613–615) is located near the middle stretch of residues forming the 3′ side of the PH (3′PH). Hence, residues contained within both the untranslated regions and ORF of the g11 (+)RNA participate in the formation of the panhandle and therefore have potential significance to the juxtapositioning of the 5′CS and the 3′CS.
FIGURE 1.
Structure and location of cis-acting replication signals in the g11 (+)RNA. (A) The best energy and consensus structure of the g11 (+)RNA predicted from the MPGAfold using runtime efn2 coaxial stacking rules. Relevant features include the panhandle (PH), the 5′ and 3′ stem loops (SL), and the 5′ and 3′-consensus sequences (CS). Dots and arrowheads denote the 5′CS and 3′CS, respectively. The beginning and end of the NSP5 ORF in the g11 (+)RNA are indicated with small arrows. (B) Template activity of g11 (+) RNAs containing deletions was determined using the open core replication system. Radiolabeled dsRNA products were resolved by gel electrophoresis and quantified with a PhosphorImager. (C) The results of two independent experiments were averaged and normalized to 100% for reaction mixtures containing the wild-type template. The bars indicate the position and the size of the deletion for each template.
To assess the importance of the predicted secondary structures in the g11 (+)RNA on (−) strand synthesis, a series of overlapping deletions were introduced into the RNA extending from the 5′ to the 3′-end. The template activity of the mutant RNAs was evaluated in cell-free replication assays that used disrupted (open) virion-derived cores as a source of replicase activity (Tortorici et al. 2003). The 32P-labeled dsRNA products of the assays were resolved by gel electrophoresis and quantified using a PhosphorImager. Overall, the analysis showed that deletion of residues at or near the ends of the g11 (+)RNA caused significant reductions in dsRNA synthesis, whereas deletion of residues located near the center of the RNA had little or no effect (less than or equal to twofold) (Fig. 1B,C ▶). Thus, based on this assay system, cis-acting signals critical to (−) strand synthesis are limited to the terminal regions of the g11 (+)RNA.
Deletion of the last 15 nt of the RNA (650–664), a region that includes the 3′CS, had the greatest inhibitory effect on g11 replication, reducing levels by ~20-fold (Fig. 1 ▶). This finding is consistent with earlier results showing that the 3′CS contains a cis-acting signal essential for efficient replication. Deletion of 50- and 100-nt stretches immediately upstream of the last 15 nt, regions that included residues located within the 3′SL and 3′PH, also reduced dsRNA synthesis but to an intermediate level (approximately three- to fourfold) (Fig. 1C ▶).
Deletion of either residues 2–50 or 2–100 of the g11 (+)RNA removed the 5′CS, 5′SL, and 5′PH residues and caused an ~10-fold reduction in the level of dsRNA synthesis (Fig. 1 ▶). Thus, although not as important as the 3′-terminal 15 nt in promoting g11 replication, cis-acting signals at the 5′-end are more significant for this process than those located in the 3′SL or the 3′PH. Because deletion of residues 50–150 reduced replication by approximately two- to threefold, much less than the effect of deleting residues from 2 to 50 or 2 to 100, it may be inferred that the key 5′-terminal signals promoting (−) strand synthesis are located within the first 50 nt of the RNA.
Effect of the 5′-end of the g11 (+)RNA on replication
To gain further insight into the nature of the 5′-terminal cis-acting signals in the g11 (+)RNA, mutant RNAs were prepared by in vitro transcription that lacked the first 12 (g11Δ12), 23 (g11Δ23), or 39 (g11Δ39) residues of the wild-type sequence. Nontemplated G additions and C > A replacements were included at the 5′-ends of the mutant RNAs to enhance their transcription by T7 and SP6 RNA polymerases, respectively. The activity of the mutant RNAs as templates for dsRNA synthesis was compared with that of wild-type g11 (g11wt) (+)RNA using the open core replication system. The results showed that the 12-nt deletion introduced into the g11Δ12 RNA reduced its template efficiency by ~50% (Fig. 2A,B ▶). Comparison of the predicted structure of the g11Δ12 RNA with that of the g11wt RNA suggested that the mutant RNA may have been a less efficient template for dsRNA synthesis because it lacked the 5′SL or the 5′CS (Fig. 2C ▶). However, the g11Δ12 RNA retained several of the same structural features predicted for the wild-type RNA including the full-length PH, the 3′SL, and an open 5′–3′ terminus containing an un-base-paired 3′CS.
FIGURE 2.
Effect of progressive deletions from the 5′-end on g11 replication. Wild-type g11 (+)RNA and derivatives with 5′-truncations of 12, 23, or 39 residues were assayed for template activity using open cores (A) or rVP1 and rVP2 (B). Radiolabeled products were resolved by gel electrophoresis and quantified with a PhosphorImager. For this and subsequent figures, levels of dsRNA product for duplicate independent assays were averaged, normalized to 100%, and plotted along with standard errors. (C) Secondary structures of the indicated RNAs were predicted using MPGAfold. Details for the g11wt RNA are presented in Figure 1 ▶. The structures shown for variants g11-Δ12, -Δ23, and -Δ39 all represent the best fit prediction (lowest energy) and the consensus structures with an appearance rate of >50% in 20 runs of each RNA. Energy values (ΔG° in kcal/mol) are also included.
The 23- and 39-nt deletions introduced into the g11Δ23 and g11Δ39 RNAs decreased their template efficiencies by ~75% and 90%, respectively, and thus made them poorer templates for dsRNA synthesis than the g11Δ12 RNA (Fig. 2 ▶). The 23- and 39-nt deletions also had greater effects on the predicted g11 structure than the 12-nt deletion of the g11Δ12 RNA. Specifically, the 23- and 39-nt deletions not only caused a loss of the 5′CS and the 5′SL, but also reduced the openness of the 5′–3′ terminus due in part to base-pairing involving residues of the 3′CS. In addition, the 23-and 39-nt deletions led to an extension of the stem length of the 3′SL. For both the 23- and 39-nt deletions, the PH became truncated, much more radically in the case of the g11Δ39 RNA. Hence, although the decreased template activity of the g11Δ12 RNA could be correlated with either the loss of the 5′CS or the 5′SL, the decreased activity of the g11Δ23 and g11Δ39 RNAs could also be correlated with the loss of the 5′CS or changes to a number of structural elements. Earlier studies indicating that base-pairing of residues making up the 3′CS reduces the template efficiency of rotavirus (+)RNAs provides a possible explanation for why g11Δ23 and g11Δ39 RNAs were replicated less efficiently than the g11Δ12 RNA (Chen and Patton 1998).
Residues of the 5′CS contributing to replication
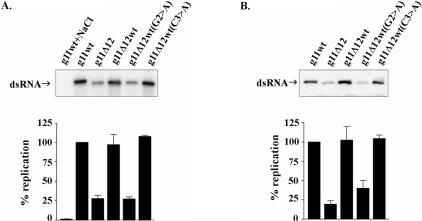
To further investigate the role of the 5′CS and 5′SL in the efficient replication of the g11 (+)RNA, we replaced the first six residues of the g11Δ12 RNA (GAUACA) with residues corresponding to the first six residues of the g11 5′CS (GGCUUU) (Fig. 3A ▶). MPGAfold-based analysis of the resultant RNA, g11Δ12wt RNA, indicated that it was structurally similar to the g11Δ12 RNA and differed from the wild-type RNA primarily in lacking the 5′SL (Fig. 3C ▶). The template activity of the g11Δ12wt RNA was compared with that of the g11wt and g11Δ12 RNAs using the open core replication system. The analysis revealed that the g11Δ12wt RNA replicated several times more efficiently than the g11Δ12 RNA, reaching levels of template activity equivalent to that of g11wt (+)RNA (Fig. 3B ▶). These results indicate that the GGCUUU portion of the 5′CS contains a cis-acting signal promoting genome replication. The fact that the g11Δ12wt RNA, though lacking a 5′SL, promoted minus-strand synthesis as efficiently at the wild-type RNA suggests that the 5′SL does not represent a cis-acting replication signal.
FIGURE 3.
Effect of 5′-terminal point mutations on the replication of the g11 (+)RNA. (A) Wild-type g11 (+)RNA and derivatives containing mutations in the 5′CS (GGCUUU) were assayed for template activity in the open core replication system. Point mutations introduced into the g11Δ12wt RNA are indicated by asterisks. (B) Radiolabeled products were resolved by gel electrophoresis and quantified with a PhosphorImager. Normalized levels of dsRNA product in reaction mixtures are plotted. (C) The secondary structures of the indicated RNAs were predicted using the MPGAfold program. Structures depicted represent the 5′–3′-end of the lowest energy or very near-lowest energy predictions generated at a 16K population. Exceptions to this are the structure associated with g11Δ12(G2 > A), which represents the best energy structure at 32K, and the structure associated with g11Δ12(U4 > A), which represents a structure that is 0.7 kcal above the best structure, generated at 32K. 32K populations were used in these instances because the results at 16K were somewhat mixed. Arrows denote positions of mutations. Differences in template activities between experiments (cf. g11Δ12 RNA results shown here with those in Fig. 2 ▶) may stem from one or more factors, including variations in the enzymatic activities associated with different preparations of open cores, minor inaccuracies in the quality and the quantity of different RNA preparations, and/or imprecision in the quantification of band intensities by PhosphorImager analysis.
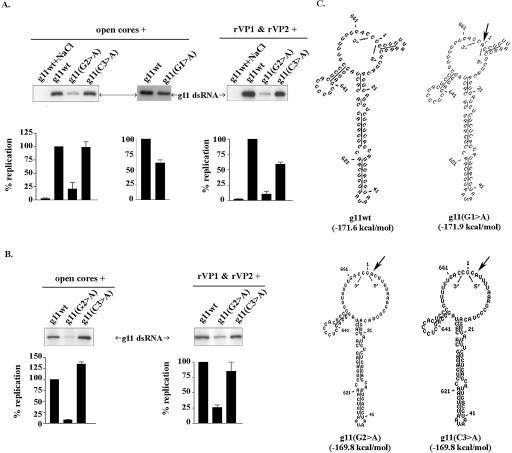
Because MPGAfold modeling indicated that the 5′-terminal GGCUUU was single-stranded and not part of any predicted secondary structure in the g11Δ12wt RNA, we explored the possibility that the GGCUUU stimulated dsRNA synthesis via a sequence-specific mechanism. This was approached by making derivatives of the g11Δ12wt RNA in which residues 2–6 of its 5′-terminal GGCUUU were individually replaced with adenine (Fig. 3A ▶). Interestingly, we found that when the second G (G2) of the sequence was changed to an A (g11Δ12wt(G2 > A)), the ability of the RNA to function as a template for dsRNA synthesis in vitro was reduced by approximately fourfold (Fig. 3B ▶). Based on the similarity of the proposed structures for the g11Δ12wt(G2 > A) and g11Δ12wt RNAs, the effect of the G2 > A replacement on template activity was not likely due to any an effect on secondary structure (Fig. 3C ▶). Hence, these results indicate that G2 enhances dsRNA synthesis through a base (G)-specific mechanism.
In contrast to the strongly negative effect of the G2 > A replacement on replication, the C3 > A replacement in the g11Δ12wt(C3 > A) RNA had little or no effect on dsRNA synthesis (Fig. 3 ▶). This replacement also had no effect on the predicted structure of the RNA molecule, relative to either the g11Δ12wt(G2 > A) RNA or its parental g11Δ12wt RNA. Thus, despite being un-base-paired and adjacent to the replication-enhancing G2 residue, C3 appears not to contribute to the efficient replication of the g11 RNA. Similarly, replacement of U4, U5, or U6 with adenine yielded RNAs [g11Δ12wt(U4 > A), g11Δ12wt(U5 > A), and g11Δ12wt-(U6 > A)] that exhibited little reduction (less than twofold) in template activity as compared with the g11Δ12wt RNA. Overall, the predicted structures of the three U > A mutant RNAs were like that of the parental g11Δ12wt RNA. The single exception was the slight reduction in size of the open 5′–3′ end and the increased PH length associated with the g11Δ12wt(U6 > A) RNA. The combined deletion of U4, U5, and U6 produced an RNA (g11Δ12(ΔU)) that replicated ~50% as efficiently as the parental g11Δ12wt RNA (Fig. 3 ▶). Despite the deletion of the three U residues, the predicted structure of the g11Δ12(ΔU) RNA remained much the same as the g11Δ12wt RNA. The only difference between the two was that the size of the open 5′–3′ end was slightly smaller in the case of the g11Δ12(ΔU) RNA, due to a decrease in the length of the un-base-paired 5′-tail from six to three residues. Thus, the decreased template efficiency of the g11Δ12(ΔU) RNA could be correlated with changes to the juxtapositioning of the 5′ and 3′-ends, brought about by truncation of the single-stranded 5′-end. Replacement of the three U residues with the sequence ACA yielded an RNA g11Δ12(ΔU > ACA) RNA that replicated as poorly as the g11Δ12 RNA (Fig. 3 ▶). The effect of this replacement on dsRNA synthesis likely resulted from structural changes that caused the loss of the single-stranded 5′-end and the partial base-pairing of the 3′CS. Taken together, the results indicate that residues U4, U5, and U6 appear to contribute minimally to efficient replication, with roles for the U residues possibly tied to providing the necessary length of un-base-paired sequence at the 5′-end for the efficient function of the G2 enhancement signal.
The importance of the 5′CS, particularly residues G2 and C3, on dsRNA synthesis was also addressed by mutagenesis of the full-length g11 (+)RNA. The analysis showed that the template activity of the full-length molecule was reduced by approximately fourfold when its G2 was mutated to an adenine and was not affected when its C3 was similarly mutated (Fig. 4A ▶). Given that the mutant forms of the full-length RNA were predicted to have identical open 5′–3′ ends (Fig. 4C ▶), it seems unlikely that the differences in the template activities of the mutant RNAs were due to structural differences. Instead, the findings support the hypothesis that base-specific recognition of G2, and not C3, is required for efficient replication of the g11 (+)RNA.
FIGURE 4.
Importance of the conserved G2 residue in g11 replication. (A) Wild-type g11 (+)RNA and derivatives with G1 > A, G2 > A, and C3 > A mutations were assayed for template activity in reaction mixtures containing open cores or rVP1 and rVP2. Initiation-complex formation was blocked in some reaction mixtures by including 250 mM NaCl (Chen and Patton 2000). (B) Initiation assays were performed by preincubating the RNAs in reaction mixtures identical to A except for lacking all nucleotides but GTP. Subsequently, NaCl was added to block further formation of initiation complexes, and the missing NTPs were added to promote (−) strand elongation from preformed initiation complexes. Radiolabeled dsRNA products were detected by gel electrophoresis and quantified with a PhosphorImager. (C) The secondary structures of the indicated RNAs are the lowest energy predictions obtained from 20 runs of MPGAfold at a 16K population. Arrows denote positions of mutations.
To gain insight into the contribution of the G1 residue on the template activity of the full-length g11(+)RNA, we used a T7 class II promoter (Huang 2003) to prepare g11 transcripts initiating with an adenine instead of a guanine. Analysis of the g11(G1 > A) RNA in open core replication assays showed that the mutant RNA was replicated to a level that was ~60% of that of wild-type RNA (Fig. 4A ▶). This suggests a contribution of the G1 residue to template activity, but to an extent less than that of the G2 residue. The similarity in the predicted structures of the g11wt and g11(G1 > A) (+)RNAs supports an interpretation that the contribution of the G1 residues to replication is through a base-specific mechanism (Fig. 4C ▶).
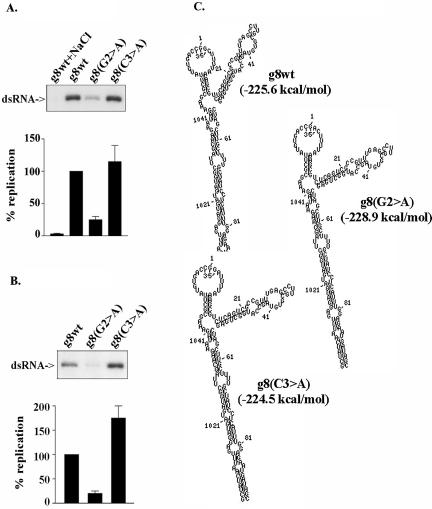
Because the 5′CS is a common element among rotavirus (+)RNAs (Desselberger and McCrae 1994), we also addressed the possibility that the G2 residue was important for the replication of other than just the g11 (+)RNA. To accomplish this, we prepared mutant g8 (+)RNAs containing G2 > A or C3 > A replacements and compared their template activity to wild-type g8 (+)RNA using the open core replication system. According to MPGAfold-based predictions, these mutations did not alter the predicted structure of the g8 RNAs from that of the wild-type species (Fig. 5C ▶). Interestingly, the g8 RNAs contained many of the same structural features predicted for the g11 (+)RNA including a stable PH, an open 5′–3′ end with both a single-stranded 3′CS and 5′GGC, and a 5′SL. Replication assays of the mutant g8 (+)RNAs showed that the G2 > A mutation reduced the template activity of the g8 (+)RNA by approximately fourfold, while the C3 > A mutation had no significant effect (Fig. 5A ▶). These results indicate that base-specific recognition of the G2 residue is required for the efficient replication of rotavirus (+)RNAs in general and does not apply just to the g11 (+)RNA.
FIGURE 5.
Common effect of the conserved G2 residue on template activity of viral (+)RNAs. (A) Wild-type g8 (+)RNA and derivatives with G2 > A and 3 C3 > A mutations were assayed for template activity in reaction mixtures containing open cores. Initiation-complex formation was blocked in a control reaction by including 250 mM NaCl. (B) Initiation assays were performed by preincubating the g8 RNAs in reaction mixtures identical to A except lacking all nucleotides but GTP. Additional initiation-complex formation was then blocked by adding NaCl, and missing NTPs were added to promote (−) strand elongation from preformed initiation complexes. Radiolabeled dsRNA products were detected by gel electrophoresis and quantified with a PhosphorImager. (C) The secondary structures of the indicated RNAs are the lowest or near-lowest energy predictions obtained from 20 runs of the MPGAfold at 32K and 64K populations.
Importance of the 5′CS to (−) strand initiation
Although it is apparent that the 5′-end of rotavirus (+)RNAs enhances (−) strand synthesis, it is not clear whether this effect is exerted at the level of initiation, elongation, and/or the recycling of the RdRP from one template to another. Earlier studies have demonstrated that the formation of the functional initiation complex for (−) strand synthesis is a salt-sensitive process that requires five components: viral (+)RNA, VP1, VP2, GTP, and Mg2+ (Tortorici et al. 2003). Once formed, the (−) strand initiation complex is stable even upon exposure to high concentrations of salt. Under these same high salt conditions, the RdRP of the initiation complex can transition to an elongation function when all four NTPs are present.
To investigate the importance of the 5′-end in the formation of the (−) strand initiation complex, g11wt (+)RNA and various forms of the g11Δ12 (+)RNA were incubated with open cores, GTP, and Mg2+ under conditions (low salt) allowing formation of initiation complexes. Afterward, the reaction mixtures were brought to 250 mM NaCl to prevent further de novo formation of initiation complexes. To promote (−) strand elongation from the preformed initiation complexes, the reaction mixtures were supplemented with 32P-UTP and cold ATP, CTP, and UTP. The level of 32P-labeled dsRNA made in these initiation assays was taken as a measure of the efficiency of the g11 (+)RNAs in supporting formation of (−) strand initiation complexes. The analysis revealed that the ability of the template RNAs to support dsRNA synthesis closely paralleled their ability to support initiation (Fig. 6 ▶). Specifically, the same mutations in the g11Δ12 and g11Δ12wt(G2 > A) RNAs that led to a reduction of dsRNA synthesis in the open core replication system led to a quantitatively similar reduction in the formation of (−) strand initiation complexes. Mutations associated with the g11Δ12wt and g11Δ12(C3 > A) RNAs had no effect on dsRNA synthesis in the open core system and, likewise, had no effect on the formation of initiation complexes. These results indicate that the 5′-end of the g11 (+)RNA contains an enhancement signal that is crucial for the efficient formation of the (−) strand initiation complex. The activity of the signal requires the contribution of the first 12 nt of the RNA, notably residue G2.
FIGURE 6.
Role of the 5′-end of the template RNA on formation of the (−)strand initiation complex. (A) Wild-type g11 (+)RNA and mutant species were assayed for template activity in reaction mixtures containing open cores. Initiation-complex formation was blocked in a control reaction by including 250 mM NaCl. (B) Initiation assays were performed by pre-incubating the RNAs in the same reaction mixtures used in A except lacking all nucleotides but GTP. Additional initiation-complex formation was then blocked by adding NaCl, and missing NTPs were added to promote (−) strand elongation from preformed initiation complexes. Radiolabeled dsRNA products were detected by gel electrophoresis and quantified with a PhosphorImager.
Assays performed with mutant species of full-length g11 (+)RNAs provided additional evidence of the importance of G2 in the formation of the (−) strand initiation complex. In particular, the results showed that a G2 > A substitution in the RNA reduced initiation-complex formation by several fold, while a C3 > A substitution had no significant effect (Fig. 4B ▶). Assays performed with mutant species of the g8 (+)RNA confirmed the importance of the G2 position in formation of the (−) strand initiation complex and supported the idea of a direct connection between levels of initiation-complex formation and levels of dsRNA synthesis in open core assays (Fig. 5B ▶). Thus, mutations at the 5′-end appear not to have an effect on (−) strand elongation.
VP3-independent role of the 5′-terminal cis-acting replication signal
VP3, a minor protein component of the core, has multiple activities related to the capping of the 5′-end of viral (+)RNAs including guanylyltransferase and methyltransferase activities and affinity for RNA (Lui et al. 1992; Chen et al. 1999; Patton and Chen 1999). To address the possibility that the activity of the 5′-enhancement signal was mediated via the interaction of VP3 with the 5′-end of the RNA, the template activity of the mutant g11 (+)RNAs was assayed in a cell-free replication system that contained baculovirus-expressed recombinant (r) VP1 and VP2 instead of open cores. VP1 was expressed with a C-terminal histidine tag (rVP1-His) to ease its purification. The catalytic and RNA-binding activities of rVP1-His are indistinguishable from those described earlier for the untagged form of VP1 (Tortorici et al. 2003; data not shown). The results of replication assays with rVP1-His and rVP2 showed that despite the absence of VP3, the progressive deletion of 5′-terminal residues from the g11 (+)RNA resulted in a reduction in dsRNA synthesis paralleling that which occurred in assays containing open cores (Fig. 2B ▶). Likewise, the effect of G2 > A and C3 > A mutations on the replication of the g11 (+)RNA by rVP1-His and rVP2 was similar to the effect of these mutations on the replication of the g11 (+)RNA by open cores (Fig. 2A ▶). As found using open cores, the formation of (−) strand initiation complexes in reaction mixtures containing recombinant rVP1-His and rVP2 was significantly reduced by the introduction of a G2 > A mutation in the g11 (+)RNA, but relatively unaffected by a C3 > A mutation (Fig. 4B ▶). These data exclude a role for VP3 both in the function of the 5′-enhancement signal in the formation of the (−) strand initiation complex and in the efficient synthesis of dsRNA in vitro.
DISCUSSION
Given the absence of a reverse genetics system for the rotaviruses, cell-free replication systems have been relied on heavily to identify cis-acting signals in the viral (+)RNAs that promote (−) strand synthesis (Chen et al. 1994; Patton et al. 1997). Such analyses have indicated that (−) strand synthesis is stimulated by a combination of signals, including those present at the 5′ and 3′-ends of the (+)RNA (Patton et al. 1996; Wentz et al. 1996). Whereas previous studies have indicated that the polymerase specifically interacts with multiple functionally distinct elements at the 3′-end, including RdRP-recognition signals and the (−) strand initiation sequence (Chen and Patton 2000; Tortorici et al. 2003), prior reports have not provided evidence of a specific interaction between the polymerase and the 5′-end. Indeed, heretofore, the 5′-end has been presumed to promote (−) strand synthesis simply by interacting with the 3′-end to drive the formation of a panhandle structure from which the 3′CS can extend as a polymerase-accessible single-stranded tail (Patton and Spencer 2000). However, the results of this study provide evidence that recognition of highly conserved residues at the 5′-end of rotavirus (+)RNAs is essential for efficient assembly of (−) strand initiation complexes. Thus, the role of the 5′-end may extend beyond simply cooperating with the 3′-end to form a structurally appropriate 3′-terminal RdRP-recruitment signal, to include presentation of a 5′ recognition signal that stimulates assembly of the (−) strand initiation complex. Further support of this hypothesis will require direct experimental evidence that the 5′–3′ interactions predicted for the g11 (+)RNA by MPGAfold accurately reflect structures formed by the ends of the RNA.
Mutagenesis indicates that the 5′ recognition signal is located in the group A rotavirus 5′CS (GGC(A/U)6–8), most notably being associated with the G2 residue. Interestingly, the (+)RNAs of all groups of rotavirus examined to date (groups A, B, and C) initiate with the dinucleotide GG, indicating that the 5′ recognition signal is highly conserved within this genera of viruses (Yang et al. 2004). In contrast, the C3 residue is not conserved among all virus groups and is not required for activity of the 5′ recognition signal. The U4, U5, and U6 residues exhibit a lesser role in the activity of the g11 5′ recognition signal, one that is perhaps linked to allowing the signal to extend sufficiently from the panhandle region so as to function appropriately in forming the initiation complex. Based on the MPGAfold predicted structures of mutant g11 (+)RNAs, the G2 residue can carry out its function independently of any 5′-terminal secondary structure that is upstream of the PH region. In fact, the results of earlier studies examining the impact of modifying the 5′-terminal sequences from an un-base-paired to base-paired form using complementary oligonucleotides suggest that the 5′-recognition signal needs to be single stranded for it to effectively promote replication (Barro et al. 2001).
The formation of the (−) strand initiation complex and the synthesis of full-length dsRNA by the cell-free replication system require only two proteins: the viral RdRP, VP1, and the core lattice protein, VP2 (Patton et al. 1997). Both of these proteins have affinity for RNA, but only in the case of VP1 has a sequence-dependent activity been detected (Labbe et al. 1994; Tortorici et al. 2003). Although we have successfully used electrophoretic mobility shift assays (EMSA) to identify RdRP-recognition signals at the 3′-end of rotavirus (+)RNAs, similar attempts to identify an interaction of the polymerase with the 5′-recognition signal have been unsuccessful so far (Tortorici et al. 2003; data not shown). One possible explanation for this failure is that the 5′ recognition signal is too small to produce a sufficiently stable probe-VP1 complex that can be detected by EMSA. Alternatively, the binding site for the 5′ recognition signal may not exist on the apo-polymerase but instead only manifest itself upon the interaction of VP1 with VP2 or with the other cofactors required in forming the initiation complex. We also cannot exclude the possibility that VP2, and not VP1, interacts with the 5′-recognition signal. Despite uncertainty over the mechanism of action of the 5′ recognition signal, our analysis is most consistent with the concept that (−) strand initiation is dependent on the interaction on one or more proteins of the replicase complex (i.e., VP1 and/or VP2) with both ends of the template RNA. An interaction of a viral polymerase with both ends of a template RNA has been noted previously for influenza virus (Li et al. 1998; Flick and Hobom 1999).
X-ray diffractions obtained from soaks of reovirus RdRP crystals with m7GpppG cap analog have revealed the existence of a cap-binding site on the surface of the enzyme that is positioned away from the (+)RNA entry channel (Tao et al. 2002). Given parallels in the replication strategies and core structures of rotavirus and reovirus, it may be anticipated that the rotavirus RdRP will also contain a cap binding site. Whether such a site would direct the interaction of the rotavirus RdRP with the 5′ recognition signal seems questionable though, given that the viral (+)RNAs that we have analyzed in this study lacked 5′-caps but were still recognized by the polymerase complex. Moreover, replication assays carried out with capped (+)RNAs have not so far shown any difference in template activity in comparison to uncapped RNAs (data not shown). Hence, the 5′ recognition signal of rotavirus (+)RNAs is probably not enhancing the formation of the (−) strand initiation complex by interacting with a cap-binding site on the viral polymerase. It may be that such cap binding sites are involved with (+)RNA synthesis (i.e., transcription) as opposed to (−)RNA synthesis.
An emerging theme concerning viral RNA replication is the recognition of 5′–3′ terminal interactions as an important prelude for generating template RNAs that are efficiently replicated and/or transcribed. Such quasi-circularization of viral RNAs has been proposed to occur for (+) strand RNA viruses, segmented and nonsegmented (−) strand RNA, and dsRNA viruses, and to be driven by 5′–3′ protein bridging or nucleotide complementarity, or a combination of the two (Hahn et al. 1987; Hsu et al. 1987; Pugachev and Frey 1998; Bae et al. 2001; Barton et al. 2001; Herold and Andino 2001; You et al. 2001; Barr and Wertz 2004; Gorchakov et al. 2004; Van Den Born et al. 2004). In the case of rotavirus and other members of the Reoviridae, 5′–3′ nucleotide complementarity alone appears sufficient to drive circularization of the (+) strand template for dsRNA synthesis (Anzola et al. 1987; Chapell et al. 1994). However, as (−) strand synthesis proceeds on the (+) strand template, the nucleotide complementarity required to hold the template in a cyclized conformation would be disrupted and could only be retained via the interaction of ends of the replicating RNA with the polymerase complex. Such protein-based circularization of the dsRNA product of replication is likely critical for efficient transcription of the rotavirus genome, as it would provide a mechanism for the polymerase to engage expeditiously the initiation site for (+) strand synthesis following a previous round of transcription on the template.
MATERIALS AND METHODS
Preparation of open cores
The DxRRV strain of rotavirus (Midthun et al. 1985) was propagated in MA104 cells, purified by CsCl centrifugation, and used to prepare open cores as described elsewhere (Patton and Chen 1999).
Preparation of recombinant baculoviruses
To generate a recombinant baculovirus (rBV) expressing VP1 with a C-terminal tag of six histidine residues (rVP1-His), the g1 cDNA of SA11 rotavirus in the vector pCR-Bacg1–4 (Patton et al. 1997) was amplified by polymerase chain reaction (PCR) using the forward primer, 5′-gcttaaagccgaattcgaagcttgg-3′, and the reverse primer, 5′-ctagtctatctaatggtgatggtgatgatgatcttgaaagaagttcgcg-3′. (Residues specifying the histidine tag are underlined.) The amplified product was self-ligated, yielding pCR-Bacg1C-His. This vector was cotransfected with linear Bac-N-Blue linear DNA (Invitrogen) into Sf9 cells according to the protocols of the suppliers. Recombinant viruses were selected and plaque purified in the presence of 5-bromo-4-chloro-3-indolyl-β-D-galactopyrano-side (X-Gal). A plaque-purified virus, rBVg1C-His, expressing rVP1-His, was identified by electrophoretic analysis of infected-cell lysates and by Western blot assay using VP1- and penta-His antisera. The sequence of the g1 cDNA in the virus was verified using an Applied Biosystems automated sequencer.
An rBV encoding the SA11 g2 product VP2 was prepared as described earlier (Patton et al. 1997).
Purification of rVP1 and rVP2
To prepare rVP1-His, spinner cultures of Sf21 were infected at a multiplicity of infection of two with rBVg1C-His and maintained in TNM-FH medium (Invitrogen) containing 2% fetal bovine serum and 1 μg per mL of each leupeptin and aprotinin. Three days post-infection, the cells were washed with phosphate-buffered saline, resuspended in lysis buffer (25 mM sodium phosphate at pH 7.8, 150 mM NaCl, and 1× EDTA-free protease inhibitor cocktail) (Roche), and sonicated. After removal of the membrane fraction by low-speed centrifugation, rVP1C-His was precipitated from the supernatant in a cut of 20%–25% (w/v) ammonium sulfate. The protein was dissolved in lysis buffer and applied to a cobalt-affinity (Talon) column (BD Bioscience). rVP1-His was recovered by elution with imidazole, loaded on a heparin-Sepharose column (Amersham Bioscience), and eluted with a gradient of NaCl. The peak fraction containing rVP1-His was concentrated, loaded on a Superdex-200 size exclusion column (Amersham Bioscience), and eluted with a buffer containing 25 mM HEPES (pH 7.8) and 150 mM NaCl. The eluted rVP1-His was stored at 4°C.
The expression and purification of rVP2 was described previously (Tortorici et al. 2003). The concentration of purified rVP1-His, rVP2, and open cores was determined by comparison with known amounts of BSA electrophoresed on SDS-polyacryl-amide gels and stained with Coomassie Brilliant Blue.
Transcription templates
The T7 transcription vector SP65g11cn86 contains a full-length cDNA of the rotavirus CN86 g11 RNA (Patton et al. 1999). PCR was used to introduce deletions into the g11 cDNA of SP65g11cn86 as appropriate for making the derivative vectors required for producing the T7 transcripts used in the experiment presented in Figure 1 ▶. The reaction mixtures contained Elongase DNA polymerase and the oligonucleotides indicated in Table 1 ▶. The amplification products were gel-purified, blunt-ended with T4 DNA polymerase, kinased, and self-ligated to produce the vector (Patton et al. 1999).
TABLE 1.
Primers used to introduce deletions into the g11 cDNA of SP65g11cn86a
| Deleted region | Forward primer | Reverse primer |
| 2–50 | ccggatatgcatttccctcaatttcttctagt | cggggcggccgccctatagtgagtcgtatta |
| 2–100 | ccggatatgcattcaactctttctggaaaatc | cggggcggccgccctatagtgagtcgtatta |
| 50–150 | ccggatatgcatgatgcagaagcattcaataa | cggggcggccgccgtcaatgctgagagacatc |
| 100–200 | ccggatatgcattggaccatctgattctgctt | cggggcggccgcacgttgtagaagatgattca |
| 150–250 | ccggatatgcatgatcgaatgcagttaagaca | cggggcggccgctggtgaaatgtactgttcac |
| 200–300 | ccggatatgcattcaacgcaatcacgaccttc | cggggcggccgcatatcctctggagacttcga |
| 250–350 | ccggatatgcatctccttaactaaaggtgtta | cggggcggccgctaatcgaaaagctggtgagt |
| 300–400 | ccggatatgcatcaatctcaactgattataag | cggggcggccgctgaatccatagacacgccag |
| 350–450 | ccggatatgcataaacactacccaagaattga | cggggcggccgcaaatccacttgatcgcaccc |
| 400–500 | ccggatatgcatagatgattcagacagtgatg | cggggcggccgcatatacatgaatcaagatta |
| 450–550 | ccggatatgcatagaagtatttcgcacttaga | cggggcggccgccctatttttatcctttttgg |
| 500–600 | ccggatatgcatatcgaagatttgtaatgtca | cggggcggccgcaaaacgtaatcttcggaatc |
| 550–650 | ccggatatgcatcccgttttgtgaccgcgg | cggggcggccgctcttatatttacaattctta |
| 600–650 | ccggatatgcatcccgttttgtgaccgcgg | cggggcggccgccaattgcattgcgacttgct |
| 650–664 | ccggatatgcatgaattccgtgtattctatag | cggggcggccgcagtggggagctccctagtgt |
aForward and reverse primers were used in polymerase chain reactions to delete the indicated residues in the g11 cDNA of the vector SP65g11cn86. Forward and reverse primers contained NsiI and NotI sites, respectively (underlined).
PCR was also used to introduce 5′-terminal mutations in the g8 and g11 cDNAs in the vectors SP65g8R and SP65g11cn86, respectively. The upstream primer included an SP6 or T7 promoter sequence, allowing the amplification products to be directly transcribed with the appropriate RNA polymerase. The g11(G1 > A) (+)RNA transcript was made from an amplified cDNA linked to an upstream T7 class promoter (Huang 2003) using the forward primer g11classIII (Table 2 ▶). In replication assays using the g11 (G1 > A) (+)RNA, the control g11wt (+)RNA was made from an amplified cDNA using the forward primer g11classIII. Amplification products were blunt-ended T4 DNA polymerase and gel-purified prior to transcription. Nucleotide sequences were confirmed using an Applied Biosystems automated sequencer.
TABLE 2.
Primers used in polymerase chain reactions to amplify templates for RNA synthesisa
| Primer | Sequence | Template (RPb) | RNA |
| g11(f) | taatacgactcactataggcttttaaagcgctacagtgatgctctc | SP65g11cn86 (T7) | g11wt |
| g11classIII(f) | attcagtaatacgactcactataggcttttaaagcgctacagtgatgctctc | SP65g11cn86 (T7) | g11wt |
| g11Δ12(f) | atttaggtgacactatagatacagtgatgtctctcagcattgacgtaacg | SP65g11cn86 (SP6) | g11Δ12 |
| g11Δ23(f) | taatacgactcactataggtctctcagcattgacgtaacgagtcttccctc | SP65g11cn86 (T7) | g11Δ23 |
| g11Δ39(f) | taatacgactcactatagggtaacgagtcttccctcaatttcttctagtatc | SP65g11cn86 (T7) | g11Δ39 |
| g11Δ12wt(f) | taatacgactcactataggctttgtgatgtctctcagcattgacg | SP65g11cn86 (T7) | g11Δ12wt |
| g11Δ12wt(G2>A)(f) | atttaggtgacactatagactttgtgatgtctctcagcattgacg | SP65g11cn86 (SP6) | g11Δ12wt(G2>C) |
| g11Δ12wt(C3>A)(f) | taatacgactcactataggatttgtgatgtctctcagcattgacg | SP65g11cn86 (T7) | g11Δ12wt(C3>A)) |
| g11Δ12wt(U4>A)(f) | taatacgactcactataggcattgtgatgtctctcagcattgacg | SP65g11cn86 (T7) | g11Δ12wt(U4>A) |
| g11Δ12wt(U5>A)(f) | taatacgactcactataggctatgtgatgtctctcagcattgacg | SP65g11cn86 (T7) | g11Δ12wt(U5>A) |
| g11Δ12wt(U6>A)(f) | taatacgactcactataggcttagtgatgtctctcagcattgacg | SP65g11cn86 (T7) | g11Δ12wt(U6>A) |
| g11Δ12wt(ΔU)(f) | taatacgactcactataggcgtgatgtctctcagcattgacg | SP65g11cn86 (T7) | g11Δ12wt(ΔU) |
| g11Δ12wt(ΔU>ACA)(f) | taatacgactcactataggcacagtgatgtctctcagcattgacgtaacg | SP65g11cn86 (T7) | g11Δ12wt(ΔU>ACA) |
| g11(G1>A)classII(f) | attcagtaatacgactcactattagcttttaaagcgctacagtgatgctctc | SP65g11cn86 (T7) | g11(G1>A) |
| g11(G2>A)(f) | atttaggtgacactatagacttttaaagcgctacagtgatgtctctcagc | SP65g11cn86 (SP6) | g11(G2>A) |
| g11(C3>A)(f) | taatacgactcactataggattttaaagcgctacagtgatgtctctcagc | SP65g11cn86 (T7) | g11(C3>A) |
| g8wt (f) | gtacatattgtcgttagaaccggc | SP65g8R (T7) | g8wt |
| g8(G2>A) (f) | atttaggtgacactatagacttttaaagcgtctcagtcgccg | SP65g8 SA11 (SP6) | g8(G2>A) |
| g8(C3>A) (f) | taatacgactcactataggattttaaagcgtctcagtcgccg | SP65g8 SA11 (T7) | g8(C3>A) |
| g8 (r) | ggtcacataacgctttctattcttgc | SP65g8 SA11 (T7 or SP6) | g8wt, g8(G2>C), g8(C3>A) |
| g11(r) | ggtcacaaaacgggagtggggagc | SP65g11cn86 (T7 or SP6) | g11wt, g11Δ12, g11Δ23, g11Δ39, g11Δ12wt, g11Δ12wt(G2>C), g11Δ12wt(C3>A), g11Δ12wt(U4>A), g11Δ12wt(U5>A), g11Δ12wt(U6>A), g11Δ12(ΔU), g11Δ12(ΔU>ACA), g11(G2>A), g11(C3>A) |
aForward (f) and reverse (r) primers used in amplification reactions to produce DNA templates for transcription.
bRNA polymerase (RP) used in transcription reactions.
Transcription of (+)RNAs
Wild-type and mutant species of g8 and g11 (+)RNAs were made using the Ambion T7 or SP6 MEGAscript system in the presence of [α-33P]UTP to trace label the RNA. The RNA products of reaction mixtures were purified by phenol-chloroform extraction and ethanol precipitation. The quality of the RNAs was assessed by electrophoresis on 8% polyacrylamide gels containing 7 M urea (Tortorici et al. 2003). RNA concentrations were calculated from optical densities at 260 nm and by analysis of RNA gels using a PhosphorImager.
Replication and initiation assays
Replication assays were performed as described earlier (Tortorici et al. 2003) with minor modifications. Briefly, reaction mixtures contained 50 mM Tris-HCl (pH 7.1), 1.5% polyethylene glycol, 2 mM dithiothreitol, 1.5 U of RNasin, 10 mM magnesium acetate, 1.25 mM each ATP, CTP, and UTP, 5 mM GTP, 10 μCi of [α-32P]UTP (800 Ci/mmol), 2.25 pmol of each template mRNA, and ~1 μg of open cores in a final volume of 20 μL. Some reaction mixtures contained 2 pmol of rVP1-His and 20 pmol of rVP2, instead of open cores. Reaction mixtures were then incubated for 4 h at 37°C.
The initial components of reaction mixtures used for analyzing formation of (−) strand initiation complexes were identical to reaction mixtures used for replication assays, except the former lacked ATP, CTP, UTP, and [α-32P]UTP. After incubation for 1 h at 37°C, 250 mM NaCl was added to prevent further formation of initiation complexes. ATP, CTP, UTP, and [α-32P]UTP were then added to promote (−) strand elongation. The reaction mixtures were then incubated for 4 h at 37°C. 32P-labeled dsRNA products were resolved by 12% polyacrylamide gel electrophoresis containing sodium dode-cyl sulfate. The product was detected by autoradiography and quantified with an Amersham Biosciences PhosphorImager.
RNA secondary structure
RNA secondary structure predictions were accomplished with MPGAfold (Shapiro and Navetta 1994; Shapiro and Wu 1996; Shapiro et al. 2001a,b) using previously reported free energy rules (Mathews et al. 1999) with the addition of runtime (not post-processing) efn2 coaxial stacking energy calculations. Wild-type and mutant species of the g11 RNAs were, for the most part, folded at a 16K population (two exceptions are noted). Gene 8 and its mutants were folded with 32K and 64K populations to accommodate the gene’s larger size. The algorithm was run 20 times for each sequence to determine structural properties. The population Z-score computed with the annealing mutation operator (Shapiro and Wu 1996) determined convergence of the algorithm. The best energetic structures and the consensus structures were analyzed with STRUCTURELAB using Stem Trace (Shapiro and Kasprzak 1996; Kasprzak and Shapiro 1999) to determine the motifs presented.
MPGAfold does not necessarily produce the lowest free energy folding result but rather generates a consensus or a best structure (best fit) based upon the folding characteristics of the given sequence by sampling statistically potential structures contained in a large population of maturing structures (typically 4K–64K population sizes). This is quite different from the approach used by the dynamic programming algorithm MFOLD. Multiple new motifs may be formed at the same time at a particular generation in an MPGAfold analysis, resulting in relative constancy of a structure for several generations before a transition takes place to a new structure. The intermediate and final structures produced by the MPGAfold have been shown to be representative of those found in RNA folding pathways (Shapiro and Navetta 1994; Shapiro et al. 2001a,b). The best energy structure does not necessarily mean the optimal free energy structure, but the best that the MPGAfold produced. Also the most commonly used versions of MFold do not use the efn2 coaxial stacking rules during the running of the algorithm. efn2 is only applied after the initial fold set is obtained and will only reorder the set of initial folds based on the efn2 rules. MPGAfold does take efn2 rules into account during runtime (thus allowing potential coaxial stacked helices to form during structure maturation) and thus can produce more optimal and different structures than MFOLD. As is the case with all folding algorithms, care has to be taken to not over-interpret the results. However, the results presented here seem to correlate well with experimental observations.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Allergy and Infectious Diseases, NIH. We appreciate the efforts of Xiaohui Lu and Stephen Harrison (Harvard Medical School) in developing purification procedures for His-tagged VP1 and for making the protein available for our studies.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2122606.
REFERENCES
- Anzola, J.V., Xu, Z.K., Asamizu, T., and Nuss, D.L. 1987. Segment-specific inverted repeats found adjacent to conserved terminal sequences in wound tumor virus genome and defective interfering RNAs. Proc. Natl. Acad. Sci. 84: 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, S.-H., Cheong, H.-K., Lee, J.H., Cheong, C., Kainosho, M., and Choi, B.-S. 2001. Structural features of an influenza virus promoter and their implications for viral RNA synthesis. Proc. Natl. Acad. Sci. 98: 10602–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, J.N. and Wertz, G.W. 2004. Bunyamwera bunyavirus RNA synthesis requires cooperation of 3′- and 5′-terminal sequences. J. Virol. 78: 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro, M., Mandiola, P., Chen, D., Patton, J.T., and Spencer, E. 2001. Identification of sequences in rotavirus mRNAs important for minus-strand synthesis using antisense oligonucleotides. Virology 288: 71–80. [DOI] [PubMed] [Google Scholar]
- Barton, D.J., O’Donnell, B.J., and Flanegan, J.B. 2001. 5′ Cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20: 1439–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapell, J.D., Goral, M.I., Rodgers, S.E., dePamphilis, C.W., and Dermody, T.S. 1994. Sequence diversity within the reovirus S2 gene: Reovirus genes reassort in nature, and their termini are predicted to form a panhandle motif. J. Virol. 68: 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. and Patton, J.T. 1998. Rotavirus RNA replication requires a single-stranded 3′-end for efficient minus-strand synthesis. J. Virol. 72: 7387–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. De novo synthesis of minus-strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA 6: 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Zeng, C.Q.-Y., Wentz, M.J., Gorziglia, M., Estes, M.K., and Ramig, R.F. 1994. Template dependent in vitro replication of rotavirus RNA. J. Virol. 68: 7030–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Luongo, C.L., Nibert, M.L., and Patton, J.T. 1999. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: Evidence that VP3 is a methyltransferase. Virology 265: 120–130. [DOI] [PubMed] [Google Scholar]
- Chen, D., Barros, M., Spencer, E., and Patton, J.T. 2001. Features of the 3′-consensus sequence of rotavirus mRNAs critical to minus strand synthesis. Virology 282: 221–229. [DOI] [PubMed] [Google Scholar]
- Desselberger, U. and McCrae, M.A. 1994. The rotavirus genome. Curr. Top. Microbiol. Immunol. 185: 31–66. [DOI] [PubMed] [Google Scholar]
- Flick, R. and Hobom, G. 1999. Interaction of influenza virus polymerase with viral RNA in the “corkscrew” conformation. J. Gen. Virol. 80: 2565–2572. [DOI] [PubMed] [Google Scholar]
- Gorchakov, R., Hardy, R., Rice, C.M., and Folov, I. 2004. Selection of functional 5′ cis-acting elements promoting efficient Sindbis virus genome replication. J. Virol. 78: 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, C.S., Hahn, Y.S., Rice, C.M., Lee, E., Dalgarno, L., Strauss, E.G., and Strauss, J.H. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198: 33–41. [DOI] [PubMed] [Google Scholar]
- Herold, J. and Andino, R. 2001. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell 7: 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, M.T., Parvin, J.D., Gupta, S., Krystal, M., and Palese, P. 1997. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl. Acad. Sci. 84: 8140–8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, F. 2003. Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res. 31: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, M., Akatani, K., Ikegami, N., and Furuchi, Y. 1983. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J. Virol. 47: 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzak, W. and Shapiro, B.A. 1999. Stem Trace: An interactive visual tool for comparative RNA structure analysis. Bioinformatics 15: 16–31. [DOI] [PubMed] [Google Scholar]
- Kearney, K., Chen, D., Taraporewala, Z.F., Vende, P., Hoshino, Y., Tortorici, M.A., Barro, M., and Patton, J.T. 2004. Cell-line-induced mutation of the rotavirus genome alters expression of an IRF3-interacting protein. EMBO J. 23: 4072–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe, M., Baudoux, P., Charpilienne, A., Poncet, D., and Cohen, J. 1994. Identification of the nucleic acid binding domain of rota-virus VP2 protein. J. Gen. Virol. 75: 3423–3430. [DOI] [PubMed] [Google Scholar]
- Lawton, J.A., Estes, M.K., and Prasad, B.V. 1997a. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4: 118–121. [DOI] [PubMed] [Google Scholar]
- Lawton, J.A., Zeng, C.-Q., Mukherjee, S.K., Cohen, J., Estes, M.K., and Prasad, B.V. 1997b. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal deleted VP2: Implications for the architecture of the VP2 capsid layer. J. Virol. 71: 7353–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Ramirez, B.C., and Krug, R.M. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: Different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17: 5844–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui, M., Mattion, N.M., and Estes, M.K. 1992. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology 188: 77–84. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Midthun, K., Greenberg, H.B., Hoshino, Y., Kapikian, A.Z., Wyatt, R.G., and Chanock, R.M. 1985. Reassortant rotaviruses as potential live rotavirus vaccine candidates. J. Virol. 53: 949–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar, U.D., Hummelman, E.G., Bresee, J.S., Miller, M.A., and Glass, R.I. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9: 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.T. 1996. Rotavirus VP1 alone specifically binds to the 3′-end of viral mRNA but the interaction is not sufficient to initiate minus-strand synthesis. J. Virol. 70: 7940–7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.T. and Chen, D. 1999. RNA-binding and capping activities of proteins in rotavirus open cores. J. Virol. 73: 1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.T. and Spencer, E. 2000. Genome replication and packaging of segmented double-stranded RNA viruses. Virology 277: 217–225. [DOI] [PubMed] [Google Scholar]
- ———. 2001. RNA structure and the replication of the rotavirus segmented double-stranded RNA genome. Recent Res. Devel. Virol. 3: 529–539. [Google Scholar]
- Patton, J.T., Wentz, M., Xiaobo, J., and Ramig, R.F. 1996. Cis-acting signals that promote genome replication in rotavirus mRNA. J. Virol. 70: 7833–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.T., Jones, M.T., Kalbach, A.N., He, Y., and Xiaobo, J. 1997. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 71: 9618–9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.T., Chnaiderman, J., and Spencer, E. 1999. Open reading frame in rotavirus mRNA specifically promotes synthesis of double- stranded RNA: Template size also affects replication efficiency. Virology 264: 167–180. [DOI] [PubMed] [Google Scholar]
- Prasad, B.V.V., Wang, G.J., Clerx, J.P.M., and Chiu, W. 1988. Three dimensional structure of rotavirus. J. Mol. Biol. 199: 269–275. [DOI] [PubMed] [Google Scholar]
- Prasad, B.V.V., Rothnagel, R., Zeng, C.Q., Jakana, J., Lawton, J.A., Chiu, W., and Estes, M.K. 1996. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382: 471–473. [DOI] [PubMed] [Google Scholar]
- Pugachev, K.V. and Frey, T.K. 1998. Effects of defined mutations in the 5′ nontranslated region of rubella virus genomic RNA on virus viability and macromolecule syntheseis. J. Virol. 72: 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, B.A. and Kasprzak, W. 1996. STRUCTURELAB: A heterogeneous bioinformatics system for RNA structure analysis. J. Mol. Graph. 14: 194–205. [DOI] [PubMed] [Google Scholar]
- Shapiro, B.A. and Wu, J.-C. 1996. An annealing mutation operator in the genetic algorithm for RNA folding. CABIOS 12: 171–180. [DOI] [PubMed] [Google Scholar]
- Shapiro, B.A., Wu, J.-C., Bengali, D., and Potts, M.J. 2001a. The massively parallel genetic algorithm for RNA folding: MIMD implementation and population variation. Bioinformatics 17: 137–148. [DOI] [PubMed] [Google Scholar]
- Shapiro, B.A., Bengali, D., Kasprzak, W., and Wu, J.-C. 2001b. RNA folding pathway functional intermediates: Their prediction and analysis. J. Mol. Biol. 312: 27–44. [DOI] [PubMed] [Google Scholar]
- Tao, Y., Farsetta, D.L., Nibert, M.L., and Harrison, S.C. 2002. RNA synthesis in a cage—Structural studies of reovirus polymerase λ3. Cell 111: 733–745. [DOI] [PubMed] [Google Scholar]
- Tortorici, M.A., Broering, T.J., Nibert, M.L., and Patton, J.T. 2003. Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J. Biol. Chem. 278: 32673–32682. [DOI] [PubMed] [Google Scholar]
- Van Den Born, E., Gultyaev, A.P., and Snijder, E.J. 2004. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA 10: 424–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentz, M.J., Patton, J.T., and Ramig, R.F. 1996. The 3′-terminal consensus sequence of rotavirus mRNA is the minimal promoter of negative-strand RNA synthesis. J. Virol. 70: 7833–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Makeyev, E.V., Zang, Z., Ji, S., Bamford, D.H., and van Dijk, A.A. 2004. Cloning and sequence analysis of dsRNA segments 5, 6 and 7 of a novel non-group A, B, C adult rotavirus that caused an outbreak of gastroenteritis in China. Virus Res. 106: 15–26. [DOI] [PubMed] [Google Scholar]
- You, S., Falgout, B., Markoff, L., and Padmanabhan, R. 2001. In vitro RNA synthesis from exogenous dengue viral templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276: 15581–15591. [DOI] [PubMed] [Google Scholar]